Abstract
We previously reported frequent truncating mutation of the RNA binding protein gene, La ribonucleoprotein domain family, member 7 (LARP7) in gastric cancers (GC) with frequent microsatellite instability. LARP7 negatively regulates positive transcription elongation factor-b (p-TEFb) by binding to and stabilizing 7sk RNA. p-TEFb has been linked to proliferation and de-differentiation in various tissues. Therefore, we reasoned that loss of LARP7 may contribute to gastric tumorigenesis. In the current study, we evaluated LARP7 mRNA expression in 18 GCs, their corresponding non-neoplastic gastric tissues (NGC), and 18 normal gastric tissues from healthy individuals (NN).We also assessed the effects of transient siRNA-mediated LARP7 knockdown in immortalized non-neoplastic gastric epithelial cells. LARP7 mRNA was significantly decreased in GCs (median 2.5) relative to NNs (median 14.9, p<0.01) as well as relative to their corresponding NGCs (median 8.1, p<0.01). Transfection of a siRNA directed against LARP7 (anti-LARP7 siRNA) decreased 7sk levels by 72% relative to a control siRNA (p<0.01). Furthermore, anti-LARP7 siRNA transfection increased cell proliferation by 23% (p<0.01) and cell migration by 22% (p<0.001) relative to control siRNA transfection. Taken together, these findings suggest that LARP7 downregulation occurs early during gastric tumorigenesis and may promote gastric tumorigenesis via p-TEFb deregulation.
Keywords: LARP7, gastric cancer, RNA binding protein, 7sk snRNA, p-TEFb
INTRODUCTION
Gastric adenocarcinoma (GC) is a major cause of cancer-related death worldwide, ranking second only to lung cancer (1). There are approximately 750,000 new cases diagnosed annually around the world, and 5-year overall survival rates are less than 25%(1). It is therefore important to increase our understanding of the molecular events underlying GC, not only to improve early detection, but also to aid in the development of more effective interventions. Unfortunately, precise mechanisms giving rise to the malignant transformation of gastric epithelial cells are not yet fully understood, awaiting the revelation of additional genes and pathways unique to this deadly disease(2).
We previously reported frequent frameshift mutation of a novel gene, La ribonucleoprotein domain family, member-7 (LARP7, also referred to as HDCMA18P or PIP7S), in a subset of human GC (3) . LARP7 belongs to the LARP RNA binding protein family, which includes the lupus LA antigen gene, and modulates the metabolism and function of a variety of RNA species (4). LARP7 binds to and stabilizes 7sk snRNA(5, 6). In turn, the 7sk-LARP7 complex suppresses the positive transcription elongation factor b (p-TEFb) complex. The p-TEFb complex, consisting of CDK9 and cyclin T1, is the ubiquitous and principal promoter of general mRNA elongation and processing (5, 7). p-TEFb is a crucial mediator of c-Myc-mediated transcriptional activation(8), and p-TEFb hyperactivity promotes malignant transformation of fibroblasts in vitro(9). Furthermore, LARP7 deletion has been reported to result in transformation of a cultured mamillary epithelial cell line via p-TEFb activation (5). Thus, these are possible pathways and mechanisms through which LARP7 may exert tumor-suppressive effects in the stomach.
In order to assess potential tumor-suppressive properties of the LARP7 gene in the stomach, we first evaluated LARP7 expression in non-neoplastic and neoplastic gastric tissues. We then investigated functional effects of siRNA-mediated LARP7 inhibition in immortalized non-neoplastic gastric epithelial cells.
MATERIALS AND METHODS
Materials
HFE145, an immortalized human non-neoplastic gastric epithelial cell line, was previously established by H. Ashktorab and D. T. Smoot (10, 11). In the current study, eighteen primary GCs and their corresponding non-neoplastic gastric mucosal tissues (referred to as NGC) were obtained at surgery from GC patients who had not undergone any neoadjvant therapy. Surgically resected tissues were macroscopically dissected to eliminate serosal and muscular layers prior to cryostorage. An additional 18 normal gastric mucosal tissues were obtained from individuals who underwent endoscopy for asymptomatic Barrett's esophagus surveillance and exhibited no abnormalities in the stomach. Written informed consent was obtained from all the participants according to a protocol approved by the Johns Hopkins University School of Medicine Institutional Review Board. Clinical and pathological characteristics are summarized in Table 1.
Table 1.
Patient clinical and demographical data
| Case class | Gastric cancer | Control | *p |
|---|---|---|---|
| Number: | 18 | 18 | |
| Age: mean (SD) | 66.3 (16.2) | 56.2 (19.6) | P=0.10 |
| Gender: | P=1.00 | ||
| F | 8 | 9 | |
| M | 10 | 9 | |
| Race: | P=0.38 | ||
| Black | 4 | 2 | |
| White | 11 | 15 | |
| Other/unavailable | 3 | 1 | |
| Tumor stage: | NA | ||
| I | 2 | NA | |
| II | 4 | NA | |
| III | 4 | NA | |
| IV | 6 | NA | |
| unavailable | 2 | NA | |
| Tumor histology: | NA | ||
| Intestinal | 11 | NA | |
| Diffuse | 6 | NA | |
| unavailable | 1 | NA | |
p: Student's T-test p-value for age, and Fisher's exact test p-values for gender and race comparison. NA, not applicable.
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) and treated with RNase-free DNase. First-strand cDNA was synthesized from 1 μg of total RNA using a RevertAid™ cDNA synthesis kit (Fermentas, Glen Burnie, MD). qRT-PCR was performed using iQ SYBR Green Supermix or iQ Probe Supermix on an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA). β-actin and GAPDH were used as normalization controls, for LARP7 mRNA and 7sk snRNA measurement, respectively. Methods for the calculation of relative RNA expression levels using the respective normalization control gene expression data were described previously(12). Duplicate experiments were performed for each LARP7 mRNA measurement. For 7sk snRNA measurement, two independent experiments were performed, each of which was carried out in triplicate. The sequences of all primers and probes are shown in the Table 2.
Table 2.
Primer and probe sequence
| Gene | Forward primer sequence (5’-3’) | Reverse primer sequence(5’-3’) | TagMan probe sequence(5’-3’) |
|---|---|---|---|
| LARP7 | TTTGCGTTTGTGGAATTTGA | AGGCTGGAATGGGCTTATTT | |
| 7sK | GACATCTGTCACCCCATTGA | GCGCAGCTACTCGTATACCC | CGATAGAGGAGGACCGGTCT |
| GAPDH | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA | |
| β-actin | ACCATGGATGATGATATCGCC | GCCTTGCACATGCCGG | CGCTCGTCGTCGACAACGACGGC |
| LARP7 siRNA Target Sequence | N.N.G.A.A.G.A.A.A.G.G.C.C.G.A.A.U.G.A.A.A | ||
| Sense Sequence | N.N.G.A.A.G.A.A.A.G.G.C.C.G.A.A.U.G.A.A.A | ||
| Antisense Sequence | U.U.U.C.A.U.U.C.G.G.C.C.U.U.U.C.U.U.C.U.U. | ||
Immunohistochemical staining
5-μm-thick sections of formaldehyde-fixed and paraffin-embedded gastric tissues were used for immunohistochemical staining. Antigen retrieval was performed in citrate buffer (pH 6.0) according to the standard protocol. Immunohistochemical staining was carried out using rabbit polyclonal anti-human LARP7 antibody (1:400 dilution; LARP7-101AP, Fab Gennix Inc, Frisco , TX ) or negative control antibody (normal rabbit IgG, Cell Signaling, Danvers, MA ) as the primary antibody. Visualization of the protein was carried out using the UltraVision Detection System Anti-Rabbit HRP kit (Lab Vision Corporation, Kalamazoo, MI) and an anti-rabbi IgG secondary antibody (PowerVision Poly HRP anti-rabbit IgG; Leica Microsystems, Buffalo Grove, IL) according to the manufacturer's protocol. Slides were then counterstained with hematoxylin and examined with an Olympus BX41 photomicroscope (Olympus, Center Valley, PA). All immunohistochemically analyzed cases were included in the cohort used for the qRT-PCR analysis.
siRNA transfection
A small interfering RNA (siRNA) duplex directed against LARP7 (anti-LARP7 siRNA) as well as a non-targeting control siRNA (control siRNA, D-001210-05; both from Dharmacon, Lafayette, CO, USA) were used. HFE-145 cells were seeded onto 6-well plates at a density of 2 × 105 cells /well. After 24 hours of culture, siRNA was transfected at a final concentration of 60nM using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA), according to the manufacturers protocol. LARP7 and 7sk snRNA expression was measured at 48 hours after transfection. Three independent siRNA transfections and triplicate RNA measurements were carried out for each experimental condition.
Western Blotting
Cells were lysed in 100 μl of cell lysis buffer (NaCl: 149mM, Nonidet P-40: 0.01%, Tris: 50 mM, pH 7.8, and protease inhibitor cocktail: 0.5% (Sigma, Saint Louis, MO). Protein concentration was determined using a BCA Protein Assay Kit (Pierce, Rockford, IL) with human serum albumin as a standard. Thirty μg of each sample was loaded into each well of a 10%TRIS-HCL gel (Bio Rad, Hercules, CA). After electrophoresis, protein was transferred onto a PVDF membrane (Millipore corp, Bedford, MA). The membranes were immunoblotted with TBS containing 5% nonfat dry milk, washed with 0.1% TBST, and probed with 1:2000 anti-human LARP7 rabbit polyclonal antibody (LARP7-101AP, Fab Gennix Inc, Frisco , TX) and 1:10000 anti-human β-actin mouse monoclonal antibody (cat#A3854: Sigma-Aldrich, Bedford, MA, USA) respectively. Horseradish peroxidase-conjugated anti-rabbit goat IgG (1:5000) (Calbiochem, Cat# 401393, San Diego, CA) and anti-mouse IgG (Invitrogen, Cat# 626520, Camarillo, CA) and an ECL Western Blotting detection kit (Amersham Pharmacia Biotech, Piscataway, NJ) were used for the target protein visualization. LARP7 protein levels were measured by densitometory and normalized to the beta-actin protein levels.
Cell proliferation assay
HFE-145 cells were re-seeded onto 96 well plates at 24 hours after siRNA transfection at a density of 1000 cells/well (day 0). Cell proliferation was assessed at day 0, day 2, and day 5, using the cell proliferation reagent WST-1 (Roche, Mannheim, Germany). 10ul of reagent was added to each well, incubated at 37°C for 1 hour, and optical density was measured at 660nm (background) and 440nm (signal) using a plate reader (Molecular Devices, Sunnyvale, CA, USA). Independent experiments were repeated three times, with 6 replicates in each experiment. Three independent siRNA transfections and triplicate measurements were carried out for each experimental condition.
Cell migration and invasion assays
Cell migration was measured by using transwell chamber plates (24-well format, BD Biosciences, St. Louis, MO, USA), as described previously(13). Briefly, 5 × 104 cells were seeded onto a Transwell insert at 48 hours after siRNA transfection. Twenty percent fetal bovine serum was used as a chemoattractant. After 22 hours’ incubation at 37 °C, cells that did not migrate through the pores of the transwell inserts were manually removed with a cotton swab. Cells present at the bottom of the membrane were fixed in cold methanol for 10 min and then stained with 0.01% crystal violet in 20% ethanol. After 10 min of incubation, the filters were washed thoroughly in water and suspended in 300ul of 5% acetic acid and 5% methanol. Colorimetric readings were taken at OD 595 nm. Independent experiments were repeated three times, with triplicates in each experiment. Invasion assays were performed in a similar fashion as migration assays, except that cells were seeded onto matrigel-coated transwell chamber plates (14). Two independent experiments were performed, with triplicates in each experiment.
Statistical Analysis
Mean and standard error values are shown as representative values for data in figures, unless otherwise stated. Quantitative data for transfection experiments were normalized to the mean value of mock transfectants, unless otherwise stated, Student's t-test or one-way ANOVA test was used for statistical evaluation of continuous variables, unless otherwise noted. Fisher's exact test or chi-squared testing was used for statistical evaluation of categorical data. LARP7 mRNA expression in tissues did not follow a normal distribution, thus statistical assessment in tissues was carried out using the non-parametric Mann-Whitney U-test, Kruskal-Wallis test, or Spearman's rank correlation. P-values less than 0.05 were used as a statistical significance cut-off.
RESULTS
LARP7 mRNA is significantly downregulated in cancerous gastric mucosa relative to nonneoplastic gastric mucosae
We first investigated the potential association of LARP7 mRNA levels with neoplastic status of human gastric mucosal tissues. For this purpose, we analyzed 18 gastric cancers (GC), their corresponding nonneoplastic gastric mucosal tissues (NGC), and 18 normal gastric mucosal tissues from healthy individuals (NN). Cancer patients tended to be older than healthy individuals, but this tendency did not reach statistical significance (66.3 +/- 16.2 versus 56.2 +/- 19.6 years of age, p=0.1; Table 1). No significant difference in gender or race was observed between cancer cases and healthy individuals (Table 1).
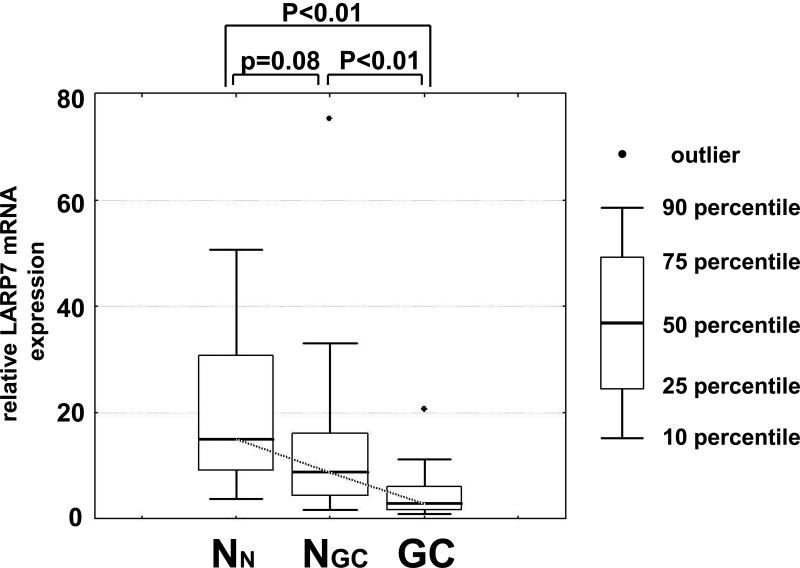
LARP7 mRNA expression was significantly reduced in GCs (median 2.5) relative to NGCs (median 8.1, p<0.01) or NNs (median 14.9, p<0.01; Fig. 1A). LARP7 mRNA expression also showed a trend toward reduction in NGCs 8.1) vs. NNs (14.9, p=0.08; Fig. 1A). LARP7 mRNA expression in GCs was not significantly associated with histological subtype (i.e., intestinal versus diffuse type) or distant metastasis. LARP7 mRNA expression was significantly and inversely correlated with age in NGCs (Spearman rank correlation coefficient -0.51, p=0.02) but not in NNs (Spearman rank correlation coefficient -0.36, p=0.14) or GCs (Spearman rank correlation coefficient -0.30, p=0.22). Because there was a non-significant but large difference in age between NGCs and NNs, we further assessed only cases older than 50 years of age. In this age-matched comparison, the nonsignificant trend toward decreased LARP7 mRNA expression in NGCs was not detected (p=0.42). LARP7 mRNA expression was not significantly correlated with gender or race in any of the three tissue categories (data not shown).
Figure 1A. LARP7 mRNA levels are downregulated in primary gastric cancers.
The LARP7 mRNA levels were assessed in 18 normal gastric mucosae from healthy individuals (NN) as well as 18 paired gastric cancer tumor tissue (GC) and non-neoplastic gastric mucosae (NGC) by qRTPCR. There were significant differences in LARP7 mRNA expression between these three tissue groups p<0.01, Kruskal-Wallis test). LARP7 mRNA levels relative to the average of the NN specimens are shown in this graph. LARP7 mRNA was significantly reduced in GCs relative to NGCs as well as to NNs (p<0.01 and p<0.01, respectively). There was an insignificant trend toward LARP7 mRNA downregulation in NGCs relative to NNs (p=0.08).
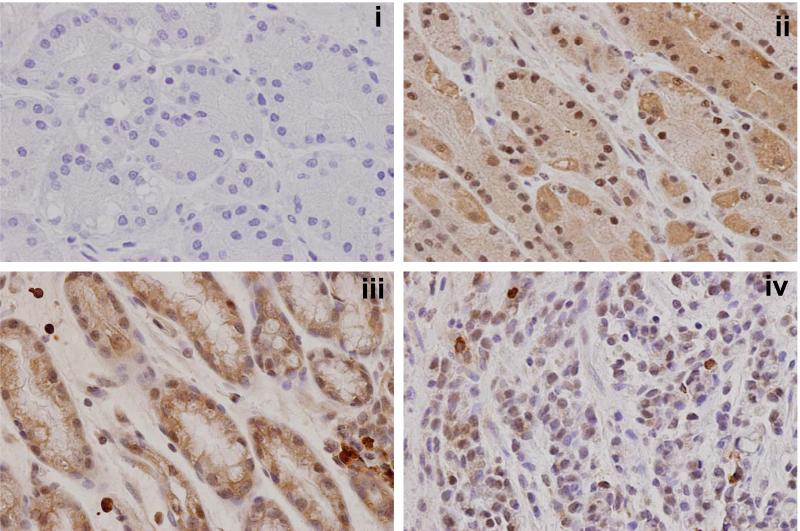
Immunohistochemical staining confirmed LARP7 protein expression in gastric epithelial cells
We further examined LARP7 protein expression patterns in the stomach by immunohistochemical staining (Figure 1B). Normal gastric mucosae from noncancer patients (i.e., NN) showed abundant nuclear and cytoplasmic LARP7 protein staining in epithelial cells, while stromal cells rarely demonstrated any LARP7 staining. Reduced LARP7 protein levels were observed in adenocarcinomas (i.e., GCs) relative to their corresponding non-cancerous gastric mucosae (i.e., NGCs), consistent with results shown in Figure 1A. As in NNs, stromal cells in NGCs did not demonstrate marked LARP7 staining.
Figure 1B. Immunohistochemical staining of LARP7 protein.
Immunohistochemical staining of LARP7 in representative gastric mucosal specimens (400x). Panel i: normal mucosa from a noncancer patient (NN) stained with negative control antibody. This slide shows no staining, assuring the specificity of the anti-LARP7 antibody (Panels ii-iv). Panel ii: the same normal mucosal specimen shown in Panel i stained with anti-LARP7 antibody. This slide displays abundant nuclear and cytoplasmic LARP7 protein expression that is restricted to gastric epithelial cells. Panel iii: non-cancerous gastric mucosa from a cancer patient (NGC) stained with anti-LARP7 antibody. This slide shows abundant LARP7 staining of normal gastric epithelial cells. As in Panel ii, very little LARP7 staining is observed in stromal cells. Panel iv: infiltrating poorly differentiated adenocarcinoma from the same patient as Panel iii stained with LARP7 antibody. This slide shows markedly reduced LARP7 staining in cancer relative to matching non-cancerous gastric mucosa from the same patient.
LARP7 knockdown decreases 7sk snRNA quantity in gastric epithelial cells
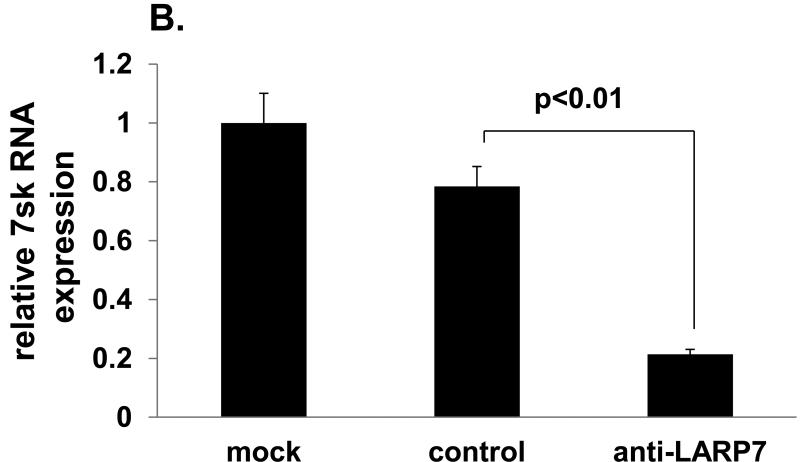
The putative tumor suppressive function of LARP7 is repression of the tumor-promoting transcriptional elongation complex, p-TEFb: LARP7 binds to and stabilizes 7sk snRNA, thus constituting the principal negative regulator of p-TEFb, 7sk snRNP (15). In order to verify the involvement of LARP7 in the negative regulation of p-TEFb via 7sk snRNA stabilization in the gastric epithelium, we analyzed the impact of siRNA-mediated LARP7 knockdown on 7sk snRNA quantity in an immortalized nonneoplastic gastric epithelial cell line, HFE145. LARP7 protein level was reduced by 95% in anti-LARP7 siRNA vs. control siRNA transfectants (Fig. 2A). 7sK mRNA levels were significantly reduced in anti-LARP7 siRNA transfectants (0.21+/-0.02) relative to control siRNA transfectants (0.78+/-0.07, p<0.01; Fig. 2B).
Figure 2. LARP7 knockdown decreases the 7sk snRNA quantity in gastric epithelial cells.
A. LARP7 protein levels were assessed by Western Blot in HFE145 cells. A marked decrease in LARP7 is evident in anti-LARP7 siRNA transfectants relative to mock- and control siRNA transfectants. B. 7sK snRNA levels were measured for mock-, anti-LARP7 siRNA-, and control siRNA-transfected HFE145 cells. The 7sk snRNA data relative to the average of the mock transfectant data are shown in this graph. A significant reduction of 7sK level was observed in anti-LARP7 siRNA transfectants relative to control siRNA and mock transfected cells (p<0.01 and p<0.01, respectively).
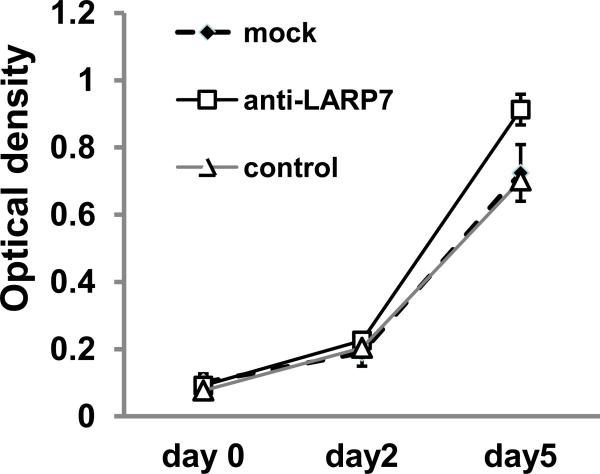
LARP7 knockdown increases gastric epithelial cell proliferation in vitro
We then evaluated effects of LARP7 knockdown on HFE145 cell proliferation by using WST-1 assays. Cells were transfected with anti-LARP7 siRNA, control siRNA, or mock reagents on day -1, and the WST-1 assay was performed on day 0, day 2 and day 5. On day 5, we observed a significant increase in cell proliferation in anti-LARP7 siRNA transfectants (0.91+/-0.05) relative to control siRNA transfectants (0.70+/-0.03, p<0.001; Fig.3).
Figure 3. LARP7 knockdown increases gastric epithelial cell proliferation in vitro.
Cell proliferation levels were measured by the WST-1 assay. There was a significant difference in cell proliferation among mock-, non-specific control siRNA-, and anti-LARP siRNA transfected cells (p<0.01, one-way ANOVA test). Anti-LARP7 siRNA significantly increase growth rates in HFE145 cells, compare to no specific control siRNA and mock condition (p<0.01 and p<0.01 respectively).
LARP7 knockdown promotes gastric epithelial cell migration in vitro
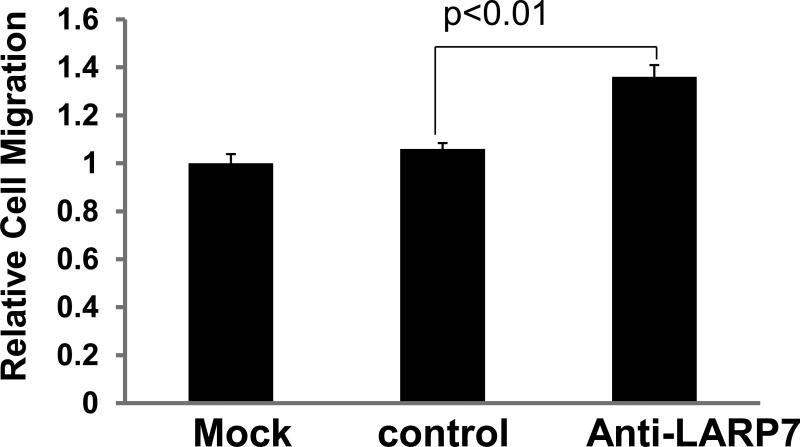
Increased cell motility is one of the properties shared by cancer cells arising in multiple organs. Therefore, we also investigated whether LARP7 knockdown influences gastric epithelial cell motility. For this purpose, we conducted transwell assays of HFE145 cells at 48 hours after transfection with anti-LARP7 siRNA, control siRNA, and mock reagent transfection. As shown in Figure 4, anti-LARP7 siRNA transfectants demonstrated significantly more migrating cells (1.36+/-0.21,) than did control siRNA transfectants (1.06+/-0.09, p<0.001). We also assessed the impact of LARP7 knockdown on HFE 145 cell invasion. However, there was no significant difference in invasion between anti-LARP7 siRNA transfectants (0.089+/-0.01,) and control siRNA transfectants (0.088+/-0.01, p=0.78).
Figure 4. LARP7 knockdown promotes gastric epithelial cell migration in vitro.
Cell migration levels were assessed for mock-, anti-LARP7 siRNA-, and control siRNA-transfected HFE145 cells at 48 hours after transfection using the transwell assay. There was a significant difference among these three groups of cells (p<0.01, one-way ANOVA test). Cell migration increased by 1.4 fold in anti-LARP7 siRNA transfectants relative to control transfectants (p<0.01).
DISCUSSION
We previously revealed frequent truncating mutations of the LARP7 gene in microsatellite –unstable (MSI-H) gastric cancers(3). In order to further evaluate the potential tumor-suppressive properties of this gene in gastric cancer, we measured LARP7 expression levels in gastric mucosae of normal and cancerous tissues, as well as the cell biological effects of LARP7 knockdown in vitro. Our data, to our knowledge, are the first to reveal that LARP7 mRNA expression is significantly reduced in human cancer relative to normal tissues, supporting our hypothesis that LARP7 is a tumor suppressor gene. In our immunohistochemical analysis, LARP7 protein staining was overwhelmingly localized to gastric epithelial cells, rather than to stromal cells including inflammatory infiltrate. Therefore, the LARP7 downregulation in gastric cancers relative to their corresponding noncancerous gastric mucosae observed in the snap-frozen tissue analysis likely reflects LARP7 downregulation in cancerous gastric epithelial cells. We observed nuclear and cytoplasmic LARP7 protein expression in normal gastric epithelial cells, consistent with LARP7's known function and reported immunohistochemical staining pattern (Human Protein Atlas)(16). We also performed transient LARP7 knockdown in a non-neoplastic gastric epithelial cell line, which resulted in a significant increase in cell proliferation and migration. Taken together, these data suggest that LARP7 downregulation contributes to the genesis of human gastric cancer.
LARP7 constitutes the specific and principal binding partner of 7sk snRNA, capturing more than 90% of 7sk snRNA(17). In the current study, we verified that loss of LARP7 results in significant and substantial loss of 7sk snRNA, and by extrapolation, loss of p-TEFb repression in gastric epithelial cells. LARP7 frameshift mutation, the prevalent mutant in MSI-H gastric cancers reported by us(3), lacks the 21 C-terminal amino-acids, whose loss abolishes LARP7's 7SK binding capacity (17, 18). Therefore, MSI-H gastric cancers also are likely to lose LARP7-mediated p-TEFb repression and, consequently, to acquire a more malignant phenotype.
Notably, multiple agents that target p-TEFb are currently being tested as anticancer drugs, with promising outcomes. Examples include a Phase I clinical trial of Seliciclib in advanced solid tumors (19), a Phase I clinical trial of Flavopiridol in advanced solid tumors including gastric cancers (20), and a Phase II clinical trial of Flavopiridol in leukemia (21). Thus, it appears that LARP7 may also become a potential therapeutic target in gastric cancer.
We observed a trend toward LARP7 downregulation in non-neoplastic gastric mucosae from gastric cancer patients vs. normal stomach from healthy individuals. However, this trend was undetectable in an age-matched comparison, suggesting that the observed LARP7 downregulation in non-cancerous gastric mucosae from gastric cancer patients may be an aging-associated event. Nevertheless, this aging-associated event may still constitute a promoting factor at early stages of carcinogenesis, because aging increases gastric cancer incidence according to the incidence rate by age (SEER17; National Cancer Institute, Surveillance, Epidemiology, and End Results Program; http://seer.cancer.gov/faststats/).
The current study has several limitations. Its relatively small cohort size provides limited power, which may lead to type II error, especially in analyses of associations between LARP7 expression and tumor stage or histological subtype. Data on intestinal metaplasia or H. pylori status of gastric cancer patients were unavailable in the majority of cases, thus we were unable to correlate these critical phenotypes with LARP7 expression. Finally, cell biologic studies were carried out on a single cell line, HFE145, since to our knowledge, no other human non-neoplastic gastric epithelial cells were known to be reliable in transfection experiments.
In summary, our molecular biological and cell biological analyses of the LARP7 gene suggest that this gene possesses characteristics typical of a gastrointestinal tumor suppressor gene. The precise molecular functions of LARP7 are not yet fully understood, but its protein domain structure suggests potential roles as an RNA binding- and -regulatory protein in the nucleus. Further studies are warranted to explore the precise antitumor mechanisms of this novel gene and its potential application to the clinical care of patients.
Acknowledgements
We thank all nurses and technicians at the Johns Hopkins Greenspring Station Endoscopy Suite, the johns Hopkins Outpatient Center Endoscopy Suite, and the Johns Hopkins Bayview Medical Center Operating Room for their assistance with sample acquisition.
Funding
U01CA084986 (National Institute of Cancer; Y.M.), Senior Research Award (Crohn's and Colitis Foundation of America; Y.M.), Wendy Will Case Cancer Fund (Wendy Will Case Cancer Fund Inc.; Y.M.), R01CA133012 and R01CA146799 (National Cancer Institute; S.J.M.).
Footnotes
Declarations of interest:
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton JP, Sato F, Jin Z, et al. Reprimo methylation is a potential biomarker of Barrett's-Associated esophageal neoplastic progression. Clin Cancer Res. 2006;12(22):6637–6642. doi: 10.1158/1078-0432.CCR-06-1781. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602(2):151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 4.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 1799(5-6):365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A. 2009;106(19):7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swarbreck D, Wilks C, Lamesch P, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36(Database issue):D1009–1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20(3):334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22(36):5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 9.Moiola C, De Luca P, Gardner K, Vazquez E, De Siervi A. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle. 9(15):3119–3126. doi: 10.4161/cc.9.15.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smoot DTAC, Barnes P, Brown M, Phadnis S, Gold B, Ashktorab H. Human gastric epithelial cell lines derived from primary cultures of normal gastric epithelial cells. Gastroenterology. 2000;118(4):A540. [Google Scholar]
- 11.Akhtar M, Cheng Y, Magno RM, et al. Promoter methylation regulates Helicobacter pylori-stimulated cyclooxygenase-2 expression in gastric epithelial cells. Cancer Res. 2001;61(6):2399–2403. [PubMed] [Google Scholar]
- 12.Mori Y, Cai K, Cheng Y, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131(3):797–808. doi: 10.1053/j.gastro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Adiseshaiah P, Lindner DJ, Kalvakolanu DV, Reddy SP. FRA-1 proto-oncogene induces lung epithelial cell invasion and anchorage-independent growth in vitro, but is insufficient to promote tumor growth in vivo. Cancer Res. 2007;67(13):6204–6211. doi: 10.1158/0008-5472.CAN-06-4687. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal R, Mori Y, Cheng Y, et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4(11):e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He N, Jahchan NS, Hong E, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29(5):588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Biewenga P, Buist MR, Moerland PD, et al. Gene expression in early stage cervical cancer. Gynecol Oncol. 2008;108(3):520–526. doi: 10.1016/j.ygyno.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 18.Markert A, Grimm M, Martinez J, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9(6):569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Tourneau C, Faivre S, Laurence V, et al. Phase I evaluation of seliciclib (R-roscovitine), a novel oral cyclin-dependent kinase inhibitor, in patients with advanced malignancies. Eur J Cancer. 46(18):3243–3250. doi: 10.1016/j.ejca.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Rathkopf D, Dickson MA, Feldman DR, et al. Phase I study of flavopiridol with oxaliplatin and fluorouracil/leucovorin in advanced solid tumors. Clin Cancer Res. 2009;15(23):7405–7411. doi: 10.1158/1078-0432.CCR-09-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karp JE, Smith BD, Levis MJ, et al. Sequential flavopiridol, cytosine arabinoside, and mitoxantrone: a phase II trial in adults with poor-risk acute myelogenous leukemia. Clin Cancer Res. 2007;13(15 Pt 1):4467–4473. doi: 10.1158/1078-0432.CCR-07-0381. [DOI] [PubMed] [Google Scholar]