Abstract
Using the PROSITE database and search tools, we conducted a comprehensive bioinformatic analysis of the predicted protein sequences of the flatworm parasites Schistosoma mansoni and S. japonicum and seven other animal genomes in order to identify novel schistosome-specific features. Our analyses revealed a relative paucity of proline-rich domains in schistosomes in comparison with their human host and a corresponding enrichment in schistosomes of asparagine-rich, serine-rich, and threonine-rich domains. Domain types found in both schistosome species but not in human included the two-component system sensor histidine kinase/response regulator; C83 family peptidase; DyP-type peroxidase; and densovirus NS1-type domain. Unique features of the schistosome proteome may help guide development of new drugs, while the presence of a densovirus-derived protein in S. mansoni suggests that this species may be infected by a virus of this group, which might be useful as a biological control agent.
1. Introduction
Schistosomiasis or bilharzia, caused by flatworm parasites (schistosomes) of the genus Schistosoma (mainly S. mansoni, S. japonicum, and S. haematobium), affects some 200 million people worldwide, exacting a substantial toll in mortality and morbidity, particularly in sub-Saharan Africa [1–2]. Current therapy for schistosomiasis is based primarily on praziquantel [3], raising concerns that schistosomes may eventually evolve resistance to this drug [4–6]. Development of alternative treatments is considered one of the potential benefits of the information provided by the recently completed draft genome sequences of S. mansoni [7] and S. japonicum [8].
One way that genome sequence data can guide development of new treatments is by revealing previously unknown aspects of the biology of a parasite. Here we conduct a comprehensive bioinformatic analysis of the predicted protein sequences of S. mansoni and S. japonicum in order to uncover schistosome-specific features. We apply proteome-wide searches for conserved sequence domains using PROSITE, which identifies sequence signatures associated with known, functionally characterized protein families or sequence domain types [9]. By comparing the results of these searches applied to the two schistosome species, to their human host, and to other animal species, we identify protein sequence patterns unique to schistosomes. This approach is complementary to approaches based on protein sequence similarity search [10], since PROSITE can identify known functionally important sequence motifs that may not be identified on the basis of sequence homology alone.
2. Methods
The complete sets of predicted protein sequences for the following genomes were obtained from the Ensembl database (http://www.ensembl.org/)[11, 12]: the mosquito Aedes aegypti (version AaegL1); the pufferfish Takifugu rubripes (v. FUGU4.57); the human Homo sapiens (v. GRCh37.57); the sea squirt Ciona intestinalis (v. JGI2.58); the fruitfly Drosophila melanogaster (v. BDGP5.13.58); and the tick Ixodes scapularis (v. IscaW1). The complete set of predicted protein sequences for Schistosoma mansoni (v. 4.0h) was obtained from the Sanger Institute (http://www.sanger.ac.uk/) FTP site, and that of Schistosoma japonicum (isolate Anhui) from the Chinese National Human Genome Center at Shanghai web site (http://www.chgc.sh.cn/en/). Lastly, the complete set of predicted protein sequences the honeybee Apis mellifera (v. pre-release 2) was obtained from the BeeBase web site hosted by the Elsik Computational Genomics Laboratory at Georgetown University (http://genomes.arc.georgetown.edu/drupal/beebase/). In all cases the verifiable mitochrondrially encoded sequences were removed from the data set by use of annotations and homology with other known mitochondrially encoded sequences. Genomes other than schistomosomes and human were chosen so as to provide a representative sampling of available chordate and arthropod genomes.
The PROSITE database and search tool [9, 13] were utilized to identify amino acid sequence motifs among all predicted protein sequences in our data set, using default values for the search criteria. The PROSITE database focuses on protein sequence domains for which precise functional charactetization is available [9]. Perl scripts were written to parse the PROSITE output into a custom database where each sequence was associated with its PROSITE motifs. In our initial search for domain types, we did not attempt to filter out multiple transcripts encoded by the same genetic locus, because different transcripts might include different domain types. However, in quantitative comparisons between S. mansoni and human, we chose only a single transcript per genetic locus. For loci with more than one transcript, if transcripts differed in the number of domain types recognized by PROSITE, we chose the transcript with the highest number of domain types. In other cases, one transcript was chosen at random.
The maximum parsimony method (branch-and-bound algorithm) [14] was used to reconstruct the phylogeny of the nine animal genomes based on presence or absence of 223 phylogenetically informative PROSITE motifs. In analyses of selected protein families, amino acid sequences were aligned using CLUSTALX [15], and phylogenetic trees were constructed by the neighbor-joining (NJ) method [16] on the basis of the JTT amino acid distance [17]. The reliability of clustering patterns in trees was assessed by bootstrapping [18]; 1000 bootstrap pseudo-samples were used. Because the S. mansoni genomic sequence is more complete than that of S. japonicum, we used the former in most detailed comparisons with human, while also including comparison with the latter in some analyses.
3. Results
3.1. Protein domain types in nine animal genomes
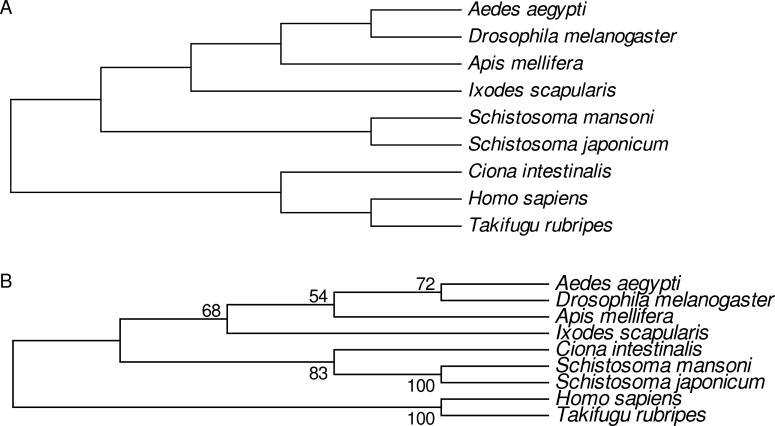
PROSITE was used to search for domain types in the predicted protein sequences from nine animal species, the currently accepted phylogenetic relationships of which [19–21] are illustrated in Fig. 1A. The numbers of different domain types identified in each species are shown in Table 1. The highest value was 662 (human) and the lowest value 528 (S. japonicum; Table 1). The second-highest value (652) was that for the pufferfish Takifugu rubripes, the species most closely related to human (Table 1). However, the only other chordate among the species examined, C. intestinalis, showed a relatively low number of domain types identified, lower than that of any of the arthropods and higher only than the values for the two schistosome species (Table 1).
Fig. 1.
(A) Currently accepted [19–21] phylogenetic relationships of the nine animal species analyzed. (B) Phylogenetic tree reconstructed by the maximum parsimony method based on presence/absence of domain types. Numbers on the branches are percentages of 1000 bootstrap pseudo-samples supporting the branch.
Table 1.
Domain types identified by PROSITE in proteomes of nine animal species.
| Species | Estimated TMRCA with human (My)1 | No. domain types | No. not found in human |
|---|---|---|---|
| Homo sapiens | -- | 662 | 10 |
| Takifugu rubripes | 455 | 652 | 6 |
| Ciona intestinalis | 774 | 558 | 6 |
| Ixodes scapularis | 910 | 610 | 19 |
| Apis mellifera | 910 | 613 | 19 |
| Aedes aegypti | 910 | 589 | 9 |
| Drosophila melanogaster | 910 | 605 | 6 |
| Schistosoma mansoni | 910 | 548 | 7 |
| Schistosoma japonicum | 910 | 528 | 4 |
TMRCA = time of most recent common ancestor, based on estimates in ref. [21]
When 223 phylogenetically informative sites were used to reconstruct the phylogenetic relationships among these species, the resulting tree captured some aspects of the true phylogenetic relationships. The four arthropod species (one tick and three insects) clustered together, although with relatively low (68%) bootstrap support (Fig. 1B). Within the insects, the two Diptera (Ae. aegypti and D. melanogaster) clustered together, although again the bootstrap support (72%) was modest (Fig. 1B). However, the urochordate Ciona intestinalis, did not cluster with the vertebrates (human and pufferfish) but with the two schistosome species (Fig. 1B). The inaccurate phylogenetic signal regarding the phylogenetic position of C. intestinalis (Fig. 1) was consistent with the relatively low number of domain types identified in that species (Table 1).
Thus, although sharing of PROSITE domain types provided some phylogenetic information, it did not provide a completely accurate picture of phylogenetic relationships. As reflected by the greater numbers of domain types identified in vertebrates and arthropods (Table 1), the identification of domain types appeared to show a certain bias toward domain types found in those groups. In spite of this bias, certain domain types not found in human were identified in each of the eight non-human species, including both schistosomes (Table 1).
3.2. Domain types shared by schistosome and human
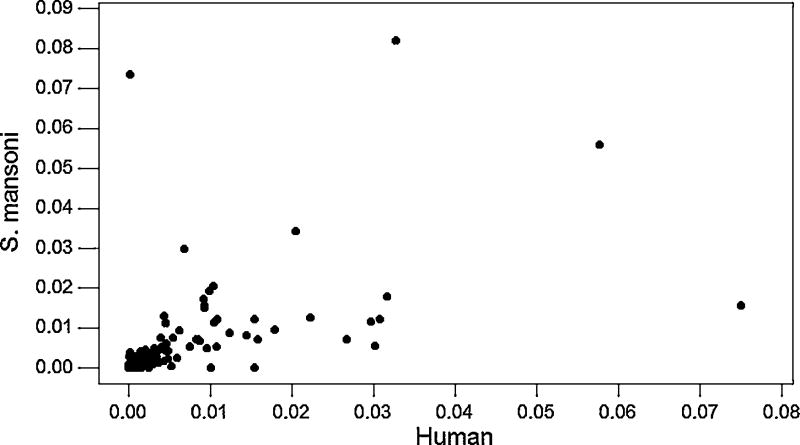
There were 539 domain types found in both human and S. mansoni. We computed the proportion of occurrence of each of these domain types in S. mansoni and the proportion of occurrence of each of these domain types in human in a data set using only one transcript per genetic locus (based on 4,582 S. mansoni loci and 12, 724 human loci). When the proportion of occurrence of each domain type in S. mansoni was plotted against its proportion of occurrence in human, the two proportions were found to be significantly positively correlated (r = 0.601; P < 0.001; Fig. 2). However, several points were outliers from the linear relationship (Fig. 2). We used studentized residuals (deleted-t residuals) to identify significant outliers; the deleted-t residual amounts to a statistical test of the improvement in the linear relationship obtained by removing a given point. Using the Bonferroni correction for multiple testing, five significant outliers were identified (Table 2).
Fig. 2.
For 539 domain types found in both human and S. mansoni, plots of the proportion of occurrence of each domain types in S. mansoni vs. that in human (r = 0.601; P < 0.001).
Table 2.
Domain types with significant deleted-t residuals in regression of proportion in Schistosoma mansoni vs. proportion in human.
| PROSITE ID | Domain type | Proportion in S. mansoni | Proportion in human | Deleted-t residual (P; Bonferroni-corrected) |
|---|---|---|---|---|
| PS50011 | Protein kinase | 0.0342 | 0.0218 | 4.17 (P < 0.05) |
| PS50099 | Proline-rich region | 0.0156 | 0.0798 | −9.11 (P < 0.001) |
| PS50321 | Asparagine- rich region | 0.0735 | 0.0003 | 19.13 (P< 0.001) |
| PS50324 | Serine-rich region | 0.0822 | 0.0345 | 14.87 (P < 0.001) |
| PS50325 | Threonine- rich region | 0.0299 | 0.0073 | 5.13 (P < 0.001) |
One of the significant outliers (P <0.05), the protein kinase domain (PROSITE domain type PS50011; Table 2), represents a domain type abundant in all eukaryotes [22]. The slightly higher frequency of occurrence of protein kinase domains in S. mansoni than in humans (Table 2) probably reflects nothing more than the relative lack of knowledge of functional protein domains in S. mansoni, leading to a slight over-representation of this well-known domain type among those identified by PROSITE in S. mansoni. The other four significant outliers (P < 0.001 in each case) involved domain-types rich in a given amino acid residue (Table 2). Only one of these domain types, the proline-rich region, was significantly over-represented in human compared to S. mansoni (Table 2). By contrast, regions rich in asparagine, serine, and threonine were significantly over-represented in S. mansoni in comparison to human (Table 2).
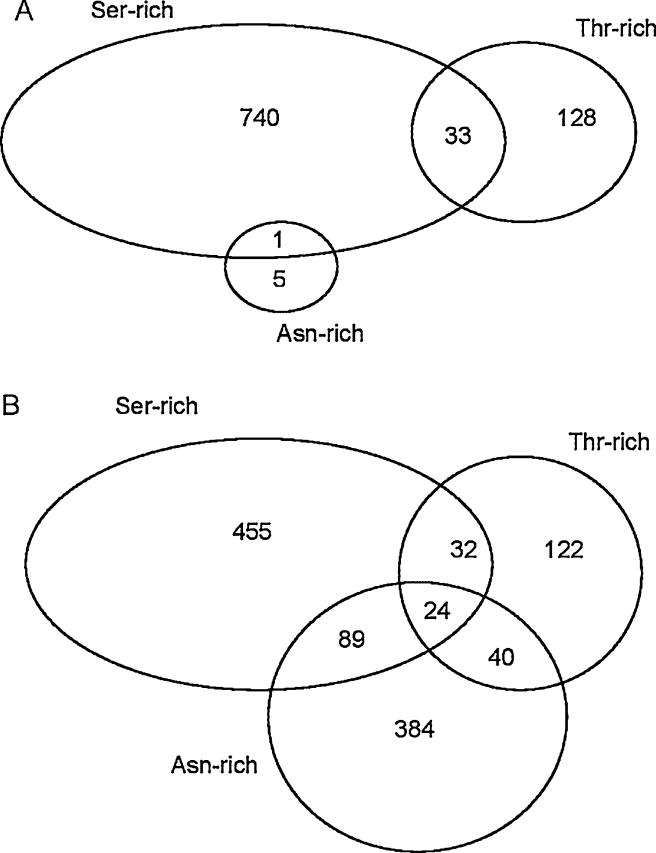
Further examination of these domain types showed that human and S. mansoni differed in the pattern of co-occurrence of these domain types in the same protein. In the human proteome, asparagine-rich domains were very rare, with only 6 in our data set (Fig. 3A). In human, only one asparagine-rich domain was found in the same protein as a serine-rich domain, and none was found in the same protein as a threonin-rich domain (Fig. 3A). We applied a log-linear model to the 2460 human proteins including at least one of the five domain types listed in Table 2, in order to test for partial association among serine-rich, threonine-rich, and asparagine-rich domains. There was no significant association (χ2 = 1.15; 1 d.f.; n.s.).
Fig. 3.
Venn diagrams indicating the numbers of proteins containing Asn-rich, Ser-rich, and Thr-rich domains and the co-occurrence of these domains in the same protein, in the predicted proteins sets of (A) human; and (B) S. mansoni.
By contrast, in S. mansoni, asparagine-rich domains were much more abundant, being found in 537 proteins (Fig. 3B). Moreover, 24 proteins of S. mansoni included serine-rich, threonine-rich, and asparagine-rich domains (Fig. 3B). When we applied a log-linear model to the 1226 S. mansoni proteins including at least one of the five domain types listed in Table 2, there was a highly significant partial association among serine-rich, threonine-rich, and asparagine-rich domains (χ2 = 201.26; 1 d.f.; P < 0.001). Thus, although unassociated in humans, these three domain types were found to be positively associated in with one another in the proteins of S. mansoni to a greater extent than expected by chance.
3.2. Domain types found in schistosome but not in human
Seven domain types found in either or both of the two schistosomes but not in human are summarized in Table 3. Two domain types found in S. mansoni but not S. japonicum, transcription regulator cysB and preprotein translocase subunit secA, occurred in proteins whose closest known homologs were bacterial (Table 3). Smp_106360, the putative homolog of cysB, was encoded by a predicted intronless open reading frame; and the predicted protein showed 100% amino acid sequence identity and 100% nucleotide sequence identity with a gene of Streptococcus equi (YP_002744367). Therefore, the gene encoding Smp_106360 is almost certainly a contaminant in the S. mansoni genome assembly.
Table 3.
Domain types found in Schistosoma mansoni and/or S. japonicum but not in human.
| PROSITE ID | S. mansoni | S. japonicum | Protein function | Homologs |
|---|---|---|---|---|
| PS50110 | Smp_112220 | Sjp_0129540 | Two-component system sensor histidine kinase/response regulator | Invertebrate animals, Bacteria |
| PS50931 | Smp_106360 | -- | Transcription regulator cysB (LysR family transcriptional regularory protein) | Bacteria (likely contaminant) |
| PS51029 | Smp_186380 | -- | MADF trinucleotide repeat-binding domain | Invertebrate animals |
| PS51196 | Smp_109540 | -- | Preprotein translocase subunit secA | Bacteria (likely contaminant) |
| PS51206 | Smp_194070 | -- | NS1 of densoviruses | Invertebrate animals, Densoviruses |
| PS51404 | Smp-172830, Smp_160550, Smp_160560 | Sjp_0057590, Sjp_0057600, Sjp_0081350 | DyP-type peroxidase | Invertebrate animals, other Eukaryotes, Bacteria |
| PS51443 | Smp_072740 | Sjp_0039680, Sjp_0132750 | C83 family peptidase | Invertebrate animals, Bacteria |
Smp_109540, the secA homolog, was encoded by a predicted reading frame consisting of four separate exons (accession FN363463.1; supercontig Smp_scaff006172, sites 99–244, 443–585, 807–1190, and 1221–1525). By BLAST search, the most similar bacterial sequence was secA of Mycoplasma agalactiae (YP_03515458; Supplementary Figure S1A). The latter sequence is much longer (837 amino acids) than Smp-109540 (325 amino acids). The region of amino acid sequence similarity between the two sequences corresponded to the third and fourth exons of the Smp_109540 gene (Supplementary Figure S1A). However, nucleotide sequence similarity also involved introns 2 and 3 of the predicted S. mansoni gene (Supplementary Figure S1B). Over the region of nucleotide sequence similarity, the percent nucleotide sequence identity between the predicted S. mansoni sequence and the M. agalactiae sequence was 66.4% (Supplementary Figure S1B). Thus the predicted gene encoding Smp_109540 probably represents a chimeric gene consisting in part of S. mansoni sequence and in part of contaminant sequence of bacterial origin.
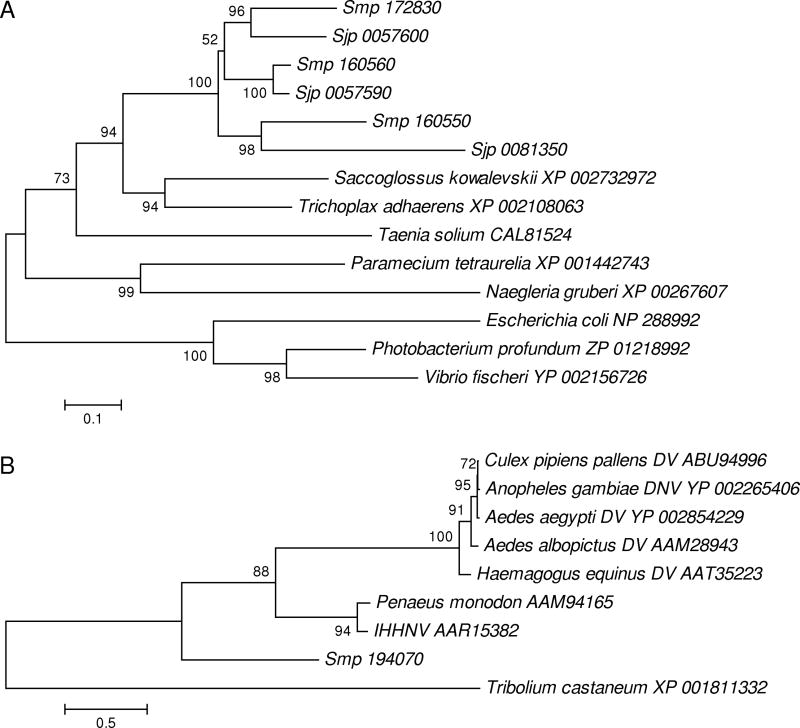
The remaining domain types found in schistosomes but not humans occurred in proteins with animal homologs (Table 3). The MADF trinucleotide repeat-binding domain was found in S. mansoni but not S. japonicum (Table 3). There were three domain types found in both S. mansoni and S. japonicum but not in human: the DyP-type peroxidase, the two-component system sensor histidine kinase/response regulator and C83 family peptidase (Table 3). The DyP-type peroxidases, though found in bacteria, also were found by database similarity search to include homologs in several invertebrate animal species and other eukaryotes (Fig. 4a). In a phylogenetic tree, each DyP-type peroxidase sequence from S. mansoni clustered with one from S. japonicum, usually with strong bootstrap support (Fig. 4A). This topology indicated that the three DyP-type peroxidase paralogs arose by gene duplication prior to the most recent common ancestor of S. mansoni and S. japonicum.
Fig. 4.
(A) NJ tree of amino acid sequences of selected DyP-type peroxidases of animals, protists, and bacteria. (B) NJ tree of amino acid sequences of densovirus NS1 protein and animal homologs. In each tree, sequences from S. mansoni are identified by the prefix “Smp,” while those from S. japonicum are identified by the prefix “Sjp.” Other sequences are identified by Genbank protein accession numbers. DV = densovirus; DNV = densonucleovirus; IHHNV = infectious hypodermal and hematopoietic necrosis virus. Numbers on the branches are percentages of 1000 bootstrap pseudo-samples supporting the branch.
Among the domain types found in S. mansoni but not S. japonicum was a domain corresponding to the NS1 protein of densoviruses, homologs of which have been found in other invertebrate animals (Supplementary Figure S2). In a phylogenetic tree, he NS1 homolog of S. mansoni did not cluster more closely with those of densoviruses than it did with those of other animal species (Figure 4b).
4. Discussion
We used proteome-wide searches for protein domain types in animal genomes using PROSITE in order to identify distinctive biological features of schistosomes. This method showed a certain bias toward domain types characteristic of vertebrates and arthropods, probably reflecting the more intense research into protein function in model organisms belonging to these taxa. Presumably because of this bias, the number of schistosome-specific domain types identified by the PROSITE search was fewer than the number of platyhelminth-specific genes identified by a previous protein homology search [10]. Unlike certain other protein domain databases such as Pfam [23] which provide a broad coverage of protein families identified by sequence similarity, the PROSITE database focuses intentionally on a curated set of protein domains for which functional information is already available [9]. Because of the functional information provided by PROSITE annotations, our analyses revealed several features of schistosome proteomes that have not previously been reported. By examining the occurrence and abundance of protein domain types in schistosomes, our analyses suggested several lines of investigation that may be promising for future empirical investigation and possible drug discovery.
We found a relative paucity of proline-rich domains in schistosomes in comparison with their human host, along with a corresponding enrichment in schistosomes of asparagine-rich, serine-rich, and threonine-rich domains. Moreover, the latter three domain types were found to co-occur in the same protein in S. mansoni to a greater extent than expected on the basis of their frequency of occurrence in the proteome, whereas they were not found to co-occur in any human protein. Proline-rich domains are known to be important in protein-protein interactions, particularly in signaling proteins in a wide variety of organisms [24]. There is also evidence that serine- and threone-rich and glutamine- and asparagine-rich domains can be important for protein-protein interactions [25–27]. Unfortunately, there is no available information at present regarding the physiological function of such domains in schistosomes. However, the difference between S. mansoni and its human host with respect to the abundance of these domain types suggests that there may be important differences between these two species with regard to the mechanisms of protein-interaction and signaling, and that empirical investigation of the role of these domains may prove rewarding.
Two domain types found in both schistosome species but not in human, the two-component system sensor histidine kinase/response regulator and C83 family peptidase, are found in bacterial proteins involved in intercellular signaling systems [28–29], suggesting a possible similar role in schistosomes and other animals. The DyP-type peroxidase domain was represented in three paralogs in each of the two schistosome genomes, but was not found in the human proteome, as previously reported [10]. The functions of DyP-type peroxidases in the schistosomes are not known; but, since DyP-type peroxidases typically have wide substrate specificity [30], it is possible that the three paralogs have specialized to some extent with regard to substrate. They may also differ with respect to expression patterns across the parasite’s life-cycle; indeed, one of the S. mansoni DyP-type peroxidases (Smp_160550) was shown to have increased expression when the parasite is infecting the intermediate host [10].
Among the domain types found in S. mansoni but not in human or in S. japonicum was the MADF trinucleotide repeat-binding domain (Table 3), which as been described in Drosophila melangaster [31]. In S. mansoni but not S. japonicum, we also found two domain types representing protein families widespread in Eubacteria: the transcriptional regulator cysB and the preprotein translocase subunit secA (Table 3). Because the predicted cysB homolog of S. mansoni was 100% identical to a gene of Streptococcus equi, the hypothesis of contamination by DNA from S. equi or a closely related species of Streptococcus seems probable. The predicted secA homolog did not show such high similarity to any known bacterial gene. However, it seemed likely that this gene is a chimera consisting in part of S. mansoni DNA and in part of a secA gene derived from an unknown bacterial source.
Another domain type found in S. mansoni but not in human or in S. japonicum showed homology to the NS1 protein of densoviruses (Table 3). Densoviruses (family Parvoviridae, subfamily Densovirinae) are single-stranded DNA viruses known to infect insects and other arthropods, particularly mosquitoes (Diptera; Culicidae) [32–33]. The genome of the black tiger pawn Penaeus monodon includes an NS1-like sequence similar to that of a densovirus infecting that species, infectious hypodermal and hematopoietic necrosis virus (IHHNV) [33]. Our phylogenetic analysis indicated that NS1-like gene of P. modonon is very closely related to that of IHHNV, supporting the hypothesis that this gene has originated by recent incorporation of a viral gene into the host genome [33].
Our sequence similarity search found an NS1-related protein in the predicted proteins of Tribolium castaneum, suggesting that this gene has been acquired from a previously unreported densovirus (Fig. 4B). The sequence from T. castaneum (Coleoptera: Tenebrionidae) did not cluster close to those of available sequences of densoviruses, all of which have mosquito (Diptera: Culicidae) hosts (Fig. 4B). This suggests that the virus from which the T. castaneum gene originated is not closely related to previously sequenced densoviruses. Similarly, the occurrence of an NS1 domain related to those of densoviruses in the predicted protein set of S. mansoni (Table 3; Fig 4B) implies infection of the latter species by a virus from that group, either in the past or currently. Since densoviruses are under investigation as biological control agents of mosquitos [34], the existence of a previously unknown densovirus of schistosomes suggests that, if such a virus were isolated, it also might be a candidate for biological control.
Supplementary Material
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health to A.L.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 2.Utzinger J, Raso G, Brooker S, de Savigny D, Tanner M, Ørnbjerg N, Singer BH, N’goran EK. Schistosomiasis and neglected tropical diseases: towards integrated and sustainable control and a word of caution. Parasitology. 2009;136:1859–1874. doi: 10.1017/S0031182009991600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenwick A, Savioli L, Engels D, Bergquist NR, Todd MH. Drugs for the control of parasitic diseases: current status and development in schistosomiasis. Trends Parasitol. 2003;19:509–515. doi: 10.1016/j.pt.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Doenhoff MJ, Cioli D, Utzinger J. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Curr Opin Infect Dis. 2008;21:659–667. doi: 10.1097/QCO.0b013e328318978f. [DOI] [PubMed] [Google Scholar]
- 5.Melman SD, Steinauer ML, Cunningham C, Kubatko LS, Mwangi IN, Wynn NB, Mutuku MW, Karanja DM, Colley DG, Black CL, Secor WE, Mkoji GM, Loker ES. Reduced susceptibility to praziquantel among naturally occurring Kenyan isolates of Schistosoma mansoni. PLoS Negl Trop Dis. 2009;3(8):e504. doi: 10.1371/journal.pntd.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasinathan RS, Morgan WM, Greenberg RM. Schistosoma mansoni express higher levels of multidrug resistance-associated protein 1 (SmMRP1) in juvenile worms and in response to praziquantel. Mol Biochem Parasitol. 2010;173:25–31. doi: 10.1016/j.molbiopara.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA, Lacerda D, Macedo CD, McVeigh P, Ning Z, Oliveira G, Overington JP, Parkhill J, Pertea M, Pierce RJ, Protasio AV, Quail MA, Rajandream MA, Rogers J, Sajid M, Salzberg SL, Stanke M, Tivey AR, White O, Williams DL, Wortman J, Wu W, Zamanian M, Zerlotini A, Fraser-Liggett CM, Barrell BG, El-Sayed NM. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:362–368. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PROSITEa protein domain database for functional characterization and annotation. Nucleic Acids Research. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick JM, Peak E, Perally S, Chalmers IW, Barrett J, Yoshino TP, Ivens AC, Hoffmann KF. Anti-schistosomal intervention targets identified by lifecycle transcriptome analyses. PLoS Negl Trop Dis. 2009;3(11):e543. doi: 10.1371/journal.pntd.0000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, Down T, Eyras E, Fernandez-Suarez XM, Gane P, Gibbins B, Gilbert J, Hammond M, Hotz HR, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Lehvaslaiho H, McVicker G, Melsopp C, Meidl P, Mongin E, Pettett R, Potter S, Proctor G, Rae M, Searle S, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Ureta-Vidal A, Woodwark KC, Cameron G, Durbin R, Cox A, Hubbard T, Clamp M. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Applied Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 14.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Sinauer; Sunderland MA: 1999. [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:83–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Paps J, Baguñà J, Riutort M. Lophotrochozoa internal phylogeny: new insights from an up-to-date analysis of nuclear ribosomal genes. Proc R Soc Lond B. 2009;276:1245–1254. doi: 10.1098/rspb.2008.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourlat SJ, Nielsen C, Economou AD, Telford MJ. Testing the new animal phylogeny: a phylum level molecular analysis of the animal kingdom. Mol Phyl Evol. 2008;49:23–31. doi: 10.1016/j.ympev.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Blair JE. Animals (Metazoa) In: Hedges SB, Kumar S, editors. The Timetree of Life. New York: Oxford University Press; 2009. pp. 223–230. [Google Scholar]
- 22.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucl Acids Res. 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 25.Moro O, Lameh J, Sadée W. Serine- and threonine-rich domain regulates internalization of muscarinic cholinergic receptors. J Biol Chem. 1993;268:6862–6865. [PubMed] [Google Scholar]
- 26.Serrador JM, Vicente-Manzaranes, Calvo J, Barreiro O, Montoya MC, Schwartz-Albiez R, Furthmayr H, Lozano F, Sánchez-Madrid F. A novel serine-rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting. J Biol Chem. 2002;277:10400–10409. doi: 10.1074/jbc.M110694200. [DOI] [PubMed] [Google Scholar]
- 27.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dirix G, Monsieurs P, Dombrecht B, Daniels R, Marchal K, Vanderleyden J, Michiels J. Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides. 2004;25:1425–1440. doi: 10.1016/j.peptides.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family. J Biol Chem. 2007;282:36652–36658. doi: 10.1074/jbc.M706996200. [DOI] [PubMed] [Google Scholar]
- 31.Bhaskar V, Corey AJ. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene. 2002;299:173–184. doi: 10.1016/s0378-1119(02)01058-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhai Y, Lv X, Sun X, Fu S, Gong Z, Fen Y, Tong S, Wang Z, Tanf Q, Attoui H, Liang G. Isolation and characterization of the full coding sequence of a novel densovirus from the mosquito Culex pipiens pallens. J Gen Virol. 2008;89:195–199. doi: 10.1099/vir.0.83221-0. [DOI] [PubMed] [Google Scholar]
- 33.Tang KF, Lightner DV. Infectious hypodermal and hematopoietic necrosis virus (IHHNV)-related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res. 2006;118:185–191. doi: 10.1016/j.virusres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Carlson J, Suchman E, Buchatsky L. Densoviruses for control and genetic manipulation of mosquitoes. Adv Virus Res. 2006;68:361–392. doi: 10.1016/S0065-3527(06)68010-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.