Abstract
Cyanobacteria and algae are becoming increasingly attractive cell factories for producing renewable biofuels and chemicals due to their ability to capture solar energy and CO2 and their relatively simple genetic background for genetic manipulation. Increasing research efforts from the synthetic biology approach have been made in recent years to modify cyanobacteria and algae for various biotechnological applications. In this article, we critically review recent progresses in developing genetic tools for characterizing or manipulating cyanobacteria and algae, the applications of genetically modified strains for synthesizing renewable products such as biofuels and chemicals. In addition, the emergent challenges in the development and application of synthetic biology for cyanobacteria and algae are also discussed.
Keywords: cyanobacteria, algae, synthetic biology, biofuel, green chemistry
INTRODUCTION
Cyanobacteria and algae are endowed with the complex photosynthesis systems (Mulkidjanian et al., 2006) which can absorb solar irradiation with a broad wave length and thereafter channel the absorbed energy to other forms of energy carriers such as chemicals (Chisti, 2007; Takahashi et al., 1998; van de Meene et al., 2006) and electricity (Furukawa et al., 2006; Pisciotta et al., 2010). Solar irradiation is a clean, abundant, and renewable energy resource and, if being properly and efficiently transferred, would be more then enough to power the entire human society (Rittmann, 2008). In addition, growing cyanobacteria and algae do not require arable land, which would eventually alleviate the increasing food prices due to the growing crop-based microbial industries (Rittmann, 2008). In contrast, they can fix carbon dioxide (CO2), a type of greenhouse gas, during photosynthesis. Furthermore, cyanobacteria and algae grow faster than plants and bear relatively simple genetic background which is relatively easy to manipulate (Koksharova and Wolk, 2002).
As an emerging discipline that tackles biotechnology from a rational-design approach, synthetic biology aims to redesign existing biological systems or create artificial life (Benner, 2003; Endy, 2005; Mukherji and van Oudenaarden, 2009). In recent years, synthetic biology research has been focused on model species such as Escherichia coli and yeast, and has greatly boosted not only the in-depth understanding of the biological mechanisms in these cells, but also the capability and efficiency of these systems in biological production of various useful products (Benner, 2003; Lee and Lee, 2003; Martin et al., 2003; Isaacs et al., 2004; Ro et al., 2006; Dwyer et al., 2007; Atsumi et al., 2008b; Inui et al., 2008; Keasling, 2008; Prather and Martin, 2008; Zhang et al., 2008, 2012; Bayer et al., 2009; Ma et al., 2009; Mukherji and van Oudenaarden, 2009; Steen et al., 2010; Yim et al., 2011). However, with over 40 cyanobacterial genome sequences1 and more than 60 algal genome sequences2 being completed and published, application of synthetic biology in cyanobacteria and algae has significantly lagged behind those in E. coli and yeast. Considering the aforementioned inherent merits of the photosynthetic microbes, we believe it would be of great scientific and application values to further develop synthetic biology tools and apply them in cyanobacteria and algae. We herein review the recent progresses and the challenges in developing and applying synthetic biology for cyanobacteria and algae.
TOOLS FOR SYNTHETIC BIOLOGY IN CYANOBACTERIA AND ALGAE
DEVELOPMENT OF “BIOBRICKS” FOR CYANOBACTERIA AND ALGAE
“BioBricks” stand for standardized DNA parts with common interface and can be assembled in living organisms. They are the basic interchangeable elements for regulating the genetics3. Here we focus on the development of the most common BioBricks for cyanobacteria and algae (i.e., promoters, transtriptional terminators, ribosome binding sites, and other regulatory factors).
Promoters
Both native and foreign promoters have been evaluated in cyanobacteria, mostly using Synechococcus elongatus PCC 7942 (hereafter Synechococcus 7942) and Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) as model species (Table 1). The native promoters used are usually from genes essential to photosynthesis such as CO2 fixation (PrbcL, Pcmp, Psbt), photosystem I (PSI; PpsaA, PpsaD), PSII (PpsbA1, PpsbA2), and photosynthesis antenna protein phycocyanin (Pcpc). A native nickel-inducible promoter, PnrsB, has also been successfully utilized to express phage lysis genes in Synechocystis 6803 (Liu and Curtiss, 2009). Besides the native promoters, a limited number of foreign promoters have also been characterized in cyanobacteria. The chimeric Ptac/Ptrc promoter, a strong promoter in E. coli, has been used in Synechococcus and Synechocystis species to initiate high-level expressions of the interest genes (Geerts et al., 1995; Ng et al., 2000; Atsumi et al., 2009; Huang et al., 2010; Niederholtmeyer et al., 2010; Lan and Liao, 2011). It is noteworthy that the composition of the cyanobacterial holopolymerase is quite different from those in most bacteria (including E. coli), so commonly used E. coli promoters might perform differently when introduced into cyanobacteria (Heidorn et al., 2011). A recent study on gene expression analysis in Synechocystis 6803 showed that the strength of Ptrc1O (a version of the Ptrc/Ptac promoter) was more than fourfold higher than all versions of the promoter of native ribulose bisphosphate carboxylase/oxygenase (RuBisCO) large subunit, P, whereas the common E. coli promoters Plac, Ptet, and λ PR exhibited very low or no detectable activities in the same system (Huang et al., 2010). Since currently very little is known about the performance of various native and foreign promoters in cyanobacteria, a systematic investigation on behaviors of various promoters in cyanobacteria would be important.
TABLE 1.
Selected promoters used in cyanobacteria.
| Promoters Sources | Gene(s) | Expression hosts | Reference | |
|---|---|---|---|---|
| Prbc | Synechococcus 6301, Synechocystis 6803, Synechococcus 7942 | firefly luciferase, pdc, adh, far, accBCDA, accD, accA, fatB2 | Synechococcus 6301, Synechocystis 6803, Synechococcus 7942 | Takeshima et al. (1994), Deng and Coleman (1999), Liu et al. (2011b), Tan et al. (2011) |
| PpetE | Synechocystis 6803 | far, far1, far2, accBCDA | Synechocystis 6803 | Tan et al. (2011) |
| PpsbA1 | Synechococcus 7942 | efe, hydEF, hydG, cvrbcLS | Synechococcus 7942 | Sakai et al. (1997), Takahama et al. (2003), Ducat et al. (2011a) |
| PpsbA2 | Synechocystis 6803 | pdc, adh, cvrbcLS, ispS, tesA, fatB1, fatB2 | Synechocystis 6803, Synechococcus 7942 | Iwaki et al. (2006), Dexter and Fu (2009), Lindberg et al. (2010), Liu et al. (2011b) |
| PpsaA | Synechocystis 6803 | luxAB | Synechocystis 6803 | Muramatsu and Hihara (2006) |
| PpsaD | Synechocystis 6803 | luxAB | Synechocystis 6803 | Muramatsu and Hihara (2007) |
| Pcpc | Synechocystis 6714 | luxAB, accB, accC | Synechococcus 7942 | Imashimizu et al. (2003) |
| PrnpB | Synechocystis 6803 | GFP | Synechocystis 6803 | Huang et al. (2010) |
| Pcmp | Synechocystis 6803 | fol, gpl, shl | Synechocystis 6803 | Liu et al. (2011a) |
| Psbt | Synechocystis 6803 | gpl | Synechocystis 6803 | Liu et al. (2011a) |
| PnrsB | Synechocystis 6803 | holin, endolysin, auxiliary lysis enzyme | Synechocystis 6803 | Liu and Curtiss (2009) |
| PT7 | Coliphage T7 | luxAB | Anabaena sp. 7120 | Wolk et al. (1993) |
| Plac | E. coli | atoB, adhE2, ter, hbd, crt, hydA, alsS, ilvC, ilvD, | Synechococcus 7942 | Atsumi et al. (2009), Ducat et al. (2011a), Lan and Liao (2011) |
| Ptrc/Ptac | E. coli | petE, atoB, adhE2, ter, hbd, crt, kivd, rbcLS, invA, glf, ldhA, lldP, phrA, GFP, EYFP | Synechococcus 7942, Synechocystis 6803 | Geerts et al. (1995), Ng et al. (2000), Atsumi et al. (2009), Huang et al. (2010), Niederholtmeyer et al. (2010), Lan and Liao (2011) |
| Ptet | E. coli | GFP | Synechocystis 6803 | Huang et al. (2010) |
In algae, CaMV 35S and SV40 promoters from viruses have been used to express target genes (Benfey et al., 1990; Wang et al., 2010). However, the most effective promoters have been derived from highly expressed algal genes. For example, the widely used promoters for Chlamydomonas transformation have been derived from the 5′ untranslated region of the Chlamydomonas reinhardtii RuBisCO small subunit gene (rbcS2 ;Stevens et al., 1996), Chlamydomonas heat shock protein 70A gene hsp70A (Schroda et al., 2000), marine diatom fucoxanthin-chlorophyll a/c binding protein gene fcp (Apt et al., 1996; Miyagawa-Yamaguchi et al., 2011), Dunaliella duplicated carbonic anhydrase 1 (DCA1; Li et al., 2010; Lu et al., 2011), Porphyra yezoensis actin1 gene (PyAct1; Takahashi et al., 2010), and two Nannochloropsis unlinked violaxanthin/chlorophyll a-binding protein (VCP) genes, VCP1 and VCP2 (Kilian et al., 2011).
Transcriptional terminators
Placing a transcription terminator downstream of the introduced genes will prevent effects on the expression of genes adjacent to the insertion loci; meanwhile, placing a terminator upstream of the promoter of an introduced gene will also prevent any background transcription effect on the upstream genes (Adhya and Gottesman, 1982). So far only a few native and foreign terminators have been utilized in cyanobacteria, including the cyanobacterial RuBisCO terminator (Takeshima et al., 1994) and strong E. coli terminators such as rrnB terminator (Geerts et al., 1995; Takahashi et al., 1998; Atsumi et al., 2009), bacteriophage T7 terminator (Lang and Haselkorn, 1991; Argueta et al., 2004), and rrnBT1-T7TE double terminator4 (Huang et al., 2010). Very little work has been conducted to characterize the termination efficiencies in cyanobacteria and algae.
Ribosome binding sites
The ribosome binding sites (RBS) play a crucial role in initiating the translation of downstream target genes. Upon translation initiation, the 3′-terminal sequence of the 16S rRNA interacts with the core Shine–Dalgarno (SD) sequence of RBS by complementary pairing of the nucleic acids. For example, in cyanobacterium Synechocystis 6803, the 3′-terminal sequence of the 16S rRNA is AUCACCUCCUUU (Kaneko et al., 1996; Ma et al., 2002) and therefore the optimal complementary SD sequence should be AAAGGAGGUGAU (core SD sequence underlined). Heidorn et al. (2011) studied the efficiencies of different RBS in expressing GFP in Synechocystis 6803 and found that the RBS sequence UAGUGGAGGU gave about twofold higher translation efficiency than a RBS sequence AUUAAAGAGGAGAAA and about fourfold higher than those of sequences UCACACAGGAAAG and AAAGAGGAGAAA. However, it is found that the efficiency of the same RBS might vary across species, such as E. coli vs. Synechocystis (Heidorn et al., 2011). The efficiency of a given RBS also depends on the surrounding nucleotide sequence that may result in secondary structures and the spacing between the SD sequence and the translation start codon AUG (de Smit and van Duin, 1990; Chen et al., 1994; Pfleger et al., 2006). In order to predict the translation efficiency of a given RBS in various genetic contexts, Salis et al. (2009) have established a thermodynamic model that calculates the impact from the SD sequence, the start codon, the spacing between the SD sequence and the start codon, and the mRNA secondary structure; the model can accurately predict protein expression levels within a factor of 2.3 over a range of 100,000-fold in E. coli. Similar model should be employed to optimize the RBS for gene expression in cyanobacteria.
Negative regulation of gene expression
The down-regulation of the target gene expression has been studied at transcriptional, translational, and post-translational levels in cyanobacteria. The negative transcriptional factor, LacI, has been utilized as a repressor in regulating the Ptac/Ptrc controlled target gene expression (Geerts et al., 1995; Atsumi et al., 2009; Huang et al., 2010; Niederholtmeyer et al., 2010; Ducat et al., 2011a; Lan and Liao, 2011). However, lacI-Ptac/Ptrc expression system can result in severe leaky expression of target genes (Huang et al., 2010). By placing dual lac operators upstream of Ptac/Ptrc promoter the leaky expression of downstream target genes was significantly repressed; however, this also resulted in a limited induction of the promoter with presence of the inducer IPTG (Huang et al., 2010). Degradation tags which can be fused to the target proteins through genetic engineering have also been investigated in cyanobacteria. Three ssrA protease degradation tags including ASV, AAV, and LVA were fused to EYFP and expressed in Synechocystis 6803. The results indicated that LVA is the strongest degradation tag, AAV is the weaker one and ASV is the weakest (Huang et al., 2010). Recently, it has been discovered that antisense RNAs (asRNAs) play an important role in cyanobacterial gene regulation (Hernández et al., 2006; Georg et al., 2009; Cerutti et al., 2011; Mitschke et al., 2011). asRNAs thus provide another approach for gene silencing in cyanobacteria. For example, Mussgnug et al. (2007) have successfully down-regulated the expression of light-harvesting antenna complexes through RNA interference.
Endogenous enhancers
The transcription of an interest gene can be positively affected by placing in the gene cluster an enhancer, a short DNA fragment which interacts with certain proteins to enhance the transcription. In cyanobacteria, some light-responsive elements exhibit enhancer activities. For example, the 5′-untranslated regions of the psbAII and psbAIII genes of Synechococcus 7942 have been found as enhancers which can increase the expression of downstream genes by 4- to 11-fold when combined with an E. coli promoter (conII) in the Synechococcus 7942 host strain (Li and Golden, 1993). A recent study by Eichler-Stahlberg et al. (2009) showed that inserting three introns from the native alga C. reinhardtii RBCS2 gene into the recombinant luciferase and erythropoietin resulted in up to fourfold increase of the expression levels. By fusing the recombinant luciferase with the endogenous RuBisCO LSU protein, Muto et al. (2009) has achieved enhanced luciferase expression by 33-fold.
PLASMID VECTORS
Both integrative and replicative plasmids have been developed for cyanobacteria. In cyanobacteria, integrative plasmids are usually utilized as vectors to integrate foreign genes into the cyanobacterial genomes via homologous recombination (Golden et al., 1987; Eaton-Rye, 2004; Heidorn et al., 2011). Integrative plasmids usually cannot replicate themselves and would eventually be eliminated through cell division. Replicative plasmids are those which can replicate in host cyanobacteria and the replication properties can be descended to daughter cells. Replicative cyanobacterial plasmids can be classified into two types: those with replicons of broad-host range plasmids (Mermet-Bouvier et al., 1993; Mermet-Bouvier and Chauvat, 1994; Ng et al., 2000; Huang et al., 2010) and those derived from endogenous cryptic plasmids (Reaston et al., 1982; Wolk et al., 1984; Lang and Haselkorn, 1991; Summers et al., 1995; Deng and Coleman, 1999; Argueta et al., 2004; Iwaki et al., 2006). Representative shuttle vectors for cyanobacteria are listed in Table 2. The copy numbers of the broad-host range RSF1010-derived plasmids have been reported as about 10 per chromosome in E. coli cells and 10–30 per cell in Synechocystis (Ng et al., 2000; Huang et al., 2010) which is slightly higher than the average copy number (approximately 10) of the Synechocystis chromosome (Eaton-Rye, 2004). Due to lack of an active partitioning mechanism, RSF1010-derived plasmids tend to be eliminated in cells and thus antibiotic selection pressure is required for the maintenance (Becker and Meyer, 1997; Meyer, 2009).
TABLE 2.
Representative shuttle vectors for cyanobacteria.
| Cyanobacterial replicons | E. coli replicons | Representative vectors | Host cyanobacteria | Reference |
|---|---|---|---|---|
| pDU1 | pMB1 | pRL1, pJL3 | Anabaena 7120, Anabaena 7118, Anabaena M-131, Nostoc 7524 | Reaston et al. (1982), Wolk et al. (1984), Lang and Haselkorn (1991) |
| pDC1 | pMB1 | pSCR119/202, pSUN119/202 | Nostoc sp. MAC 8009, Nostoc punctiforme, Nostoc ATCC 29133 | Lambert and Carr (1983), Summers et al. (1995), Argueta et al. (2004) |
| pUH24 | P15A | pUC303 | Synechococcus 7942 | Kuhlemeier et al. (1983), Iwaki et al. (2006) |
| pUH24 | pMB1 | pCB4, pSG111 | Synechococcus 7942 | Golden and Sherman (1983), Luinenburg and Coleman (1993), Deng and Coleman (1999) |
| pAQ1 | pMB1 | pAQE17 | Synechococcus 7002 | Buzby et al. (1985) |
| PBA1 | pMB1 | pARUB19 | Synechococcus 6301 | Takeshima et al. (1994) |
| RSF1010 | RSF1010 | pFC1, pSL1211, pPMQAK1, | Synechosystis 6803, Synechosystis 6714, Synechococcus 7942, Synechococcus 6301, Anabaena 7120, Nostoc ATCC 29133 | Mermet-Bouvier et al. (1993), Mermet-Bouvier and Chauvat (1994), Ng et al. (2000), Huang et al. (2010) |
Plasmid vectors have been developed to transform algae (León-Bañares et al., 2004). Recombinant eukaryotic algal viruses as transformation vectors (Langridge et al., 1986) and Agrobacterium tumefaciens-mediated method (Kumar et al., 2004) were also successfully developed for both marine and freshwater algae (Wang et al., 2010).
CODON USAGE
Since different organisms usually bear particular codon usage patterns, when a gene is cloned from one species and expressed in a second organism, some codons might become rare codons in the new host, leading to poor translation efficiency (Kane, 1995). Genes in cyanobacteria show a bias in use of synonymous codons (Campbell and Gowri, 1990; Nakamura et al., 2000; Beck et al., 2012; Yu et al., 2012); it is thus very important to examine the difference in codon usage of a heterologous gene before it is expressed in cyanobacteria. In a recent study, Lindberg et al. (2010) studied the effects of codon usage on heterologous expression of kudzu IspS gene (encoding the isoprene synthase) in Synechocystis 6803. The results showed that the codon-optimized IspS showed remarkable improved expression, 10-fold higher than that of the native IspS gene under control of the same promoter. The importance of codon optimization in algal genetic applications is also increasingly acknowledged. For instance, it has been shown that codon bias significantly affects the GFP expression in C. reinhardtii (Heitzer et al., 2007). As a result, in recent transgenic research, the codon-optimized luciferase gene was used in a green alga Gonium pectorale (Lerche and Hallmann, 2009) and the codon-modified β-glucuronidase gene was transformed in a red seaweed P. yezoensis (Takahashi et al., 2010).
TRANSFORMATION OF CYANOBACTERIA AND ALGAE
Methods to introduce DNA into cyanobacteria include conjugation (Thiel and Wolk, 1987; Elhai and Wolk, 1988), electroporation (Zang et al., 2007), and natural transformation (Shestakov and Khyen, 1970; Grigorieva and Shestakov, 1982; Kuhlemeier and van Arkel, 1987). The methods have been well summarized in several recent reviews (Eaton-Rye, 2004; Koksharova and Wolk, 2002; Heidorn et al., 2011) and we suggest readers to refer these excellent reviews for details. Compared to cyanobacteria, transformation methods for algae are less developed and more complicated. Since the chloroplast and nucleus of alga C. reinhardtii were stably transformed more than two decades ago (Boynton et al., 1988; Debuchy et al., 1989; Fernández et al., 1989), different methods have been employed in algal transformation which include, but not limited to, particle bombardment, glass bead agitation, microinjection, electroporation and A. tumefaciens-mediated transformation (León-Bañares et al., 2004; Coll, 2006; León and Fernández, 2007; Potvin and Zhang, 2010). Specifically, bombardment of target cells with DNA-coated metal particles turns to be an effective and highly reproducible method to transform algae. This method has been so far applied in the transformation of nuclear and chloroplast of many algal species such as C. reinhardtii, Volvox carteri, Chlorella sorokiniana, Chlorella ellipsoidea, Chlorella kessleri, Haematococcus pluvialis, Phaeodactylum tricornutum, and G. pectorale (Boynton and Gillham, 1993; Potvin and Zhang, 2010). In addition, agitation of the cell wall-deficient algal cells with glass beads, polyethylene glycol (PEG) and foreign DNA has been used to transform algae such as C. reinhardtii, Dunaliella salina, and red alga Porphyra haitanensis (Kindle, 1990; Feng et al., 2009; Wang et al., 2010). Microinjection of the viral SV40 DNA or the chimeric construction pSV2neo into the marine unicellular green alga Acetabularia mediterranea also resulted in a high yield and stable nuclear transformation (Neuhaus et al., 1986); nevertheless, it is hard to operate and the throughput of transformation is low. Agrobacterium tumefaciens has been used to mediate the transformation of C. reinhardtii (Kumar et al., 2004) and H. pluvialis (Kathiresan et al., 2009). Recently, it was discovered that the industrially relevant oil-producing alga Nannochloropsis sp. is haploid and can be transformed with high efficiency using high electric field electroporation. It has also been found that efficient stable transformation of this species via homologous recombination requires using linear DNA fragment rather than circular plasmid DNA (Kilian et al., 2011). However, the mechanism for the high homologous recombination efficiency is to be elucidated.
APPLICATIONS OF MODIFIED CYANOBACTERIA AND ALGAE
We focus here on recent progress in producing biofuels and other useful chemicals using genetically modified cyanobacteria and algae. For other applications, readers can refer to several other excellent reviews published recently (Radakovits et al., 2010; Ruffing, 2011; Qin et al., 2012).
BIOFUELS
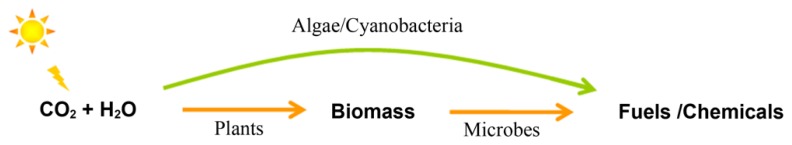
United States consumed 13.3 million barrels of petroleum per day for transportation purposes in 2009, accounting for 71% of all petroleum used (Energy Information Administration, 2010). Many alternatives to current liquid fuels have been proposed, including ethanol, 1-butanol, isobutanol, short-chain alcohols, short-chain alkanes, biodiesel (FAME, fatty acid methyl esters), fatty alcohols, alkanes, linear and cyclic isoprenoids (Lee et al., 2008; Connor and Atsumi, 2010). Current routes for biological production of fuels and chemicals are summarized in Figure 1. Traditionally people follow a two-step route to firstly collect plant biomass and then convert biomass to fuels by microbial fermentation (Stephanopoulos, 2007); whereas recently interest in harnessing photosynthetic microbes to directly convert CO2 to fuels has been dramatically increased (Chisti, 2007; Lu, 2010). Compared to crops, the per-hectare oil yield of cyanobacteria or microalgae is about two orders of magnitude higher and the cultivation land needed is around two orders of magnitude less (Chisti, 2007). It is estimated that more than $1 billion has been invested in the algae-to-biofuel research and development since 2007 in US alone (Mascarelli, 2009).
FIGURE 1.
Routes for biological production of fuels and chemicals. Arrows indicate the carbon and energy flow between different carriers.
Biodiesel
Cyanobacteria and algae are rich in energy stock compounds, such as diacylglycerol (DAG), triacylglycerol (TAG) and starch, which can be extracted and used for biodiesel production (van de Meene et al., 2006; Chisti, 2007; Radakovits et al., 2010; Sheng et al., 2011). To further increase the oil contents in the cells, effects have been made to block metabolic pathways as well as to overexpress genes of limiting steps. For example, two different starch-deficient strains of C. reinhardtii, the sta6 and sta7 mutants that carries gene knockout in the ADP-glucose pyrophosphorylase and isoamylase genes, respectively, have been isolated (Mouille et al., 1996; Posewitz et al., 2005); and these mutants accumulated increased levels of TAG during nitrogen deprivation (Wang et al., 2009). Another starchless mutant of Chlorella pyrenoidosa has also been reported that the lipid content of this mutant has been elevated by nearly twofold relative to the wild-type under nitrogen limitation culture conditions (Ramazanov and Ramazanov, 2006). It indicated that blocking the starch biosynthesis may be an effective way to increase lipid, and thus potentially biodiesel, production.
Nevertheless, lipid extraction process is energy-intensive and significant amount of glycerol as a byproduct have been two of the major hurdles for commercial production of biodiesel (Chisti, 2007; Fernando et al., 2007; Liu and Curtiss, 2009). Efforts have been made from both process engineering and genetic engineering approaches to facilitate the lipid extraction (Liu and Curtiss, 2009; Liu et al., 2011a; Sheng et al., 2011). Specifically, Liu and colleagues have constructed inducible systems to conditionally express phage lysis genes and lipolytic enzyme genes in Synechocystis 6803 to trigger the cell lysis upon harvest and thus help lipid extraction from this species (Liu and Curtiss, 2009; Liu et al., 2011a). To produce secretable biofuels from a synthetic biology approach is another way to resolve above issues.
Free fatty acids
Enhanced production of free fatty acid (FFA) has already been achieved in E. coli through a series of genetic engineering (Davis et al., 2000; Lu et al., 2008). In a recent study, Liu et al. (2011b) engineered cyanobacterium Synechocystis strains to produce and secret FFAs to up to 197 mg/L at a cell density of 1.0 × 109 cells/mL. The acetyl-CoA carboxylase (ACC) was overexpressed to drive the metabolic flux toward FFAs, while the fatty acid activation gene aas (slr1609) was deleted to inactivate the FFAs degradation. Poly-β-hydroxybutyrate (PHB) synthesis genes (slr1993 and slr1994) and the phosphotransacetylase gene pta (slr2132) were deleted to block competitive pathways. Particularly, two genetic modifications turned to significantly increase the FFAs production and secretion: overexpression of thioesterases and weakening the polar peptidoglycan layer of the cell wall of Synechocystis 6803.
Alkanes and alkenes
Although it was known that some cyanobacteria can synthesize alkanes, the molecular mechanism had been mysterious until recently an alkane/alkene biosynthetic pathways were identified in cyanobacteria (Steen et al., 2010; Mendez-Perez et al., 2011). Steen et al. (2010) identified an alkane/alkene biosynthetic pathway that two successive biochemical reactions catalyzed by an acyl-ACP reductase and an aldehyde decarbonylase, respectively, converts acyl-ACP (intermediates of fatty acid metabolism) to alkanes/alkenes. In order to increase the alkane production in cyanobacteria, heterologous expression of acyl-ACP reductase and aldehyde decarbonylase genes (from Synechococcus 7942) has been achieved in Synechococcus 7002, which led to a total intracellular accumulation of n-alkane to up to 5% of the dry cell weight (Reppas and Ridley, 2010). In another research, Mendez-Perez et al. (2011) identified the genes responsible for α-olefin biosynthesis in Synechococcus 7002. In addition, overexpression of the accBCDA operon (which encodes ACC) in Synechocystis was also reported to enhance alkane/alkene production (Tan et al., 2011), consistent with the aforementioned results of FFAs production. Although it is believed there are certain alkane/alkene secretion pathways, the specific mechanisms are still under exploration (Radakovits et al., 2010).
Ethanol
Ethanol production via microbial fermentation has undergone a sharp increasing in the past decade for its utility as supplement in transportation fuel (Stephanopoulos, 2007; Energy Information Administration, 2010). In 1999, photosynthetic production of up to 230 mg/L ethanol has been reported using genetically engineered cyanobacterium Synechococcus 7942, in which an artificial operon of pdc-adh (genes originally from Zymomonas mobilis) was expressed under a Plac and Prbc promoters via a shuttle vector pCB4 (Deng and Coleman, 1999). In a recent study, the pdc-adh expression cassette was integrated into the chromosome of Synechocystis 6803 at the psbA2 locus. Driven by the light-inducible strong PpsbA2 promoter, expression of pdc/adh resulted in ~550 mg/L ethanol production by the engineered Synechocystis under high light (~1000 μE/m2/s) conditions (Dexter and Fu, 2009). In algae, although many species have fermentative pathways to produce ethanol, the pathways are only functional under dark and anaerobic conditions (Hirayama et al., 1998). Algal ethanol is currently produced via heterotrophic fermentation of algal biomass using heterotrophs such as yeast and E. coli (Nguyen et al., 2009; Harun et al., 2010; Wargacki et al., 2012), which follows the two-step route (Figure 1). Direct photosynthetic production of ethanol by algae would be possible using a similar approach as being demonstrated in cyanobacteria by expressing foreign ethanol biosynthesis pathways, or by tuning the native regulatory pathways in algae.
Isobutanol and 1-butanol
Compared with ethanol, isobutanol and 1-butanol have much higher energy density. The energy density of butanol reaches 29.2 MJ/L, about 90% of that of gasoline, 32.5 MJ/L, and it is also less volatile and less corrosive than ethanol (Dürre, 2007). Therefore, butanol is regarded as a better gasoline substitute. Recently, significant progress has been achieved for photosynthetic production of butanol. Liao and colleagues introduced an artificial isobutanol biosynthesis pathway into Synechococcus 7942 and the engineered strains were able to photosynthetically produce isobutyraldehyde and isobutanol at titers of 1100 and 450 mg/L, respectively (Atsumi et al., 2009). In contrast, photosynthetic production of 1-butanol in oxygenic cyanobacteria or algae has been hard because the intrinsic oxygen-sensitivity and NADH-dependence of the 1-butanol biosynthetic pathway are conflict with the photo-oxygenisis and lack of NADH cofactors in cyanobacteria (Atsumi et al., 2008a; Inui et al., 2008; Lan and Liao, 2011). When a 1-butanol pathway was overexpressed in Synechococcus 7942, the 1-butanol was barely detectable (~1 mg/L) after 2 weeks cultivation under photosynthetic conditions. Up to 14.5 mg/L 1-butanol has been achieved in Synechococcus 7942 under an anoxic condition (Lan and Liao, 2011). Further analysis revealed that the reversible acetyl-CoA condensation reaction catalyzed by thiolase (encoded by atoB) strongly favors the thiolysis of acetoacetyl-CoA rather than the condensation of two acetyl-CoA molecules, and thus AtoB may be insufficient to drive the flux from acetyl-CoA pool toward 1-butanol biosynthesis under photosynthetic conditions (Lan and Liao, 2012). To this end, an alternate ATP-driven acetoacetyl-CoA biosynthetic pathway was constructed by overexpressing an acetoacetyl-CoA synthase (NphT7) which instead condenses malonyl-CoA and acetyl-CoA in Synechococcus. With co-expressing the downstream NADH-dependent 1-butanol biosynthetic pathway, 6.5 mg/L 1-butanol has been produced under photosynthetic conditions. After the NADH-dependent bifunctional aldehyde/alcohol dehydrogenase (AdhE2) was further replaced with separate NADPH-dependent butyraldehyde dehydrogenase (Bldh) and alcohol dehydrogenase (YqhD), the 1-butanol production was increased by fourfold, up to ~30 mg/L, under the same photosynthetic condition (Lan and Liao, 2012).
Longer carbon chain fatty alcohols
In order to produce long-chain alcohols, Lu and colleagues heterologously expressed fatty acetyl-CoA reductases from different sources in Synechocystis and the resultant strains achieved production of fatty alcohols, including hexadecanol (C16) and octadecanol (C18; Tan et al., 2011). Although the titer was very low (about 0.2 mg/L), it is amenable for further improvement via further enhancing upstream pathways and addressing secretion issues as that in the engineering of Synechocystis 6803 for enhanced fatty acid production (Liu et al., 2011b). Production of the intermediate-chain alcohols (C5 to C10) in E. coli has been well summarized by Lamsen and Atsumi (2012). Briefly, C5 to C10 alcohols have been successfully biosynthesized via the expanded 1-butanol pathway (Dekishima et al., 2011), the engineered reversal of the β-oxidation pathway (Dellomonaco et al., 2011) and the 2-keto acid metabolic pathways (Atsumi et al., 2008b; Zhang et al., 2008). Since cyanobacteria and algae share with E. coli the most chassis metabolic pathways required for longer-chain alcohol biosynthesis, it is believed that similar approaches can be used to achieve the biosynthesis of alcohols with carbon chain length >5 in cyanobacteria and algae.
Hydrogen
Besides liquid biofuels, production of hydrogen – a gaseous, carbon-free, and high-energy-content fuel – in algae and cyanobacteria has also gained increasing attention in recent years (Melis et al., 2000; Kruse et al., 2005; Ghirardi et al., 2007, 2009; Hankamer et al., 2007; Hemschemeier et al., 2009; Lee et al., 2010; Srirangan et al., 2011). Many cyanobacteria and algae naturally produce hydrogen as a secondary metabolite to balance the redox energetics. In order to fortify the hydrogen production, endeavor has been made to augment the electron flux, instead of the carbon flux, toward H2 biosynthesis catalyzed by hydrogenases (2H+ + 2e– → H2). In alga C. reinhardtii, for instance, blocking the cyclic electron transfer around PSI turned to eliminate the possible electron competition for electron with hydrogenase; as a result, the H2 evolution rate increased 5–13 times under a range of conditions (Kruse et al., 2005). Hydrogenase has been tethered to the PSI to obtain a much greater electron throughput and thus H2 evolution rate (Ihara et al., 2006; Schwarze et al., 2010; Lubner et al., 2011). However, to date these experiments were all conducted in vitro and efforts need to be made from a synthetic biology approach to validate the concept in vivo. In another study, expression of an exogenous ferredoxin from Clostridium acetobutylicum in addition to the native ferredoxin could fortify the electron flow toward the hydrogenase HydA via siphoning electrons from the fermentation of internal reducing equivalents (such as glycogen). As a result, the hydrogen production was enhanced by approximately twofold (Ducat et al., 2011a) under light-dependent anoxic conditions. On the other hand, efforts have been made to block pathways competitive for reductant consumption to facilitate the H2 production. For example, after the ldhA gene (which is responsible for NADH consumption in lactate production) was inactivated in Synechococcus 7002, the NADH/NAD+ ratio increased markedly and therefore the hydrogen production by the native bidirectional [NiFe] hydrogenase was increased fivefold under anoxic dark conditions (McNeely et al., 2010). The oxygen-sensitivity of both two major [NiFe] and [FeFe] hydrogenases is the greatest challenge to date which is discussed infra.
OTHER COMMODITY CHEMICALS
Although significant attention has been paid to photosynthetic production of fuels from CO2, the relative values (in term of USD per photon) of fuels are much lower than those of other commodity chemicals. For example, it is estimated that the relative value of a photon fixed in lactic acid is about 3.5-fold greater than that in octane (Ducat et al., 2011b). Therefore, photosynthetic production of chemicals with higher unit values than fuels is economically more desirable at least in the near term.
Ethylene
Ethylene, the simplest unsaturated alkene, is one of the most important building-blocks in synthetic chemical industry. However, its production almost exclusively relies on petroleum. To make the production sustainable, biosynthesis of ethylene from renewable resources has been explored. Sakai et al. (1997) first demonstrated photosynthetic production of gaseous ethylene from CO2 in genetically engineered Synechococcus by heterologously expressing a single efe gene of Pseudomonas syringae on a pUC303-derived shuttle vector. Later, by integrating the efe gene into the psbA1 locus of the Synechococcus 7942 genome, the research group achieved higher ethylene production with a titer of ~37 mg/L (Takahama et al., 2003). However, the engineered Synechococcus strains were not genetically stable, resulting in declined ethylene production during successive batch cultivations (Takahama et al., 2003). Since production of every two molecules of ethylenes consumes three molecules of 2-oxoglutarate and one molecule of L-arginine (Fukuda et al., 1992), the genetic instability might be attributed to the shortage in the tricarboxylic acid (TCA) cycle intermediates, leading to a severe depression on cell growth (Takahama et al., 2003). In order to sustain the ethylene production in cyanobacteria or algae, metabolic flux toward TCA cycle should be enhanced and alternative ethylene biosynthesis pathways might be considered (Yang and Hoffman, 1984; Fukuda et al., 1989; Kende, 1993; Kosugi et al., 2000).
Isoprene
Isoprene is another important feedstock in the synthetic chemistry and potentially a biofuel. Biosynthesis and emission of isoprene occurs in many plants as a way to cope with heat flecks and reactive oxygen species, and the genetic mechanism has been investigated (Sharkey et al., 2008). Lindberg et al. (2010) cloned the IspS gene (encodingisoprene synthase) from Pueraria montana and integrated into the psbA2 locus of the Synechocystis genome, conferring heterologous expression of the isoprene synthase under the light-dependent PpsbA2 promoter in Synechocystis. Codon usage turned to be a very important factor for optimal expression of the IspS gene. After codon optimization, the IspS gene expression was enhanced by about 10-fold. Isoprene was eventually produced at a rate of ~50 mg/g dry cell/day under high light (~500 μE/m2/s) culture conditions. It is noteworthy that heterologous expression of IspS by replacing the psbA2 gene did not affect photosynthesis significantly and depress the growth of the transformants (Lindberg et al., 2010), which was differed from the aforementioned ethylene-producing cyanobacteria (Sakai et al., 1997; Takahama et al., 2003).
Acetone
Acetone represents the simplest ketone which serves as a solvent and precursor for industrial chemicals (Yurieva et al., 1996). Microbial production of acetone has been achieved in fermentation of Clostridia and recombinant E. coli using sugar as feedstocks (Bermejo et al., 1998). However, the maximal yield is merely 50% with the other half carbon being released as CO2 when hexose is the sole carbon source. Recently, through a combination of co-expression of the acetoacetate decarboxylase (adc) and coenzyme A transferase (ctfAB) and deletion of the PHB polymerase (PhaEC) in Synechocystis 6803, 3–5 mg/L acetone has been produced under nitrogen and phosphate deprived, dark and anaerobic culture conditions. After deleting the phosphotransacetylase-encoding gene pta, the competitive acetate production was remarkably reduced and the acetone titer has been evidently increased to 36.0 mg/L in the culture (Zhou et al., 2012).
Poly-β-hydroxybutyrates
Cyanobacteria are the natural producers of PHB, a type of polyhydroxyalkanoates (PHAs) that serves as biodegradable plastics (Hein et al., 1998; Taroncher-Oldenburg et al., 2000). However, the yield is very low and nutrient deprivation and acetate addition are usually necessary for accumulation of PHB (Wu et al., 2001). By introducing PHB biosynthesis genes from Ralstonia eutropha into Synechococcus 7942 coupled with nitrogen starvation and acetate supplementation, the PHB biosynthesis in the recombinant cyanobacteria has reached a maximum of 25.6% of the dry cell weight (Takahashi et al., 1998). Efforts in identifying gene disruptions which might contribute to increase of PHB accumulation were also made and several gene disruptions with positive effects were discovered (Tyo et al., 2009). Nevertheless, similar with other types of macromolecules PHB can not be secreted out of cells; the required extraction process is energy-intensive and remains as one of the major hurdles for commercial applications (Chisti, 2007; Liu and Curtiss, 2009). As a result, 3-hydroxybutyrate (3HB), the monomer of PHB and a building block molecule for other PHAs, has been successfully produced and secreted by genetically engineered E. coli (Lee and Lee, 2003; Liu et al., 2007; Tseng et al., 2009). Hence, photosynthetic production of 3HB in cyanobacteria and algae might be a feasible approach to cope with the secretion problem.
Lactic acid
Lactic acid is another chemical that can serve as a building block for synthesizing biodegradable polyesters with valuable medical properties. It is also used as a preservative and acidulant in food industry, and can serve as an advanced nutrient for neuron cells (Wee et al., 2006). While conventional production of lactic acid relies on microbial fermentation of sugars (Wee et al., 2006), photosynthetic production of lactic acid using CO2 as carbon source has been recently demonstrated (Niederholtmeyer et al., 2010). Through heterologously expressing three genes, including ldhA, lldP, and udhA, in cyanobacterium Synechococcus 7942, Niederholtmeyer et al. (2010) has accomplished production of lactic acid with a titer of ~56 mg/L under photoautotrophic culture condition. While LdhA catabolizes the conversion of pyruvate to lactate, expression of the lactate transporter gene lldP turned to be essential for lactate secretion from the engineered Synechococcus strain (Niederholtmeyer et al., 2010). Repletion of NADH, a cofactor for LdhA, through expressing the NADPH/NADH transhydrogenase (encoded by udhA) greatly enhanced the lactate production but reduced the growth rate of Synechococcus (Niederholtmeyer et al., 2010).
Sugars
Fresh water cyanobacteria accumulate solutes such as glucosylglycerol and sucrose when they are exposed to salt stress (Hagemann, 2011). By knocking out the agp gene (which contributes to the biosynthesis of glucosylglycerol) from the Synechocystis 6803 genome, Miao et al. (2003) achieved sucrose accumulation of up to 44 mg/L/OD730 after 0.9 M salt shock for 96 h. In another study, overexpression of invA, glf, and galU genes in Synechococcus 7942 resulted in up to 45 mg/L total hexose production (including glucose and fructose) in the culture supplemented with 200 mM NaCl (Niederholtmeyer et al., 2010). While InvA catalyzes the conversion of sucrose to glucose and fructose, expression of the glucose or fructose transporter GLF (encoded by glf gene) was essential for glucose or fructose secretion. Additional expression of GalU enhanced the biosynthesis of intracellular precursors and thus further increased the hexose sugar production by over 30% in the culture (Niederholtmeyer et al., 2010).
CHALLENGES AND OPPORTUNITIES OF SYNTHETIC BIOLOGY IN CYANOBACTERIA AND ALGAE
Despite of promising progresses, there are challenges ahead for synthetic biology to reach its full power in modifying cyanobacteria and algae for biotechnological applications. Here we briefly discuss the challenges and possible strategies.
IMPROVING TOOLS FOR GENETIC MANIPULATION
Effective “BioBricks”
Although a few “BioBricks” have been characterized in cyanobacteria, the limit number of gene expression elements would not fulfill the need of synthetic biology in cyanobacteria. After an initial gene expression, a fine-tuning of the gene expression is usually the next step in order to further optimize the properties of the genetically engineered strains, which requires a good number of “BioBricks.” Currently most of the “BioBricks” were collected from E. coli, but the E. coli “BioBricks” might behave differently in cyanobacteria. For example, tightly regulated IPTG-inducible lacI/Ptac gene expression system does not work as well in cyanobacteria as it does in E. coli (Huang et al., 2010). Thus, systematic collection and characterization of “BioBricks” in cyanobacteria is necessary. Additionally, in contrast to various commercialized E. coli and yeast strains that have been genetically modified to serve as chassis for different purposes, there are few such cyanobacterial or algal species available nowadays. To design and construct a series of chassis strains is thus an urgent task. Moreover, to our knowledge, there has been no study of the performance of a given BioBrick in different cyanobacterial species. We assume that a defined BioBrick might behave differently across cyanobacterial species and the efficiency of the BioBrick need to be characterized for each cyanobacterial species.
Improved transformation efficiency
Standardized transformation vectors/protocols have been established for model cyanobacteria, such as Synechococcus and Synechocystis, although the transformation efficiency still needs further improvement (Eaton-Rye, 2004; Heidorn et al., 2011). However, the transformation methods for model filamentous cyanobacteria, such as Anabaena and Spirulina, are still under development (Ducat et al., 2011b), and so far no genetic engineering has been conducted in the marine N2-fixing cyanobacterium Trichodesmium despite significant interest on its ability of peaking the fixation of CO2 and N2 simultaneously during the day time (Chen et al., 1998; Berman-Frank et al., 2001). In vivo restriction activities have been demonstrated as an important barrier for introducing foreign DNA into cyanobacterial cells (Elhai et al., 1997; Koksharova and Wolk, 2002). Hence, it would be helpful to construct methylation-defect cyanobacterium host strains or to establish in vitro systems that can methylate the foreign DNA before transformation. Additionally, since the bacteriophage λ recombination system has greatly improved the E. coli transformation efficiency (Yu et al., 2000), we propose that high-efficiency homologous recombination in cyanobacterial cells might be achievable by developing a proper cyanophage recombination system.
In order to improve transformation efficiency of other algal species, endeavor could be made to uncover the mechanism behind the recently discovered truth that highly efficient homologous recombination occurs after electroporation of the industrially relevant oil-producing alga Nannochloropsis sp. (Kilian et al., 2011). Recently, through an approach of ex vivo assembly of the chloroplast genome before bombarding it into the green alga C. reinhardtii, O’Neill et al. (2012) demonstrated that simultaneous and multi-loci genetic modifications of the chloroplast of the green alga C. reinhardtii could occur after one single round of transformation, providing an alternative method to improve the efficiency of multiple-gene transfer.
IMPROVING PHOTOSYNTHESIS EFFICIENCY
Although the solar energy conversion efficiencies of algae and cyanobacteria are 2–3 folds higher than those of crop plants, the efficiencies are still low with yields around 5–7% during the growing season and around 3% in bioreactors on an annual basis (Blankenship et al., 2011). A recent study on in silico modeling of the reconstructed photosynthetic process revealed that the regulation of the photosynthesis activity is quite complex and a high degree of cooperativity of nine alternative electron flow pathways is responsible for optimized photosynthesis performance in Synechocystis 6803 (Nogales et al., 2012).
Light harvesting
The photosynthetic microorganisms in nature have been selected by the abilities to reproduce but not by the ability to produce a maximal amount of biomass or specific products. In order to thrive in the wild environment, cyanobacteria and algae have maximized their expression of pigments and antenna to compete with the competitors for sunlight. However, when monoculture was employed to produce high-density of biomass or maximal titers of specific products in photo-bioreactors, excessive photon capture by the cells in the surface layer can block the light availability to the cells underneath (Melis, 2009). To address this issue, studies have been conducted on minimizing the size of the photosystem antenna complex through various strategies, such as by expressing truncated light-harvesting antenna complex (LHC) mutants (Blankenship et al., 2011; Ort and Melis, 2011; Work et al., 2012), by down-regulating the expression of LHC through RNA interference (RNAi) and expression of LHC translation repressor in both cyanobacteria and algae (Mussgnug et al., 2007; Work et al., 2012). For example, the photosynthetic activity (measured by oxygen evolution) was about threefold higher in the alga strain Stm3LR3 (with LHC being down-regulated via RNAi) than in the parent strain Stm3 (without RNAi) after 100 min of high-light treatment; the cell growth rate also increased under high-light conditions after the LHC was down-regulated via RNAi (Mussgnug et al., 2007). Another bold proposal was to increase the photosynthesis efficiency by extending the light absorption range of the photosystems in cyanobacteria and algae (Blankenship et al., 2011). As the chlorophyll, carotenoids, and other accessory pigments in cyanobacteria and algae capture only visible region of the spectrum of solar radiation (400 to 700 nm), about 50% of the incident solar energy is dissipated and wasted during photosynthesis. Moreover, since the two photosystems compete for light with the same wavelengths, the overall efficiency is significantly reduced. Thus, it was proposed that one of the two photosystems be engineered to extend the absorption maxima to ~1100 nm, approximately doubling the solar photon capture, by heterologously expressing bacteriochlorophyll b (Blankenship et al., 2011).
CO2 fixation
RuBisCO is an essential enzyme in photosynthetic carbon fixation in Calvin–Benson–Bassham (CBB) cycle, catalyzing the combination of ribulose-1,5-bisphosphate with CO2. However, the reaction is slow. In addition, RuBisCO can also take O2 as substrate in addition to CO2 which further lower the carbon fixation efficiency. A recent study has revealed that despite slow catalytic turnover and confused CO2/O2 substrate specificity, RuBisCOs might have been nearly perfectly optimized (Tcherkez et al., 2006). In nature, cyanobacteria and some algae have evolved certain CO2-concentrating mechanisms (CCMs) to increase the CO2 fixation efficiencies. In cyanobacteria, RuBisCOs are sequestered together with carbonic anhydrous in carboxysomes, polyhedral microcompartments (MCPs) with proteinaceous shells. Anhydrase catalyzes the conversion of HCO3- to CO2 which is trapped by MCPs for RuBisCOs. Because CCMs can result in much higher CO2 concentration, and thus higher CO2 to O2 ratio, around the RuBisCOs, the carbon fixation efficiency is greatly increased (Espie and Kimber, 2011). It has been found that Synechococcus 7942 cells with more carboxysomes exhibited higher CO2 fixation rates (Savage et al., 2010). Heterologous expression of Synechococcus 6301 rbcLS (that encodes RuBisCO) in Synechococcus 7942 also led to more efficient CO2 fixation and higher yield of isobutyraldehyde in the genetically modified isobutyraldehyde-producing strain (Atsumi et al., 2009). Besides, overexpression of bicarbonate transporters has also been proposed to improve the photosynthesis efficiency (Price et al., 2011). Alternatively, RuBisCO-independent carbon fixation pathways have been posited. A recent work using in silico modeling of the recombination of existing metabolic building blocks showed that some of the proposed carbon fixation cycles have overall higher kinetic rates (Bar-Even et al., 2010). For example, by coupling the phosphoenolpyruvate carboxylase and the core of the natural C4 carbon fixation cycle, the overall CO2 fixation rate was predicted as 2–3 folds higher than that of the CBB cycle which employs RuBisCO (Bar-Even et al., 2010).
OVERCOMING THE OXIDATIVE STRESS
Since cyanobacteria and algae are oxygenic microorganisms, the abundant oxygen evolved by splitting water during photosynthesis process becomes an issue for expressing oxygen-sensitive enzymes. For example, either [NiFe] or [FeFe] hydrogenase required for biological production of H2 has low oxygen-tolerance (Lee et al., 2010); and the nitrogenases which fix N2 into NH4+ are also extremely oxygen-sensitive (Fay, 1992). From a broader prospect, this oxygen sensitivity issue could be crucial for successful expression of a large number of pathways from anaerobic microorganisms in oxygenic cyanobacteria and algae. To address the issue, efforts have been made to obtain oxygen-resistant enzymes from nature or through mutagenesis. For example, hydrogenases with better oxygen-tolerance have been found from Ralstonia eutropha H16 (Saggu et al., 2009) and Hydrogenovibrio marinus (Yoon et al., 2011); and elevated oxygen-tolerance has been made for the hydrogenase of Desulfovibrio fructosovorans by a single V74M mutation (Dementin et al., 2009). Alternatively, temporal segregation of oxygenic photosynthesis and hydrogen biosynthesis would be another option. In nature, many cyanobacterial species have evolved the mechanism to photosynthetically fix CO2 during day time while fix N2 by the oxygen-sensitive nitrogenases during night (Fay, 1992). Thereby, the solar energy can be firstly fixed into carbohydrates, such as starch, during oxygenic photosynthesis and then be utilized to power the oxygen-sensitive reactions during dark anoxic conditions. In addition, spatial segregation could be used. Hydrogenases can be localized to certain advantageous space, such as being expressed in heterocysts, to avoid the oxidative stress (Fay, 1992). Recent studies on bacterial MCPs assembling might have provided another opportunity to spatially segregate incompatible oxygenic and oxygen-sensitive processes (Fan et al., 2010; Heinhorst and Cannon, 2010; Bonacci et al., 2012). Moreover, Mehler reaction can be used to overcome the oxidative stress (Mehler, 1951; Asada, 2006). Mehler reaction has been evolved to overcome the intracellular oxidative stress by scavenging reactive oxygen species in cyanobacteria and chloroplasts (Kana, 1993; Asada, 2006). For instance, during N2-fixation period Mehler reaction consumes ~75% of gross O2 production and therefore maintains the O2 concentration at a low level (Kana, 1993; Milligan et al., 2007). However, Mehler reaction consumes reductant significantly (Asada, 2006); thus, in the future it will be of great interest and of vital importance to maintain the activity locally around the oxygen-sensitive enzymes rather than in the entire cytoplastic environment.
SYSTEMATIC APPROACHES
Functional genomics
Functional genomics, i.e., transcriptomics, proteomics, and metabolomics, would greatly promote the development of synthetic biology in cyanobacteria and algae. Albeit the genomes of some many species of cyanobacteria and algae have been sequenced1,2, a large portion of the sequenced genomes have not yet been annotated and the regulatory networks are still very poorly understood. The study of cyanobacterial and algal transcriptomes, proteomes, and metabolomes would allow for identification of new genes, pathways and regulatory networks which are essential to expand the size and diversity of the pool of genetic tools for synthetic biology. For example, recent transcriptomics studies on Synechocystis 6803 has enhanced the understanding of the transcriptional regulation in this photosynthetic microorganism which revealed that approximate two-thirds of the transcriptional start sites give rise to asRNAs and noncoding RNAs (ncRNAs), indicating that asRNAs and ncRNAs play an important role in cyanobacterial genetic regulation (Mitschke et al., 2011). We prospect that omics may be the key to collect information about the interactions and regulations to develop a sustainable green chemistry industry.
Metabolic modeling
Although most synthetic biology research in cyanobacteria and algae focus on local pathway optimization, comprehensive synthetic biology summons optimization of the genetic network and metabolic flux at the systems level. Genome-scale metabolic modeling allows theoretically evaluating the impact of genetic and environmental perturbations on the biomass yield and metabolic flux distribution and allows predicting the optimal metabolic flux profile to maximize the value of a given objective function (Shastri and Morgan, 2005; Knoop et al., 2010; Yoshikawa et al., 2011). The in silico modeling may thus provide a systematic approach to design an optimal metabolic network to maximize the production of the interest biofuel or chemical. Such genome-scale metabolic network models have been constructed for cyanobacteria and algae, and have been utilized to predict new targets to improve product yields and new pathways (Shastri and Morgan, 2005; Knoop et al., 2010; Dal’Molin et al., 2011; Yoshikawa et al., 2011). However, the reconstruction of the global metabolic networks is still in the infancy stage and the simulation results rely significantly on the included pathways. For instance, with ambiguities in metabolic networks in Synechocystis 6803, the estimated metabolic fluxes could be significantly different from the experimental results (Yoshikawa et al., 2011). In order to refine the quality of the reconstructed metabolic networks and thus the simulation of metabolic flux, it is inevitable to couple with experimental characterization of the metabolic networks in cyanobacteria and algae (Yoshikawa et al., 2011). As an example, by firstly investigating the in vitro activities of the purified relevant enzyme products (heterologously expressed in E. coli) and subsequently verifying their in vivo activities in the native host Synechococcus 7002, Zhang and Bryant (2011) reported that two enzymes could functionally compensate for the missing 2-oxoglutarate dehydrogenase in the TCA cycle. Further database searches indicated that homologs of these two enzymes occur in all cyanobacteria but Prochlorococcus and marine Synechococcus, which overturned the previously widely accepted assumption that cyanobacteria possess an incomplete TCA cycle (Meeks, 2011; Zhang and Bryant, 2011). Such discoveries would be of utter importance to reconstruct qualified in silico models for simulating the metabolic flux in the future.
CONCLUSION
Owing to the relatively simple genetic contents and the ability to capture solar energy, fix CO2, grow fast and directly synthesize specific products, cyanobacteria and algae have become excellent candidates for building autotrophic cell factories to produce renewable surrogate fuels and chemicals. With a large pool of genome sequences and improved genetic tools being available, application of synthetic biology in these photosynthetic microorganisms are highly desirable. In recent years, exciting results have been achieved not only in understanding of the fundamental molecular mechanisms but also in producing various interest products, such as biofuels and chemicals, utilizing cyanobacteria and algae as the production platforms. Nevertheless, synthetic biology in cyanobacteria and algae is still in its infancy and synthetic biologists are facing great challenges and opportunities in addressing various issues, such as improving the tools for genetic manipulation, enhancing light harvesting, increasing CO2 fixation efficiency, and overcoming of the intracellular oxidative stress. Systematic approaches, such as functional genomics and metabolic modeling, may also diversify the genetic tools and help the metabolic network design. It is doubtless that synthetic biology would be indispensable for the future success in applying cyanobacteria and algae for various biotechnological purposes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the NEPTUNE fund granted to Deirdre R. Meldrum at ASU. Weiwen Zhang is currently funded by a grant from National Basic Research Program of China (National “973” program, project No. 2011CBA00803).
Footnotes
REFERENCES
- Adhya S., Gottesman M. (1982). Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29 939–944 [DOI] [PubMed] [Google Scholar]
- Apt K. E., Kroth-Pancic P. G., Grossman A. R. (1996). Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252 572–579 [DOI] [PubMed] [Google Scholar]
- Argueta C., Yuksek K., Summers M. (2004). Construction and use of GFP reporter vectors for analysis of cell-type-specific gene expression in Nostoc punctiforme. J. Microbiol. Methods 59 181–188 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Cann A. F., Connor M. R., Shen C. R., Smith K. M., Brynildsen M. P., Chou K. J., Hanai T., Liao J. C. (2008a). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10 305–311 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J. C. (2008b). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451 86–89 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Higashide W., Liao J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27 1177–1180 [DOI] [PubMed] [Google Scholar]
- Bar-Even A., Noor E., Lewis N. E., Milo R. (2010). Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 107 8889–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T. S., Widmaier D. M., Temme K., Mirsky E. A., Santi D. V., Voigt C. A. (2009). Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 131 6508–6515 [DOI] [PubMed] [Google Scholar]
- Beck C., Knoop H., Axmann I. M., Steuer R. (2012). The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genomics 13 56 10.1186/1471-2164-13-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E., Meyer R. (1997). Acquisition of resistance genes by the IncQ plasmid R1162 is limited by its high copy number and lack of a partitioning mechanism. J. Bacteriol. 179 5947–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P. N., Ren L., Chua N. H. (1990). Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 9 1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S. (2003). Act natural. Nature 421 118 [DOI] [PubMed] [Google Scholar]
- Berman-Frank I., Lundgren P., Chen Y. B., Küpper H., Kolber Z., Bergman B., Falkowski P. (2001). Segregation of nitrogen fixation and oxygenic photosynthesis in the marine cyanobacterium Trichodesmium. Science 294 1534–1537 [DOI] [PubMed] [Google Scholar]
- Bermejo L. L., Welker N. E., Papoutsakis E. T. (1998). Expression of Clostridium acetobutylicum ATCC 824 genes in Escherichia coli for acetone production and acetate detoxification. Appl. Environ. Microbiol. 64 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship R. E., Tiede D. M., Barber J., Brudvig G. W., Fleming G., Ghirardi M., Gunner M. R., Junge W., Kramer D. M., Melis A., Moore T. A., Moser C. C., Nocera D. G., Nozik A. J., Ort D. R., Parson W. W., Prince R. C., Sayre R. T. (2011). Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science 332 805–809 [DOI] [PubMed] [Google Scholar]
- Bonacci W., Teng P. K., Afonso B., Niederholtmeyer H., Grob P., Silver P. A., Savage D. F. (2012). Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. U.S.A. 109 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W. (1993). Chloroplast transformation in Chlamydomonas. Methods Enzymol. 217 510–536 [DOI] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B., Sanford J. C. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240 1534–1538 [DOI] [PubMed] [Google Scholar]
- Buzby J. S., Porter R. D., Stevens S. E. , Jr (1985). Expression of the Escherichia coli lacZ gene on a plasmid vector in a cyanobacterium. Science 230 805–807 [DOI] [PubMed] [Google Scholar]
- Campbell W. H., Gowri G. (1990). Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 92 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Ma X., Msanne J., Repas T. (2011). RNA-mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryot. Cell 10 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Bjerknes M., Kumar R., Jay E. (1994). Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22 4953–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. B., Dominic B., Mellon M. T., Zehr J. P. (1998). Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS101. J. Bacteriol. 180 3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisti Y. (2007). Biodiesel from microalgae. Biotechnol. Adv. 25 294–306 [DOI] [PubMed] [Google Scholar]
- Coll J. M. (2006). Methodologies for transferring DNA into eukaryotic microalgae. Spanish J. Agric. Res. 4 316–330 [Google Scholar]
- Connor M., Atsumi S. (2010). Synthetic biology guides biofuel production. J. Biomed. Biotechnol. 2010 541698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal’Molin C. G., Quek L. E., Palfreyman R. W., Nielsen L. K. (2011). AlgaGEM – a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genomics 12 S5 10.1186/1471-2164-12-S4-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. S., Solbiati J., Cronan J. E., Jr. (2000). Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 275 28593–28598 [DOI] [PubMed] [Google Scholar]
- de Smit M. H, van Duin J. (1990). Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 87 7668–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J. D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekishima Y., Lan E. I., Shen C. R., Cho K. M., Liao J. C. (2011). Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J. Am. Chem. Soc. 133 11399–11401 [DOI] [PubMed] [Google Scholar]
- Dellomonaco C., Clomburg J. M., Miller E. N., Gonzalez R. (2011). Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476 355–359 [DOI] [PubMed] [Google Scholar]
- Dementin S., Leroux F., Cournac L., de Lacey A. L., Volbeda A., Léger C., Burlat B., Martinez N., Champ S., Martin L., Sanganas O., Haumann M., Fernández V. M., Guigliarelli B., Fontecilla-Camps J. C., Rousset M. (2009). Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant. J. Am. Chem. Soc. 131 10156–10164 [DOI] [PubMed] [Google Scholar]
- Deng M. D., Coleman J. R. (1999). Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J., Fu P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energ. Environ. Sci. 2 857–864 [Google Scholar]
- Ducat D. C., Sachdeva G., Silver P. A. (2011a). Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108 3941–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat D. C., Way J. C., Silver P. A. (2011b). Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 29 95–103 [DOI] [PubMed] [Google Scholar]
- Dürre P. (2007). Biobutanol: an attractive biofuel. Biotechnol. J. 2 1525–1534 [DOI] [PubMed] [Google Scholar]
- Dwyer D. J., Kohanski M. A., Hayete B., Collins J. J. (2007). Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton-Rye J. J. (2004). The construction of gene knockouts in the cyanobacterium Synechocystis sp. PCC 6803. Methods Mol. Biol. 274 309–324 [DOI] [PubMed] [Google Scholar]
- Eichler-Stahlberg A., Weisheit W., Ruecker O., Heitzer M. (2009). Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229 873–883 [DOI] [PubMed] [Google Scholar]
- Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. (1997). Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179 1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J., Wolk C. P. (1988). Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167 747–754 [DOI] [PubMed] [Google Scholar]
- Endy D. (2005). Foundations for engineering biology. Nature 438 449–453 [DOI] [PubMed] [Google Scholar]
- Energy Information Administration. (2010). Annual Energy Review 2009, U.S. [Google Scholar]
- Espie G. S., Kimber M. S. (2011). Carboxysomes: cyanobacterial RubisCO comes in small packages. Photosynth. Res. 109 7–20 [DOI] [PubMed] [Google Scholar]
- Fan C., Cheng S., Liu Y., Escobar C. M., Crowley C. S., Jefferson R. E., Yeates T. O., Bobik T. A. (2010). Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 107 7509–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay P. (1992). Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56 340–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Xue L., Liu H., Lu P. (2009). Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 36 1433–1439 [DOI] [PubMed] [Google Scholar]
- Fernando G., Adhikari S., Kota K., Bandi R. (2007). Glycerol based automotive fuels from future biorefineries. Fuel 86 2806–2809 [Google Scholar]
- Fernández E., Schnell R., Ranum L. P., Hussey S. C., Silflow C. D., Lefebvre P. A. (1989). Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 86 6449–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Ogawa T., Tazaki M., Nagahama K., Fujii T., Tanase S., Morino Y. (1992). Two reactions are simultaneously catalyzed by a single enzyme: the arginine-dependent simultaneous formation of two products, ethylene and succinate, from 2-oxoglutarate by an enzyme from Pseudomonas syringae. Biochem. Biophys. Res. Commun. 188 483–489 [DOI] [PubMed] [Google Scholar]
- Fukuda H., Takahashi M., Fujii T., Tazaki M., Ogawa T. (1989). An NADH:Fe(III)EDTA oxidoreductase from Cryptococcus albidus: an enzyme involved in ethylene production in vivo? FEMS Microbiol. Lett. 51 107–111 [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Moriuchi T., Morishima K. (2006). Design principle and prototyping of a direct photosynthetic/metabolic biofuel cell (DPMFC). J. Micromech. Microeng. 16 S220–S225 [Google Scholar]
- Geerts D., Bovy A., de Vrieze G., Borrias M., Weisbeek P. (1995). Inducible expression of heterologous genes targeted to a chromosomal platform in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology 141 831–841 [DOI] [PubMed] [Google Scholar]
- Georg J., Voss B., Scholz I., Mitschke J., Wilde A., Hess W. R. (2009). Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M. L., Dubini A., Yu J., Maness P. C. (2009). Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38 52–61 [DOI] [PubMed] [Google Scholar]
- Ghirardi M. L., Posewitz M. C., Maness P. C., Dubini A., Yu J., Seibert M. (2007). Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58 71–91 [DOI] [PubMed] [Google Scholar]
- Golden S. S., Brusslan J., Haselkorn R. (1987). Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 153 215–231 [DOI] [PubMed] [Google Scholar]
- Golden S. S., Sherman L. A. (1983). A hybrid plasmid is a stable cloning vector for the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 155 966–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieva G., Shestakov S. (1982). Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 13 367–370 [Google Scholar]
- Hagemann M. (2011). Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 35 87–123 [DOI] [PubMed] [Google Scholar]
- Hankamer B., Lehr F., Rupprecht J., Mussgnug J. H., Posten C., Kruse O. (2007). Photosynthetic biomass and H-2 production by green algae: from bioengineering to bioreactor scale-up. Physiol. Plant. 131 10–21 [DOI] [PubMed] [Google Scholar]
- Harun R., Danquah M. K., Forde G. M. (2010). Microalgal biomass as a fermentation feedstock for bioethanol production. J. Chem. Technol. Biotechnol. 85 199–203 [Google Scholar]
- Heidorn T., Camsund D., Huang H. H., Lindberg P., Oliveira P., Stensjö K., Lindblad P. (2011). Synthetic biology in cyanobacteria: engineering and analyzing novel functions. Methods Enzymol. 497 539–579 [DOI] [PubMed] [Google Scholar]
- Hein S., Tran H, Steinbüchel A. (1998). Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch. Microbiol. 170 162–170 [DOI] [PubMed] [Google Scholar]
- Heinhorst S., Cannon G. (2010). Addressing microbial organelles: a short peptide directs enzymes to the interior. Proc. Natl. Acad. Sci. U.S.A. 107 7627–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer M., Eckert A., Fuhrmann M., Griesbeck C. (2007). Influence of codon bias on the expression of foreign genes in microalgae. Adv. Exp. Med. Biol. 616 46–53 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A., Melis A., Happe T. (2009). Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth. Res. 102 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J. A., Muro-Pastor A. M., Flores E., Bes M. T., Peleato M. L., Fillat M. F. (2006). Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J. Mol. Biol. 355 325–334 [DOI] [PubMed] [Google Scholar]
- Hirayama S., Ueda R., Ogushi Y., Hirano A., Samejima Y., Hon-Nami K., Kunito S. (1998). Ethanol production from carbon dioxide by fermentative microalgae. Stud. Surf. Sci. Catal. 114 657–660 [Google Scholar]
- Huang H. H., Camsund D., Lindblad P., Heidorn T. (2010). Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 38 2577–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M., Nishihara H., Yoon K. S., Lenz O., Friedrich B., Nakamoto H., Kojima K., Honma D., Kamachi T., Okura I. (2006). Light-driven hydrogen production by a hybrid complex of a [NiFe]-hydrogenase and the cyanobacterial photosystem I. Photochem. Photobiol. 82 676–682 [DOI] [PubMed] [Google Scholar]
- Imashimizu M., Fujiwara S., Tanigawa R., Tanaka K., Hirokawa T., Nakajima Y., Higo J., Tsuzuki M. (2003). Thymine at -5 is crucial for cpc promoter activity of Synechocystis sp. strain PCC 6714. J. Bacteriol. 185 6477–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M., Suda M., Kimura S., Yasuda K., Suzuki H., Toda H., Yamamoto S., Okino S., Suzuki N., Yukawa H. (2008). Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77 1305–1316 [DOI] [PubMed] [Google Scholar]
- Isaacs F. J., Dwyer D. J., Ding C., Pervouchine D. D., Cantor C. R., Collins J. J. (2004). Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22 841–847 [DOI] [PubMed] [Google Scholar]
- Iwaki T., Haranoh K., Inoue N., Kojima K., Satoh R., Nishino T., Wada S., Ihara H., Tsuyama S., Kobayashi H., Wadano A. (2006). Expression of foreign type I ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) stimulates photosynthesis in cyanobacterium Synechococcus PCC7942 cells. Photosynth. Res. 88 287–297 [DOI] [PubMed] [Google Scholar]
- Kana T. M. (1993). Rapid oxygen cycling in Trichodesmium thiebautii. Limnol. Oceanogr. 38 18–24 [Google Scholar]
- Kane J. F. (1995). Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6 494–500 [DOI] [PubMed] [Google Scholar]
- Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., Kimura T., Hosouchi T., Matsuno A., Muraki A., Nakazaki N., Naruo K., Okumura S., Shimpo S., Takeuchi C., Wada T., Watanabe A., Yamada M., Yasuda M., Tabata S. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3 109–136 [DOI] [PubMed] [Google Scholar]
- Kathiresan S., Chandrashekar A., Ravishankar A., Sarada R. (2009). Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (Chlorophyceae, Volvocales). J. Phycol. 45 642–649 [DOI] [PubMed] [Google Scholar]
- Keasling J. (2008). Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3 64–76 [DOI] [PubMed] [Google Scholar]
- Kende H. (1993). Ethylene biosynthesis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 44 283–307 [Google Scholar]
- Kilian O., Benemann C. S., Niyogi K. K., Vick B. (2011). High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U.S.A. 108 21265–21269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H., Zilliges Y., Lockau W., Steuer R. (2010). The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol. 154 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksharova O., Wolk C. (2002). Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 58 123–137 [DOI] [PubMed] [Google Scholar]
- Kosugi Y., Shibuya K., Tsuruno N., Iwazaki Y., Mochizuki A., Yoshioka T., Hashiba T., Satoh S. (2000). Expression of genes responsible for ethylene production and wilting are differently regulated in carnation (Dianthus caryophyllus L.) petals. Plant Sci. 158 139–145 [DOI] [PubMed] [Google Scholar]
- Kruse O., Rupprecht J., Bader K. P., Thomas-Hall S., Schenk P. M., Finazzi G., Hankamer B. (2005). Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 280 34170–34177 [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C. J., Thomas A. A., van der Ende A., van Leen R. W., Borrias W. E., van den Hondel C. A, van Arkel G. A. (1983). A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid 10 156–163 [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C. J, van Arkel G. A. (1987). Host-vector systems for gene cloning in cyanobacteria. Methods Enzymol. 153 199–215 [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Misquitta R. W., Reddy V. S., Rao B. J., Rajam M. V. (2004). Genetic transformation of the green alga – Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 166 731–738 [Google Scholar]
- Lambert G., Carr N. (1983). A restriction map of plasmid pDC1 from the filamentous cyanobacterium Nostoc sp. MAC PCC 8009. Plasmid 10 196–198 [DOI] [PubMed] [Google Scholar]
- Lamsen E. N., Atsumi S. (2012). Recent progress in synthetic biology for microbial production of C3-C10 alcohols. Front. Microbiol. 3:196. 10.3389/fmicb.2012.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E. I., Liao J. C. (2011). Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 13 353–363 [DOI] [PubMed] [Google Scholar]
- Lan E. I., Liao J. C. (2012). ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 109 6018–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J. D., Haselkorn R. (1991). A vector for analysis of promoters in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173 2729–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P., Brown J. W. S., Pintortoro J. A., Feix G., Neuhaus G., Neuhausurl G., Schweiger H. G. (1986). Expression of zein genes in Acetabularia mediterranea. Eur. J. Cell Biol. 39 257–264 [Google Scholar]
- Lee H. S., Vermaas W. F., Rittmann B. E. (2010). Biological hydrogen production: prospects and challenges. Trends Biotechnol. 28 262–271 [DOI] [PubMed] [Google Scholar]
- Lee S. K., Chou H., Ham T. S., Lee T. S., Keasling J D. (2008). Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19 556–563 [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Lee Y. (2003). Metabolic engineering of Escherichia coli for production of enantiomerically pure (R)-(-)-hydroxycarboxylic acids. Appl. Environ. Microbiol. 69 3421–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León R, Fernández E. (2007). Nuclear transformation of eukaryotic microalgae: historical overview, achievements and problems. Adv. Exp. Med. Biol. 616 1–11 [DOI] [PubMed] [Google Scholar]
- León-Bañares R., González-Ballester D., Galván A, Fernández E. (2004). Transgenic microalgae as green cell-factories. Trends Biotechnol. 22 45–52 [DOI] [PubMed] [Google Scholar]
- Lerche K., Hallmann A. (2009). Stable nuclear transformation of Gonium pectorale. BMC Biotechnol. 9 64 10.1186/1472-6750-9-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu Y., Xue L., Xie H. (2010). A structurally novel salt-regulated promoter of duplicated carbonic anhydrase gene 1 from Dunaliella salina. Mol. Biol. Rep. 37 1143–1154 [DOI] [PubMed] [Google Scholar]
- Li R., Golden S. S. (1993). Enhancer activity of light-responsive regulatory elements in the untranslated leader regions of cyanobacterial psbA genes. Proc. Natl. Acad. Sci. U.S.A. 90 11678–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg P., Park S., Melis A. (2010). Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12 70–79 [DOI] [PubMed] [Google Scholar]
- Liu Q., Ouyang S., Chung A., Wu Q., Chen G. (2007). Microbial production of R-3-hydroxybutyric acid by recombinant E. coli harboring genes of phbA, phbB, and tesB. Appl. Microbiol. Biotechnol. 76 811–818 [DOI] [PubMed] [Google Scholar]
- Liu X., Curtiss R. III. (2009). Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 106 21550–21554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fallon S., Sheng J., Curtiss R. III. (2011a). CO(2)-limitation-inducible Green Recovery of fatty acids from cyanobacterial biomass. Proc. Natl. Acad. Sci. U.S.A. 108 6905–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sheng J., Curtiss R. III. (2011b). Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108 6899–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. (2010). A perspective: Photosynthetic production of fatty acid-based biofuels in genetically engineered cyanobacteria. Biotechnol. Adv. 28 742–746 [DOI] [PubMed] [Google Scholar]
- Lu X., Vora H., Khosla C. (2008). Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10 333–339 [DOI] [PubMed] [Google Scholar]
- Lu Y., Li J., Xue L., Yan H., Yuan H., Wang C. (2011). A duplicated carbonic anhydrase 1 (DCA1) promoter mediates the nitrate reductase gene switch of Dunaliella salina. J. Appl. Phycol. 23 673–680 [Google Scholar]
- Lubner C. E., Applegate A. M., Knörzer P., Ganago A., Bryant D. A., Happe T., Golbeck J. H. (2011). Solar hydrogen-producing bionanodevice outperforms natural photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108 20988–20991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luinenburg I., Coleman J. R. (1993). Expression of Escherichia coli phosphoenolpyruvate carboxylase in a cyanobacterium. Functional complementation of Synechococcus PCC 7942 ppc. Plant Physiol. 101 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Campbell A., Karlin S. (2002). Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184 5733–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S. M., Li J. W., Choi J. W., Zhou H., Lee K. K., Moorthie V. A., Xie X., Kealey J. T., Da Silva N. A., Vederas J. C., Tang Y. (2009). Complete reconstitution of a highly reducing iterative polyketide synthase. Science 326 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21 796–802 [DOI] [PubMed] [Google Scholar]
- Mascarelli A. L. (2009). Gold rush for algae. Nature 461 460–461 [DOI] [PubMed] [Google Scholar]
- McNeely K., Xu Y., Bennette N., Bryant D. A., Dismukes G. C. (2010). Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 76 5032–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks J. C. (2011). Closing the cycle. Science 334 1508–1509 [DOI] [PubMed] [Google Scholar]
- Mehler A. H. (1951). Studies on reactions of illuminated chloroplasts: I. Mechanism of the reduction of oxygen and other hill reagents. Arch. Biochem. Biophys. 33 65–77 [DOI] [PubMed] [Google Scholar]
- Melis A. (2009). Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency. Plant Sci. 177 272–280 [Google Scholar]
- Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Perez D., Begemann M. B., Pfleger B. F. (2011). Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 77 4264–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermet-Bouvier P., Cassier-Chauvat C., Marraccini P., Chauvat F. (1993). Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the general Synechocystis and Synechococcus. Curr. Microbiol. 27 323–327 [Google Scholar]
- Mermet-Bouvier P., Chauvat F. (1994). A conditional expression vector for the cyanobacteria Synechocystis sp. strains PCC6803 and PCC6714 or Synechococcus sp. strains PCC7942 and PCC6301. Curr. Microbiol. 28 145–148 [DOI] [PubMed] [Google Scholar]
- Meyer R. (2009). Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Wu Q., Wu G., Zhao N. (2003). Sucrose accumulation in salt-stressed cells of agp gene deletion-mutant in cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 218 71–77 [DOI] [PubMed] [Google Scholar]
- Milligan A. J., Berman-Frank I., Gerchman Y., Dismukes G. C., Falkowski P. G. (2007). Light-dependent oxygen consumption in nitrogen-fixing cyanobacteria plays a key role in nitrogenase protection. J. Phycol. 43 845–852 [Google Scholar]
- Mitschke J., Georg J., Scholz I., Sharma C. M., Dienst D., Bantscheff J., Voss B., Steglich C., Wilde A., Vogel J., Hess W. R. (2011). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa-Yamaguchi A., Okami T., Kira N., Yamaguchi H., Ohnishi K., Adachi M. (2011). Stable nuclear transformation of the diatom Chaetoceros sp. Phycol. Res. 59 113–119 [Google Scholar]
- Mouille G., Maddelein M. L., Libessart N., Talaga P., Decq A., Delrue B., Ball S. (1996). Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, van Oudenaarden A. (2009). Synthetic biology: understanding biological design from synthetic circuits. Nat. Rev. Genet. 10 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian A. Y., Koonin E. V., Makarova K. S., Mekhedov S. L., Sorokin A., Wolf Y. I., Dufresne A., Partensky F., Burd H., Kaznadzey D., Haselkorn R., Galperin M. Y. (2006). The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103 13126–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Hihara Y. (2006). Characterization of high-light-responsive promoters of the psaAB genes in Synechocystis sp. PCC 6803. Plant Cell Physiol. 47 878–890 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Hihara Y. (2007). Coordinated high-light response of genes encoding subunits of photosystem I is achieved by AT-rich upstream sequences in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 189 2750–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug J. H., Thomas-Hall S., Rupprecht J., Foo A., Klassen V., McDowall A., Schenk P. M., Kruse O., Hankamer B. (2007). Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 5 802–814 [DOI] [PubMed] [Google Scholar]
- Muto M., Henry R. E., Mayfield S. P. (2009). Accumulation and processing of a recombinant protein designed as a cleavable fusion to the endogenous Rubisco LSU protein in Chlamydomonas chloroplast. BMC Biotechnol. 9 26 10.1186/1472-6750-9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Gojobori T., Ikemura T. (2000). Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28 292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus G., Neuhaus-Url G., de Groot E. J., Schweiger H. G. (1986). High yield and stable transformation of the unicellular green alga Acetabularia by microinjection of SV40 DNA and pSV2neo. EMBO J. 5 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. O., Zentella R., Wang Y., Taylor J. S., Pakrasi H. B. (2000). PhrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch. Microbiol. 173 412–417 [DOI] [PubMed] [Google Scholar]
- Nguyen M. T., Choi S. P., Lee J., Lee J. H., Sim S. J. (2009). Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J. Microbiol. Biotechnol. 19 161–166 [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H., Wolfstädter B. T., Savage D. F., Silver P. A., Way J. C. (2010). Engineering cyanobacteria to synthesize and export hydrophilic products. Appl. Environ. Microbiol. 76 3462–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J., Gudmundsson S., Knight E. M., Palsson B. O., Thiele I. (2012). Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. U.S.A. 109 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill B. M., Mikkelson K. L., Gutierrez N. M., Cunningham J. L., Wolff K. L., Szyjka S. J., Yohn C. B., Redding K. E., Mendez M. J. (2012). An exogenous chloroplast genome for complex sequence manipulation in algae. Nucleic Acids Res. 40 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort D. R., Melis A. (2011). Optimizing antenna size to maximize photosynthetic efficiency. Plant Physiol. 155 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger B. F., Pitera D. J., Smolke C. D., Keasling J. D. (2006). Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24 1027–1032 [DOI] [PubMed] [Google Scholar]
- Pisciotta J. M., Zou Y., Baskakov I. V. (2010). Light-dependent electrogenic activity of cyanobacteria. PLoS ONE 5 e10821 10.1371/journal.pone.0010821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz M. C., King P. W., Smolinski S. L., Smith R. D., Ginley A. R., Ghirardi M. L., Seibert M. (2005). Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33 102–104 [DOI] [PubMed] [Google Scholar]
- Potvin G., Zhang Z. (2010). Strategies for high-level recombinant protein expression in transgenic microalgae: a review. Biotechnol. Adv. 28 910–918 [DOI] [PubMed] [Google Scholar]
- Prather K. L., Martin C. H. (2008). De novo biosynthetic pathways: rational design of microbial chemical factories. Curr. Opin. Biotechnol. 19 468–474 [DOI] [PubMed] [Google Scholar]
- Price G. D., Badger M. R, von Caemmerer S. (2011). The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol. 155 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Lin H., Jiang P. (2012). Advances in genetic engineering of marine algae. Biotechnol. Adv. 10.1016/j.biotechadv.2012.05.004. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Radakovits R., Jinkerson R. E., Darzins A., Posewitz M. C. (2010). Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazanov A., Ramazanov Z. (2006). Isolation and characterization of a starchless mutant of Chlorella pyrenoidosa STL-PI with a high growth rate, and high protein and polyunsaturated fatty acid content. Phycol. Res. 54 255–259 [Google Scholar]
- Reaston J., van den Hondel C. A., van Arkel G. A., Stewart W. D. (1982). A physical map of plasmid pDU1 from the cyanobacterium Nostoc PCC 7524. Plasmid 7 101–104 [DOI] [PubMed] [Google Scholar]
- Reppas N. B., Ridley C. P. (2010). Methods and Compositions for the Recombinant Biosynthesis of n-Alkanes. Joule Unlimited, Inc., US Patent No. 7794969 [Google Scholar]
- Rittmann B. E. (2008). Opportunities for renewable bioenergy using microorganisms. Biotechnol. Bioeng. 100 203–212 [DOI] [PubMed] [Google Scholar]
- Ro D. K., Paradise E. M., Ouellet M., Fisher K. J., Newman K. L., Ndungu J. M., Ho K. A., Eachus R. A., Ham T. S., Kirby J., Chang M. C., Withers S. T., Shiba Y., Sarpong R., Keasling J. D. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440 940–943 [DOI] [PubMed] [Google Scholar]
- Ruffing A. M. (2011). Engineering cyanobacteria: teaching and old bug new tricks. Bioeng. Bugs 2 136–149 [DOI] [PubMed] [Google Scholar]
- Saggu M., Zebger I., Ludwig M., Lenz O., Friedrich B., Hildebrandt P., Lendzian F. (2009). Spectroscopic insights into the oxygen-tolerant membrane-associated [NiFe] hydrogenase of Ralstonia eutropha H16. J. Biol. Chem. 284 16264–16276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M., Ogawa T., Matsuoka M., Fukuda H. (1997). Photosynthetic conversion of carbon dioxide to ethylene by the recombinant cyanobacterium, Synechococcus sp. PCC 7942, which harbors a gene for the ethylene-forming enzyme of Pseudomonas syringae. J. Ferment. Bioeng. 84 434–443 [Google Scholar]
- Salis H. M., Mirsky E. A., Voigt C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27 946–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D. F., Afonso B., Chen A. H., Silver P. A. (2010). Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327 1258–1261 [DOI] [PubMed] [Google Scholar]
- Schroda M., Blöcker D., Beck C. F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21 121–131 [DOI] [PubMed] [Google Scholar]
- Schwarze A., Kopczak M. J., Rögner M., Lenz O. (2010). Requirements for construction of a functional hybrid complex of photosystem I and [NiFe]-hydrogenase. Appl. Environ. Microbiol. 76 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T. D., Wiberley A. E., Donohue A. R. (2008). Isoprene emission from plants: why and how. Ann. Bot. 101 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri A. A., Morgan J. A. (2005). Flux balance analysis of photoautotrophic metabolism. Biotechnol. Prog. 21 1617–1626 [DOI] [PubMed] [Google Scholar]
- Sheng J., Vannela R., Rittmann B. E. (2011). Evaluation of methods to extract and quantify lipids from Synechocystis PCC 6803. Bioresour. Technol. 102 1697–1703 [DOI] [PubMed] [Google Scholar]
- Shestakov S. V., Khyen N. T. (1970). Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol. Gen. Genet. 107 372–375 [DOI] [PubMed] [Google Scholar]
- Srirangan K., Pyne M. E, Perry Chou C. (2011). Biochemical and genetic engineering strategies to enhance hydrogen production in photosynthetic algae and cyanobacteria. Bioresour. Technol. 102 8589–8604 [DOI] [PubMed] [Google Scholar]
- Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463 559–562 [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. (2007). Challenges in engineering microbes for biofuels production. Science 315 801–804 [DOI] [PubMed] [Google Scholar]
- Stevens D. R., Rochaix J. D., Purton S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251 23–30 [DOI] [PubMed] [Google Scholar]
- Summers M. L., Wallis J. G., Campbell E. L., Meeks J. C. (1995). Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 177 6184–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K., Matsuoka M., Nagahama K., Ogawa T. (2003). Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 95 302–305 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Miyake M., Tokiwa Y., Asada Y. (1998). Improved accumulation of poly-3-hydroxybutyrate by a recombinant cyanobacterium. Biotechnol. Lett. 20 183–186 [Google Scholar]
- Takahashi M., Uji T., Saga N., Mikami K. (2010). Isolation and regeneration of transiently transformed protoplasts from gametophytic blades of the marine red alga Porphyra yezoensis. Electron. J. Biotechnol. 13 2 [Google Scholar]
- Takeshima Y., Sugiura M., Hagiwara H. (1994). A novel expression vector for the cyanobacterium, Synechococcus PCC 6301. DNA Res. 1 181–189 [DOI] [PubMed] [Google Scholar]
- Tan X., Yao L., Gao Q., Wang W., Qi F., Lu X. (2011). Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 13 169–176 [DOI] [PubMed] [Google Scholar]
- Taroncher-Oldenburg G., Nishina K., Stephanopoulos G. (2000). Identification and analysis of the polyhydroxyalkanoate-specific beta-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 66 4440–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G. G., Farquhar G. D., Andrews T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 103 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T., Wolk C. P. (1987). Conjugal transfer of plasmids to cyanobacteria. Methods Enzymol. 153 232–243 [DOI] [PubMed] [Google Scholar]
- Tseng H., Martin C., Nielsen D., Prather K. (2009). Metabolic engineering of Escherichia coli for enhanced production of (R)- and (S)-3-hydroxybutyrate. Appl. Environ. Microb. 75 3137–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo K. E., Jin Y. S., Espinoza F. A., Stephanopoulos G. (2009). Identification of gene disruptions for increased poly-3-hydroxybutyrate accumulation in Synechocystis PCC 6803. Biotechnol. Prog. 25 1236–1243 [DOI] [PubMed] [Google Scholar]
- van de Meene A. M., Hohmann-Marriott M. F., Vermaas W. F., Roberson R. W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184 259–270 [DOI] [PubMed] [Google Scholar]
- Wang J., Jiang P., Cui Y., Guan X., Qin S. (2010). Gene transfer into conchospores of Porphyra haitanensis (Bangiales, Rhodophyta) by glass bead agitation. Phycologia 49 355–360 [Google Scholar]
- Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009). Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargacki A. J., Leonard E., Win M. N., Regitsky D. D., Santos C. N., Kim P. B., Cooper S. R., Raisner R. M., Herman A., Sivitz A. B., Lakshmanaswamy A., Kashiyama Y., Baker D., Yoshikuni Y. (2012). An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335 308–313 [DOI] [PubMed] [Google Scholar]
- Wee Y. J., Kim J. N., Ryu H. W. (2006). Biotechnological production of lactic acid and its recent applications. Food Technol. Biotech. 44 163–172 [Google Scholar]
- Wolk C. P., Elhai J., Kuritz T., Holland D. (1993). Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol. Microbiol. 7 441–445 [DOI] [PubMed] [Google Scholar]
- Wolk C. P., Vonshak A., Kehoe P., Elhai J. (1984). Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 81 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work V. H., D’Adamo S., Radakovits R., Jinkerson R. E., Posewitz M. C. (2012). Improving photosynthesis and metabolic networks for the competitive production of phototroph-derived biofuels. Curr. Opin. Biotechnol. 23 290–297 [DOI] [PubMed] [Google Scholar]
- Wu G. F., Wu Q. Y., Shen Z. Y. (2001). Accumulation of poly-beta-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresour. Technol. 76 85–90 [DOI] [PubMed] [Google Scholar]
- Yang S. F., Hoffman N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Ann. Rev. Plant Physiol. 35 155–189 [Google Scholar]
- Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J. D., Osterhout R. E., Stephen R., Estadilla J., Teisan S., Schreyer H. B., Andrae S., Yang T. H., Lee S. Y., Burk M. J, Van Dien S. (2011). Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 7 445–452 [DOI] [PubMed] [Google Scholar]
- Yoon K. S., Fukuda K., Fujisawa K., Nishihara H. (2011). Purification and characterization of a highly thermostable, oxygen-resistant, respiratory [NiFe]-hydrogenase from a marine, aerobic hydrogen-oxidizing bacterium Hydrogenovibrio marinus. Int. J. Hydrogen Energ. 36 7081–7088 [Google Scholar]
- Yoshikawa K., Kojima Y., Nakajima T., Furusawa C., Hirasawa T., Shimizu H. (2011). Reconstruction and verification of a genome-scale metabolic model for Synechocystis sp. PCC6803. Appl. Microbiol. Biotechnol. 92 347–358 [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Li J., Yang Y., Qi L., Chen B., Zhao F., Bao Q., Wu J. (2012). Codon usage patterns and adaptive evolution of marine unicellular cyanobacteria Synechococcus and Prochlorococcus. Mol. Phylogenet Evol. 62 206–213 [DOI] [PubMed] [Google Scholar]
- Yurieva T. M., Plyasova L. M., Makarova O. V., Krieger T. A. (1996). Mechanisms for hydrogenation of acetone to isopropanol and of carbon oxides to methanol over copper-containing oxide catalysts. J. Mol. Catal. A Chem. 113 455–468 [Google Scholar]
- Zang X., Liu B., Liu S., Arunakumara K. K., Zhang X. (2007). Optimum conditions for transformation of Synechocystis sp. PCC 6803. J. Microbiol. 45 241–245 [PubMed] [Google Scholar]
- Zhang S., Bryant D. A. (2011). The tricarboxylic acid cycle in cyanobacteria. Science 334 1551–1553 [DOI] [PubMed] [Google Scholar]
- Zhang F., Carothers J. M., Keasling J. D. (2012). Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30 354–359 [DOI] [PubMed] [Google Scholar]
- Zhang K., Sawaya M. R., Eisenberg D. S., Liao J. C. (2008). Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U.S.A. 105 20653–20658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhang H., Zhang Y., Li Y., Ma Y. (2012). Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 14 394–400 [DOI] [PubMed] [Google Scholar]