Abstract
Purpose
Hepatocellular carcinoma (HCC) is a heterogeneous cancer with active Wnt-signaling. Underlying biological mechanisms remain unclear and no drug targeting this pathway has been approved to date. We aimed to characterize Wnt-pathway aberrations in HCC patients, and to investigate sorafenib as a potential Wnt modulator in experimental models of liver cancer.
Experimental Design
The Wnt-pathway was assessed using mRNA (642 HCCs and 21 liver cancer cell lines) and miRNA expression data (89 HCCs), immunohistochemistry (108 HCCs) and CTNNB1-mutation data (91 HCCs). Effects of sorafenib on Wnt-signaling were evaluated in four liver cancer cell lines with active Wnt signaling and a tumor xenograft model.
Results
Evidence for Wnt activation was observed for 315 (49.1%) cases, and was further classified as CTNNB1-class [138 cases (21.5%)] or Wnt-TGFβ-class [177 cases (27.6%)]. CTNNB1-class was characterized by up-regulation of liver-specific Wnt-targets, nuclear β-catenin and glutamine-synthetase immunostaining, and enrichment of CTNNB1-mutation-signature, while Wnt-TGFβ-class was characterized by dysregulation of classical Wnt-targets and the absence of nuclear β-catenin. Sorafenib decreased Wnt-signaling and β-catenin protein in HepG2 (CTNNB1-class), SNU387 (Wnt-TGFβ-class), SNU398 (CTNNB1-mutation) and Huh7 (Lithium-chloride-pathway activation) cell lines. Additionally, sorafenib attenuated expression of liver-related Wnt-targets GLUL, LGR5, and TBX3. The suppressive effect on CTNNB1-class-specific Wnt-pathway activation was validated in vivo using HepG2 xenografts in nude mice, accompanied by decreased tumor volume and increased survival of treated animals.
Conclusions
Distinct dysregulation of Wnt-pathway constituents characterize two different Wnt-related molecular classes (CTNNB1 and Wnt-TGFβ), accounting for half of all HCC patients. Sorafenib modulates β-catenin/Wnt-signaling in experimental models that harbor the CTNNB1-class-signature.
Keywords: CTNNB1, Wnt-TGFβ, β-catenin, molecular classification, molecular therapies
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and its incidence is increasing worldwide (1). As the third leading cause of cancer-related death and the most common cause of death among cirrhotic patients, it is emerging as a major global health problem (1). Hepatitis B and C viral infections, and alcohol abuse are the main risk factors for HCC development. Curative treatments (e.g. resection, transplantation or local ablation) are available only to patients with early stage disease, but are limited by high recurrence rates, which impair patient outcomes. At advanced stages, the multi-kinase inhibitor sorafenib is the only effective treatment, although vigorous efforts are underway to better characterize the molecular pathogenesis of liver cancer in order to refine the efficacy of other molecular targeted therapies (2, 3).
Wnt signaling is involved in multiple physiological processes, embryonic development and cancer (4). The pathway is activated upon Wnt-ligand binding to frizzled receptors (FZD) followed by cytosolic accumulation of β-catenin through prevention of glycogen synthase kinase three β (GSK3β) mediated phosphorylation of the β-catenin Ser/Thr domain. Cytosolic β-catenin can translocate to the nucleus to initiate transcription of target-genes through interaction with T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors (4). Aberrant activation of Wnt can also result from mutations in the β-catenin gene (CTNNB1), which is the second most frequent mutation observed in HCC after the p53 tumor suppressor (TP53) (5).
Several genomic studies have identified molecular subclasses of hepatocellular carcinoma using unsupervised clustering of mRNA or miRNA expression data. However, there is no universally accepted molecular classification for this disease. Nonetheless, Wnt-signaling associated molecular classes have been reported by different investigators emphasizing the importance of Wnt-aberrations in liver cancer (5, 6). We recently reported a molecular classification of HCC, which includes a CTNNB1-class characterized by over-expression of liver-related Wnt-target-genes (which have been described in the liver, but are also found in other tissues, e.g. glutamine synthetase, GLUL, and leucine-rich repeat-containing G protein-coupled receptor 5, LGR5), enrichment in nuclear β-catenin staining and CTNNB1-mutations (6). This subclass strongly overlapped with Bouyault’s Wnt-related G5-6 classes, which were also characterized by mutations in CTNNB1 and significant over-expression of liver-related Wnt-target-genes (5, 6). The biological similarity of HCC tumors within CTNNB1-class was further strengthened by a significant overlap with our recently published miRNA-class A (7). In parallel, we characterized the molecular subclass S1 (Wnt-TGFβ) related to TGFβ-activated Wnt-signaling through a meta-analysis of 603 HCC patients (8). With no association to CTNNB1-mutations or over-expression of liver-related Wnt-targets, this new molecular class was defined by activation of commonly reported Wnt-target-genes (e.g. CCND1 and MYC) (8). Wnt-signaling was also reportedly activated in a hepatic stem-cell-like HCC class showing a close correlation with EpCAM expression and induction of known Wnt-target-genes BAMBI and DKK1(9, 10). Even though the dysregulation of Wnt-signaling in hepatocellular cancer is described (3, 11), the simultaneous presence of multiple Wnt-related molecular profiles in cancer is a new concept that requires thorough characterization. It remains unclear to what extent different Wnt-targets, their transcriptional regulators and their downstream signals contribute to Wnt-signaling diversity. In addition, efforts to develop anti-tumoral agents specifically targeting this cascade have not yet been developed.
Herein, we demonstrate that distinct differential expression of Wnt-pathway-members, up-regulation of Wnt-target-genes, enrichment of CTNNB1-mutation-signature, and β-catenin and glutamine synthetase immunostaining characterize the two coexisting but distinct Wnt-related classes, CTNNB1 and Wnt-TGFβ, in HCC. We also provide evidence that sorafenib modulates Wnt/β-catenin signaling in experimental models of liver cancer recapitulating the human CTNNB1-class.
Materials and Methods
Human samples, mRNA and miRNA expression arrays data
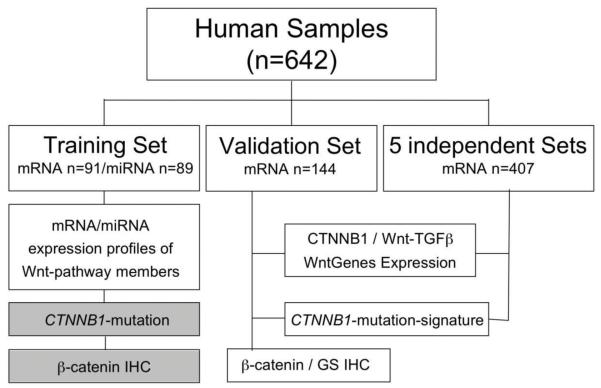
Gene expression profiling of a total of 642 human samples was analyzed for this study (Figure 1). We used as training (91 HCV-related HCC; GSE9843 and GSE20594) and independent validation sets (144 mixed etiology HCC; GSE19977 without overlapping samples with the training set), samples previously profiled in the studies of our HCC Genomic Consortium (Suppl. Table 1). Normal liver tissue (n=10) was obtained from patients with non-malignant diseases. Written consents from patients, and institutional review board approvals were obtained from all institutions. We further analized gene expression of 407 HCC samples of patients with mixed etiology available from five public HCC datasets (Suppl. Table 2) (8). Gene expression of 21 human liver cancer cell lines analyzed in this study is publicly available at http://www.broadinstitute.org/ccle.
Fig. 1. Flow Chart of Study Design.
Flow Chart displaying the different analyses performed according to the datasets used. Boxes in grey represent information recently published by our group (6).
Genomic profiling of Wnt-related mRNAs and miRNAs
Wnt-pathway mRNA and miRNA lists
In order to characterize Wnt-pathway alterations in the Wnt-related subclasses of HCC, we tested gene expression of 210 genes obtained from publicly available Wnt-pathway lists (Suppl. Table 3: BIOCARTA_WNT_PATHWAY, KEGG_WNT_SIGNALING_PATHWAY,REACTOME_SIGNALING_BY_WNT,and WNT_SIGNALING), the Molecular Signatures Database (MSigDB) and the Cancer Cell Map Pathway Map. Additionally all miRNAs reported to be involved in Wnt-Signaling in the literature were summarized in a separate list of 49 miRNAs associated with Wnt-signaling (Suppl. Table 4).
Wnt-related subclasses of HCC
CTNNB1 class was assessed in the training set as previously reported (6) and determined by nearest template prediction (NTP) method (12) in 6 additional datasets (validation set and 5 publicly available independent gene expression data sets with sample size >50)(8). Wnt-TGFβ-class samples were identified as previously published (8) or NTP (validation set). Remaining samples not captured by either class were defined as non-Wnt.
Analysis of Wnt pathway alterations in Wnt-related classes
Differential mRNA and miRNA expression of Wnt-pathway members were analyzed by the GenePattern platform (www.broadinstitute.org/genepattern, Broad Institute, Boston, MA) using Comparative Marker Selection module (13), and false discovery rate (FDR) for multiple hypothesis testing correction (14).
We generated a gene signature to characterize each Wnt-related subclass, by using the training set of samples. The CTNNB1-WntGenes-signature was obtained from genes of the Wnt-pathway-mRNAs significantly altered (FDR<0.05) in CTNNB1-class samples compared to normal liver and non-Wnt HCC samples,. The same approach was used to generated the Wnt-TGFβ-WntGenes-signature. Then these two signatures were validated by NTP using 6 additional datasets. CTNNB1- and Wnt-TGFβ-class marker genes were excluded from the WntGenes-signatures before the validation analyses.
In order to characterize the miRNA profile differentially expressed in these two subclasses, we compared Wnt-pathway-miRNAs in each subclass vs non-Wnt samples in the training set. Thirty-three out of 49 Wnt-pathway-miRNAs could be analyzed through the miRNA platform used (7) and we additionally excluded probes if >60% of the samples had low expression values (cut off <30, 2/33, 6.1%).
Association of CTNNB1 mutations to CTNNB1 class
The published strategy of a signature recapitulating TP53 mutations in breast cancer was used to generate a gene-signature of CTNNB1-mutation (15). The signature was generated with genes differentially expressed in samples HCC samples harboring or not CTNNB1-mutations (FDR<0.05) in the training set. The ability of the signature to capture CTNNB1 mutated samples was validated in one HCC dataset (5). NTP analysis was used to asses overlap between CTNNB1 class and the gene signature of CTNNBI mutations in 6 additional datasets.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on 5 μm sections of formalin-fixed paraffin-embedded (FFPE) tissue by heat-induced epitope retrieval in 10 mM sodium citrate (pH=6) before blocking with 5% BSA-PBS. Samples were incubated overnight at 4°C with anti-β-catenin and anti-glutamine synthetase (GS) antibodies at 1:1000 dilution (BD Biosciences, San Jose, CA). EnVision™+ System-HRP was applied as secondary antibody (Dako, Golstrup, Denmark) and sections were counterstained with hematoxylin. Positive staining was defined according to percentage of stained cells (0=no cells, 1=1-4%, 2=5-19%, 3=20-39%, 4=40-59%, 5=60-79%, 6=80-100% of positive cells) and intensity (0-3). Percentage scores were multiplied by intensity scores to yield an overall score. Nuclear and cytoplasmic Beta-Catenin score of >3 and membranous β-catenin, and cytoplasmatic GS cytoplasmic score of >11 were considered as positive.
Cell lines and drug treatments
SNU398, SNU387 and HepG2 cells were purchased from ATCC (Manassa, VA). Huh7 (Riken Bioresource Center) and HepG2 cells were cultured in DMEM, SNU398 and SNU387 cells in RPMI medium supplemented with 10% fetal bovine serum, 1% penicillin (100 U/ml) and 1% streptomycin (100 μg/mL). Cells were plated in 12-24 well plates or 10 cm2 dishes and cultured overnight. Sorafenib (LC Laboratories, Woburn, MA) was added for 6 hours before cells were processed for chemiluminescence, western blot, immunocytofluorescence, and TCF/LEF-reporter-assay. The Wnt-activator lithium-chloride (LiCl) (16) was added to Huh7 cells 30 minutes after sorafenib treatment. Final DMSO concentration in all experiments was <0.05%. Gene expression of 21 cell lines was used to identify those that better characterize CTNNB1 and Wnt-TGFβ-classes.
TCF/LEF-reporter-assay
Cells were transduced using Transduction-ready TCF/LEF reporter (luciferase) lentiviral particles (CLS-018L, SA Biosciences, Frederick, MD) following manufacturer’s instructions. Luminescence was assayed in cells stably expressing TCF/LEF luciferase reporter using a Luciferase Assay Kit (E1501, Promega, Madison, WI) and a Veritas Microplate Luminometer (Promega, Madison, WI). Luminescence/cell number (10,000-750,000 cells) line graphs were used to quantify luciferase activity with recombinant luciferase standard (E170A, Promega).
Immunocytofluorescence and Western Blot
(standard methods were used as described in detail in supplementary methods)
Real time PCR
Total mRNA was obtained from 24h treated cells and reversed transcribed using SprintRT Complete-Double PrePrimed strips (ClonTech, Mountain View, CA). TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA) were used to measure mRNA levels of GLUL (assay ID: Hs01013056_g1), LGR5 (Hs00173664_m1), and TBX3 (Hs00255591_m1) according to manufacturer’s instructions. LightCycler480, and ΔΔCT method were used to quantify the data (normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH Hs00266705_g1) following the methodology recently described (17).
Animal Experiments
All animal studies were done upon Mount Sinai School of Medicine Institutional Animal Care and Use Committee (IACUC) approval. BALB/c female mice were injected with 5×106 HepG2 cells subcutaneously and followed as described previously (18). Sorafenib (30 mg/kg/day) was administered by gavage using canola oil as vehicle. Animals were sacrificed and tumors collected when tumor volume reached 1 cm3 according to the following formula= length × width2 × 0.4 (18).
Statistical analyses
Bars represent the mean + standard error of the mean. Comparisons between groups were made using two-tailed t-test or U-test for continuous variables, and Fisher exact test for comparison of proportions. Correlations were calculated with the non-parametric Spearman’s Coefficient (SC). Gene set enrichment analysis (GSEA) was used to assess enrichment of which the CTNNB1- and Wnt-TGFβ-class-signature in the cell lines as previously described (8). All calculations were done with the SPSS package (SPSS 15.0, Chicago, IL).
RESULTS
Molecular portrait of two human Wnt HCC subclasses: CTNNB1 and Wnt-TGFβ
Differential expression of Wnt genes in CTNNB1- and Wnt-TGFβ-classes
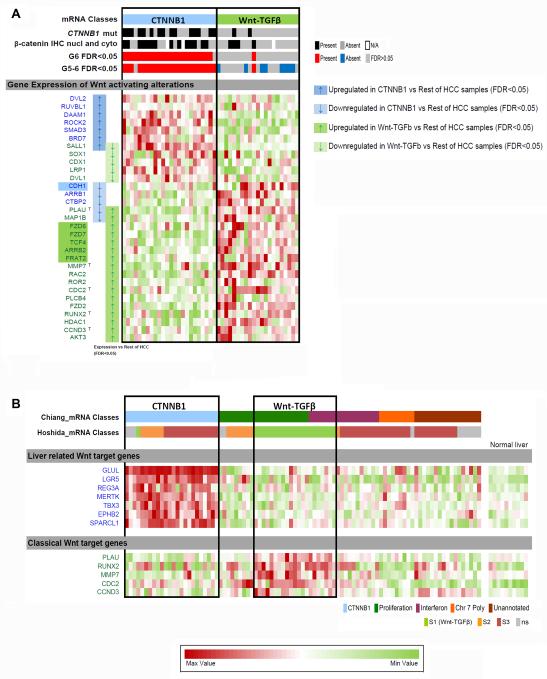
In order to identify Wnt-pathway differential alterations in CTNNB1- and Wnt-TGFβ-classes, we analyzed the gene expression of 210 Wnt-pathway-related genes in comparison to normal liver and HCCs not related to Wnt-classes in the training set. Tumors of the CTNNB1- (24/91, 26.4%) and Wnt-TGFβ-classes (23/91, 25.3%). showed significant dysregulation of 36 and 48 Wnt-pathway-mRNAs, respectively (FDR<0.05) (Suppl. Fig 1, Suppl. Table 5). Overall, both classes had only four significantly dysregulated genes in common (SALL1, PRKCD, PLAU, MAP1B) but with expression in opposite directions). Therefore each molecular class was characterized by its own specific Wnt-related mRNA expression pattern. Suppl Table 6 provides a full list of genes, subcellular localization and effect on Wnt-signaling (activation/inhibition) (19).
A higher number of activating alterations are observed in Wnt-TGFβ-class samples, among which five of them (FZD7, FZD6, TCF4, FRAT2 and ARBB2) have been related to Wnt activation in cancer (20-25) (Fig. 2A). CTNNB1-class was characterized by CDH1 down-regulation, and a significant number of potentially inhibiting alterations (Suppl. Fig 1). Several WNTs and FZDs were significantly down-regulated in both classes inhibiting Wnt-signaling (Suppl. Table 5,6).
Fig. 2. Expression alterations of Wnt pathway genes and miRNAs in two Wnt subclases of HCC, CTNNB1 (blue) and Wnt-TGFβ (green), in the training set.
A) Distribution of CTNNB1 mutations (6), β-catenin IHC nuclear and cytosolic staining(6) and G6/G5-6 molecular profile(5). Heatmap shows Wnt pathway genes differentially expressed (arrows) between CTNNB1 (blue) or Wnt-TGFβ (green) subclasses. Only genes potentially related to Wnt pathway activation are shown (see Suppl. Fig. 1 for full list of genes and non-Wnt samples). Highlighted gene names correspond to those that have been related to induction of Wnt pathway activation in cancer (20-25). B) Full display of mRNA-based molecular classifications showing CTNNB1 subclass (7) and Wnt-TGFβ subclass (9) in the training set. Heatmap shows Liver-related Wnt target genes (5) and Classical Wnt target genes (list obtained from public databases, see Methods) differentially expressed between CTNNB1 (blue) or Wnt-TGFβ (green) subclasses and the rest of HCC samples. T highlights target genes (within the list obtained from public databases) found differentially expressed between either Wnt class compared to the rest of HCC samples (B) and are summarized in panel C as Classical Wnt target genes.
In CTNNB1-class, 7 out of 9 reported liver-related Wnt-target-genes (GLUL, REG3A, LGR5, MERTK, TBX3, EPHB2 and SPARCL1) (26) were significantly up-regulated (Fig. 2B, Suppl. Table 7) and showed a significant correlation with the expression of several transcription factors dysregulated in that class (SALL1, TLE1, CTNNBIP1, TCF7, TCF7L1, SENP2, FSTL1, SMAD3, RUVBL1, CTBP2) (Suppl. Table 5,8). In contrast, the classical Wnt-targets MMP7, PLAU, RUNX2, CDC2 and CCND3 (Fig. 2B, Suppl. Table 5) were only significantly up-regulated in Wnt-TGFβ-class and showed a significant correlation to most altered transcription factors (HDAC1, CDX1, TCF4, SALL1, SOX1, TCF1, TAXIBP3) (Suppl. Table 5,8).
In summary, clear differential expression of Wnt-pathway-genes contributes to the molecular diversity of our two previously reported Wnt-related molecular classes (Suppl. Fig. 1). Interestingly, CTNNB1- and Wnt-TGFβ-class express liver-related and classical Wnt-target-genes, respectively, which are significantly correlated to the expression of class-specific transcription factors.
MiRNA array analysis also reflected key differences between both classes (Suppl. Fig. 2). While 19 Wnt-related-miRNAs were significantly dysregulated in CTNNB1-, only 4 miRNAs were differentially expressed in the Wnt-TGFβ-class. Dysregulated miRNAs were either subclass exclusive or dysregulated in opposed directions in CTNNB1-compared to Wnt-TGFβ-class. (Suppl. Table 10).
Validation of CTNNB1- and Wnt-TGFβ-class-specific expression profiles in 6 independent HCC datasets
To validate the significant differential expression of Wnt-pathway-mRNAs we generated two gene sets, named CTNNB1- and Wnt-TGFβ-WntGenes-signature, comprising the Wnt-related genes dysregulated in each class (Suppl. Fig. 1B). Instead of validating the dysregulation for each gene, we evaluated if the global dysregulation defined by the two signatures overlapped with their corresponding molecular classes. We tested the CTNNB1- and Wnt-TGFβ-WntGenes-signatures in an independent cohort of 144 samples from our Consortium, and in 5 HCC datasets reported from other groups.
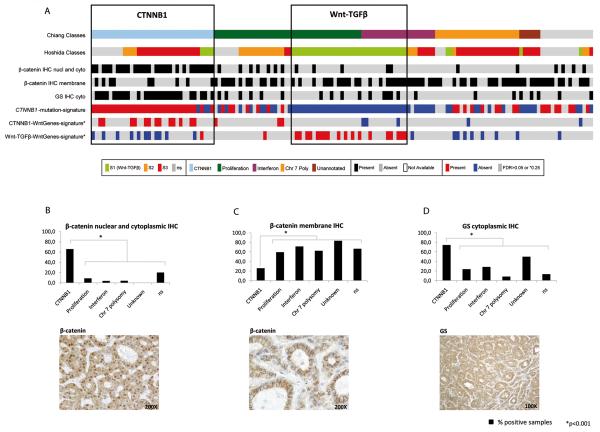
In our validation set 76/144 (52.8%) samples had active Wnt-signaling, with 35 (24.3%) belonging to the CTNNB1- and 41 (28.5%) to the Wnt-TGFβ-class. Their corresponding gene-signatures of dysregulated Wnt-pathway-mRNAs were associated with the CTNNB1- and Wnt-TGFβ-classes (p<0.001 and p=0.005, respectively) (Fig. 3). Next, the expression of liver-related Wnt-target-genes was analyzed by comparative marker selection, which remained significantly up-regulated in 7 (GLUL, REG3A, LGR5, MERTK, TBX3, EPHB2 and SPARCL1) out of 9 genes in CTNNB1-class (Suppl. Table 7). Since no mutation data were available for samples of the validation class, we generated a CTNNB1-mutation-signature with the mutated samples of the training set in order to predict the mutation status. Samples identified were significantly enriched in CTNNB1-class (p<0.001) (Fig. 3).
Fig. 3. Validation of 2 distinct Wnt-classes and their specific Wnt-related-mRNA expression profiles in an independent set of HCCs.
(A) Prediction of Chiang’s (6) and Hoshida’s (8) CTNNB1- and Wnt-TGFβ-class in our validation set using NTP (FDR<0.05). CTNNB1 class was significantly enriched for combined nuclear-cytoplasmic β-catenin (p<0.001) and cytoplasmic glutamine-synthetase (p<0.05) immunohistochemistry (IHC). Samples identified by CTNNB1-mutation-signature and CTNNB1-WntGenes-signature significantly correlated with CTNNB1-class (p<0.001), while samples identified by Wnt-TGFβ-WntGenes-signature significantly overlapped with Wnt-TGFβ-class samples (p=0.005). (B-D) β-catenin and glutamine synthetase immunohistochemistry: Percentage of positive samples in Chiang’s subclasses in the validation set and representative positive immunostaining for (A) nuclear and cytoplasmic β-catenin, (B) membrane β-catenin and (C) glutamine synthetase.
In an additional approach we interrogated 5 publicly available HCC genomic datasets representing a total of 407 patients to assess the relevance of Wnt-activation in HCC and to further confirm our class-specific Wnt-genes expression profiles. Among these 407 HCC patients, 194 (47.7%) harbored activation of the Wnt-pathway, 81 (19.9%) in CTNNB1- and 113 (27.8%) for Wnt-TGFβ-class. In 4/5 and 5/5 HCC datasets, samples identified by the CTNNB1- and the Wnt-TGFβ-WntGenes-signature, respectively, significantly correlated with the molecular classes CTNNB1 and Wnt-TGFβ, respectively (Suppl. Fig. 3, Suppl. Table 11). In all datasets analyzed, CTNNB1-class was also significantly enriched in samples called by the CTNNB1-mutation-signature (Suppl. Fig. 3, Suppl. Table 11). Specifically, CTNNB1-mutation-signature was validated in one dataset where CTNNB1 mutations were reported (5). Overall the prediction accuracy of our signature was of 91% (Specificity =87%, Sensitivity = 100%; Suppl. Fig. 4).
Overall 315/642 (49.1%) HCC samples analyzed in this study showed active Wnt-signaling belonging either to the CTNNB1- (21.5%) or the Wnt-TGFβ-class (27.6%). Particularly, Wnt-TGFβ-class was enriched in patients with non-HCV etiology (35%) compared with those with HCV-related HCC (17%). Thus, the existence of two distinct Wnt-pathway mRNA expression patterns for CTNNB1- and Wnt-TGFβ-class could be confirmed in a total of 5 and 6 independent HCC datasets, respectively.
Nuclear β-catenin and glutamine synthetase protein in the CTNNB1-class
To better characterize Wnt-activation at the protein level, immunostaining for β-catenin and glutamine synthetase as liver Wnt-target was assessed in our validation set (Fig. 3). Strong nuclear and cytoplasmic β-catenin staining was observed in 28/35 (80%) CTNNB1-class samples compared to 18/108 (16.6%) of non-Wnt HCC samples (p<0.001), while membrane staining was significantly decreased in 9/35 (25.7%) in CTNNB1-class compared to 70/108 (64.8%) of the remaining HCCs (p<0.001) (Fig. 3). β-catenin staining distribution was similar in the training and validation cohorts (Suppl. Table 9). Similarly, glutamine synthetase staining was enriched in 26/35 (74.3%) CTNNB1-class samples compared to 23/108 (21.3%) non-Wnt HCCs (p<0.001). Although both Wnt-related molecular classes show clear alteration of the Wnt-pathway at the mRNA and miRNA level, only the CTNNB1-class displayed increased cytoplasmic and nuclear β-catenin and glutamine synthetase by immunostaining.
CTNNB1- and Wnt-TGFβ-class signatures in liver cancer cell lines
We next sought to identify liver cancer cell lines with a gene expression profile similar to either CTNNB1- or Wnt-TGFβ-class. For that purpose, we obtained the gene expression data of 21 liver cancer cell lines (Suppl. Table 12; Suppl. Fig. 5) (6, 8). Analyzed by Gene Set Enrichment Analysis, HepG2 cells were significantly enriched in the CTNNB1-class-signature, while SNU387 cells were significantly enriched in the Wnt-TGFβ-class-signature (p<0.005). Therefore HepG2 cells were used as the cell line resembling the CTNNB1-class, SNU387 cells as a model for the Wnt-TGFβ-class, and SNU398 cells as a model harboring CTNNB1-mutation. Huh7 showed no clear similarity to either molecular class and was used to experimentally activate Wnt-signaling.
Sorafenib alters Wnt-signaling in vitro and in vivo
Sorafenib decreases TCF/LEF luciferase-reporter activity and β-catenin protein levels in human liver cancer cell lines
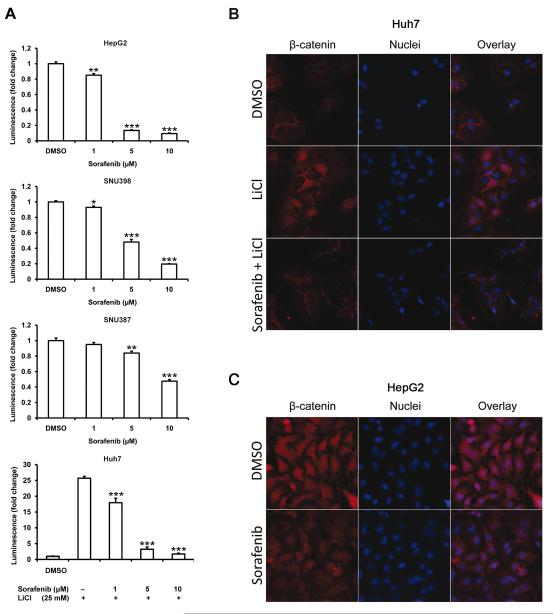
Due to clear dysregulation of Wnt-signaling in human HCC, we sought to investigate whether sorafenib can block this cascade in experimental models of liver cancer. HepG2, SNU398, SNU387 and Huh7 cells were stably transduced with luciferase-reporter of TCF/LEF β-catenin/Wnt-signaling, and according to baseline TCF/LEF luciferase reporter expression, cells could be ordered as HepG2>SNU398>Huh7>SNU387 (Suppl. Fig. 6). Baseline expression of luciferase in HepG2 was >1,000 times greater than in Huh7. These levels are consistent with the mutation status of Wnt components in these cells. HepG2 express a truncated β-catenin lacking the Ser/Thr domain that regulates its degradation whereas SNU398 have a point mutation in a key Ser in the same domain (S37C). Huh7 and SNU387 have no known mutations in β-catenin or other Wnt-related gens (27, 28). Sorafenib was effective in modulating transcriptional activity through TCF site, and in decreasing β-catenin protein levels in cells with truncated/mutated β-catenin (HepG2, SNU398), in cells with wild type β-catenin (SNU387) and in cells with wild type β-catenin and induction of Wnt-signaling by GSK3β-inhibitor LiCl (Huh7) (Fig. 4A, Suppl. Fig. 7) (16).
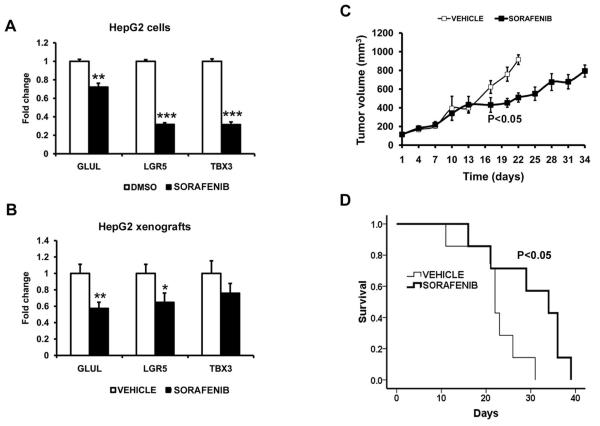
Fig. 4. Sorafenib decreases β-catenin/Wnt-signaling in 4 human liver cancer cell lines, prevents β-catenin nuclear accumulation in LiCl-stimulated Huh7 cells and decreases β-catenin protein levels in HepG2 cells.
(A) Decrease of TCF/LEF luciferase reporter activity after treatment with sorafenib (6h) in HepG2, SNU398, SNU387 and LiCl-stimulated Huh7 cells in a concentration dependent manner. (B) Immunocytofluorescence of Huh7 cells showed accumulation and nuclear localization of β-catenin after LiCl stimulation, prevented by sorafenib. (C) Sorafenib treatment decreased total β-catenin protein levels in HepG2 cells.
Immunocytofluorescence further demonstrated that treatment with sorafenib prevents β-catenin translocation to the nucleus in LiCl-treated Huh7 cells, and decreases total β-catenin levels in HepG2 cells (Fig. 4 B,C). Taken together, these data suggest that sorafenib can decrease Wnt/β-catenin signaling by modulating transcriptional activity through the TCF/LEF site and reduces β-catenin protein levels in liver cancer cells.
Sorafenib reduces liver-related Wnt-target-genes in experimental models of CTNNB1-class
As previously stated, HepG2 cells recapitulates the CTNNB1-class-signature in culture. mRNA levels of 3 known liver-related Wnt-target-genes GLUL, LGR5 and TBX3 were significant decreased following sorafenib treatment in vitro (Fig. 5A). Thus, we tested the effect of sorafenib in a subcutaneous xenograft mouse model using HepG2 cells. In this xenograft model, sorafenib treatment (30 mg/kg/day) reduced tumor volume (Fig. 5C) and extended median survival from 22 days in control animals (n=7) to 34 days in treated mice (Fig. 5D, n=7). After sorafenib treatment, GLUL and LGR5 were significantly down-regulated in HCC murine xenografts, and TBX3 showed a trend of decreased expression (Fig. 5B). Therefore, sorafenib’s effect on modulating aberrant β-catenin/Wnt-signaling is also observed in a xenograft model resembling the CTNNB1-molecular class.
Fig. 5. Sorafenib decreases liver-related Wnt-target-genes in HepG2 (CTNNB1-class) cells and modulates Wnt-signaling in HepG2 xenografts in mice.
(A) Expression of GLUL, LGR5, and TBX3 liver-related target-genes in HepG2 cells after 24 hours treatment with sorafenib. (B) Expression of GLUL, LGR5 and TBX3 genes in treated and non-treated HepG2 xenografts. (C) Decreased tumor volume in sorafenib treated mice (30 mg/kg/day), mean tumor volumes for each arm are respresented until half of mice have been sacrificed. (D) Increased median survival of sorafenib treated mice (34 days, n=7) compared to vehicle treated mice (22 days, n=7).
DISCUSSION
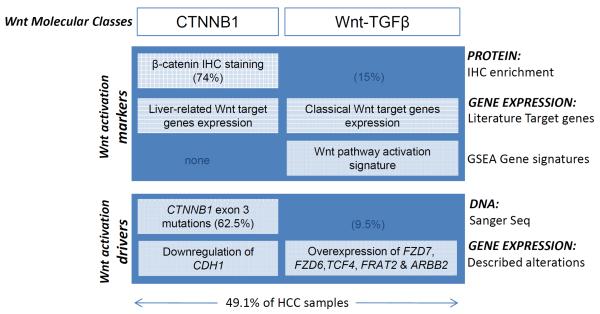
This study reports distinct Wnt-pathway alterations at the transcriptome, miRNA and protein expression level that characterize two distinct Wnt-associated molecular classes, CTNNB1 and Wnt-TGFβ, in human hepatocellular carcinoma. Markers and known drivers of Wnt activation are observed in both subclasses (Fig. 6). In addition, we demonstrate that sorafenib can modulate β-catenin/Wnt-signaling in liver cancer cells, and in CTNNB1-class-like HepG2 xenograft tumors in mice.
Fig. 6. Molecular markers and drivers of Wnt activation identified in the two Wnt-related subclasses of HCC.
Markers: CTNNB1 subclass shows the gold-standard marker of Wnt pathway activation (i.e β-catenin localization in the nucleus or cytoplasm), while common Wnt pathway target genes included in public databases (i.e Classical Wnt target genes) are enriched in Wnt-TGFβ subclass. Drivers: CTNNB1 subclass has the most commonly descrived driver of Wnt activation (i.e CTNNB1 exon3 mutations), while few other activating alterations characterize this class in comparison to Wnt-TGFβ samples which show clear activating alterations at the gene expression level (i.e overexpression of FZD7, FZD6, TCF4, FRAT2 & ARBB2).
Although several molecular classes have been associated with Wnt-signaling in HCC (5, 6, 8) and the concept of liver-related and classical Wnt-signaling has been proposed (29), no detailed characterization of underlying mechanisms has been pursued yet. Our group recently reported two fundamentally different molecular classes associated to Wnt-signaling in HCC based on expression of liver-related (CTNNB1-class) or classical (Wnt-TGFβ-class) Wnt-target-genes (6, 8). Both have been identified as distinct classes in several independent HCC datasets (8), underscoring their biological divergence. While the CTNNB1-class was correlated with CTNNB1-mutations, nuclear β-catenin staining and tumor diameter >3cm (6), Wnt-TGFβ-class was associated with TGF-β activation, cytoplasmic β-catenin staining, vascular invasion, satellitosis and greater risk of early recurrence after surgical resection (8).
The expression analysis of 210 Wnt-pathway-genes in 642 HCC samples strengthens the concept of two mutually exclusive Wnt-related molecular classes in hepatocellular carcinoma by revealing class-specific Wnt-related expression profiles in these classes. Several Wnt-pathway proteins show class-specific dysregulation at the membrane, cytoplasm and nucleus, reinforcing the presence of two different, active Wnt-pathways in liver cancer. Besides CTNNB1 mutations, we identify CDH1 down-regulation as the key contribution to Wnt-signaling activation in CTNNB1-class samples. In contrast, several genes dysregulated in Wnt-TGFβ-class characterize Wnt-signaling activation in cancer (20-25) (Fig. 6). This is the case of both FZD7(21) and TCF4(22) which upregulation has been previously described to activate Wnt signaling in the absence of β-catenin mutations. In addition, the expression of Wnt-related transcription factors appears to be highly class-specific and significantly correlates to liver-related (CTNNB1-class) or classical (Wnt-TGFβ-class) Wnt-target-genes. Regarding TCF4 over-expression, it was previously related to classical Wnt-target-genes expression (e.g. CCNDs or MMPs), even though isoform specific functions remain unclear and would require further analysis (30). Interestingly, common Wnt pathway target genes MYC and CCND1 were not found to be associated to Wnt activation in the liver, as described elsewhere (5, 31).
In accordance with our previously reported miRNA-based HCC classification that identified miRNA class A significantly enriched in the CTNNB1-class (7), we found significantly more Wnt-pathway-miRNAs dysregulated in the CTNNB1-than in the Wnt-TGFβ-class. Although several studies have implicated these Wnt-related miRNAs in Wnt-signaling, including the association of miR-375 expression to CTNNB1-mutation (32), no Wnt-class-specific miRNA expression profiles or Wnt-related miRNA-signatures have been described so far. Whether aberrant expression of miRNAs characterizing CTNNB1-class-related Wnt-signaling has functional relevance needs to be further elucidated.
Since a TP53-mutation signature reportedly predicts mutation in breast cancer accurately (15), we aimed to estimate CTNNB1-mutations with a mutation-signature generated in our training set. Its prediction accuracy was 91% when tested in an independent dataset (5) and it was enriched in CTNNB1-class in 6 independent HCC datasets, underscoring the idea that CTNNB1-Wnt is activated and driven by CTNNB1- mutations in HCC.
At the protein level we confirmed Wnt-pathway activation in CTNNB1-class by positive nuclear and cytoplasmic β-catenin immunostaining, while in Wnt-TGFβ-class β-catenin staining remained membranous. Glutamine synthetase staining as a surrogate of active β-catenin-signaling was only enriched in the CTNNB1-class, validating the existence of liver-related Wnt-target-genes at the protein level. Although the importance of Wnt-signaling for the Wnt-TGFβ-class has been proven experimentally (8), it remains unclear why pathway activation at the protein level is not detected in our set of samples. While some studies reported pathway activation without detection of nuclear β-catenin, others described negative feedback loops that activate pathway repressors or suppress pathway activators in Wnt-signaling (33). However, it has recently been reported that positive β-catenin and glutamine synthetase staining are good predictors of CTNNB1-mutation (34), further strengthening the idea that CTNNB1-class is primarily driven by CTNNB1-mutation.
The fact that half of the HCC samples displayed activation of Wnt-signaling points to the importance of this signaling cascade in hepatocarcinogenesis and reinforces the need for drugs targeting this pathway. However, all efforts to develop and translate effective and safe Wnt-pathway modulators into the clinical setting have been complicated by the multifaceted nature of Wnt-signaling (4, 15). Wnt-inhibitor toxicity has been reported for several cancers and might be intolerable in cirrhotic patients with limited hepatocyte regenerative capacity. The only FDA-approved drug for advanced HCC, sorafenib is well tolerated in cirrhotic patients (2) and inhibits several tyrosine kinases (e.g. VEGF, PDGFR, ERK/AKT, Ras, etc.), and their downstream oncogenic pathways (35). Although the Wnt-pathway has not been described as a direct target of sorafenib, there is increasing evidence of potential crosstalk between, for example, Wnt/β-catenin and PI3K/AKT signaling pathways (36, 37). The current data reveal a potential off-target effect of sorafenib, and demonstrate its ability to reduce β-catenin/Wnt-signaling in several experimental models of liver cancer. For HepG2 cells, which resemble the CTNNB1-class in their expression profile, sorafenib-mediated Wnt modulation was observed both in culture and in vivo. It remains unclear whether Wnt modulation contributes to sorafenib’s antitumoral benefits in patients with HCC, and in particular those with Wnt-pathway aberrations. Therefore, profiling of HCC patients treated with sorafenib would shed light on potential correlations between Wnt-related molecular classes and treatment response. Certainly, the mechanism by which sorafenib modulates Wnt-signaling needs to be characterized in greater detail to determine whether pathway crosstalk or a direct interaction with Wnt-pathway members leads to the observed effect on β-catenin and Wnt-target-genes.
Supplementary Material
Translational Relevance.
Hepatocellular carcinoma is a major health problem globally with increasing incidence worldwide. Despite recent advancements in the understanding of its molecular pathogenesis and treatment, the knowledge of key molecular drivers and pathways remains ill-defined. Although Wnt-signaling involvement in the pathogenesis of several malignancies is known, its specific role in HCC is unclear. Here, we dissect two major mechanisms of Wnt-activation in a large cohort of HCC samples reflecting different molecular subclasses of HCC. The Wnt-TGFβ subclass recapitulates classical Wnt-signaling as described in other cancers and is linked to a more aggressive phenotype while the CTNNB1 subclass is characterized by liver-specific Wnt-activation mediated by CTNNB1-mutations associated with a less aggressive phenotype. The fact that the multikinase inhibitor sorafenib was able to partially disrupt the activation of Wnt-signaling in a xenograft model of liver cancer, opens the path to further explore specific mechanism of action of this molecule, and to explore its role in abrogating Wnt-signaling in other cancers.
Acknowledgments
Financial support: Josep M Llovet is supported by grants from the US National Institutes of Diabetes and Digestive and Kidney Diseases (1R01DK076986-01), the European Commission’s Framework Programme 7 (HEPTROMIC, proposal no: 259744), the Samuel Waxman Cancer Research Foundation, the Spanish National Health Institute (SAF-2010-16055) and the Asociación Española Contra el Cáncer (AEEC). The study was supported by the Landon Foundation-American Association for Cancer Research Innovator Award for International Collaboration in Cancer Research. Scott Friedman has grants from the National Institutes of Health (1RO1DK37340, 1RO1DK56621), and Jordi Bruix is supported by Instituto de Salud Carlos III (PI08/0146). Sara Toffanin received a fellowship from National Cancer Institute, Milan, Italy. Clara Alsinet is supported by Instituto de Salud Carlos III (FI09/00605) and Anja Lachenmayer was supported by a fellowship from the German Research Foundation (DFG).
Abbreviations
- HCC
hepatocellular carcinoma
- TS
training set
- VS
validation set
- FFPE
formalin-fixed paraffin-embedded
- FDR
false discovery rate
- IHC
immunohistochemistry
- LiCl
lithium-chloride
- IACUC
Institutional Animal Care and Use Committee
- SC
Spearman’s Coefficient
- NTP
Nearest Template Prediction.
Footnotes
Disclosures
Josep M. Llovet, Jordi Bruix and Augusto Villanueva are consultants for Bayer Pharmaceuticals. There are no further conflicts to disclose by any of the authors.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 3.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 5.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 6.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, et al. MicroRNA-Based Classification of Hepatocellular Carcinoma and Oncogenic Role of miR-517a. Gastroenterology. 2011;140:1618–28. doi: 10.1053/j.gastro.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–92. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–61. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–9. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 11.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci. 2007;120:385–93. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 12.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 14.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 15.Coutant C, Rouzier R, Qi Y, Lehmann-Che J, Bianchini G, Iwamoto T, et al. Distinct p53 gene signatures are needed to predict prognosis and response to chemotherapy in ER-positive and ER-negative breast cancers. Clin Cancer Res. 2011 Apr 15;:2591–601. doi: 10.1158/1078-0432.CCR-10-1045. [DOI] [PubMed] [Google Scholar]
- 16.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–8. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 17.Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–67. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. doi: 10.1053/j.gastro.2008.08.008. 83 e1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bengochea A, de Souza MM, Lefrancois L, Le Roux E, Galy O, Chemin I, et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer. 2008;99:143–50. doi: 10.1038/sj.bjc.6604422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merle P, De La Monte S, Kim M, Herrmann M, Tanaka S, Von dem Bussche A, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–22. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Ravindranath A, Yuen HF, Chan KK, Grills C, Fennell DA, Lappin TR, et al. Wnt-beta-catenin-Tcf-4 signalling-modulated invasiveness is dependent on osteopontin expression in breast cancer. Brit J Cancer. 2011;105:542–51. doi: 10.1038/bjc.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnans C, Flaceliere M, Grillet F, Dantec C, Desvignes JP, Pannequin J, et al. Essential requirement for beta-arrestin2 in mouse intestinal tumors with elevated Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:3047–52. doi: 10.1073/pnas.1109457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh T, Katoh M. FRAT1 and FRAT2, clustered in human chromosome 10q24.1 region, are up-regulated in gastric cancer. Int J Oncol. 2001;19:311–5. doi: 10.3892/ijo.19.2.311. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh T, Moriwaki J, Koike J, Takagi A, Miwa T, Shiokawa K, et al. Molecular cloning and characterization of FRAT2, encoding a positive regulator of the WNT signaling pathway. Biochem Biophys Res Commun. 2001;281:815–20. doi: 10.1006/bbrc.2001.4421. [DOI] [PubMed] [Google Scholar]
- 26.Zucman-Rossi J, Benhamouche S, Godard C, Boyault S, Grimber G, Balabaud C, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–80. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 27.Yuzugullu H, Benhaj K, Ozturk N, Senturk S, Celik E, Toylu A, et al. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer. 2009;8:90. doi: 10.1186/1476-4598-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cagatay T, Ozturk M. P53 mutation as a source of aberrant beta-catenin accumulation in cancer cells. Oncogene. 2002;21:7971–80. doi: 10.1038/sj.onc.1205919. [DOI] [PubMed] [Google Scholar]
- 29.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: Sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao DH, Hong JJ, Guo SY, Yang RL, Yuan J, Wen CY, et al. Aberrant expression and function of TCF4 in the proliferation of hepatocellular carcinoma cell line BEL-7402. Cell Res. 2004;14:74–80. doi: 10.1038/sj.cr.7290205. [DOI] [PubMed] [Google Scholar]
- 31.Burke ZD, Reed KR, Phesse TJ, Sansom OJ, Clarke AR, Tosh D. Liver zonation occurs through a beta-catenin-dependent, c-Myc-independent mechanism. Gastroenterology. 2009;136:2316–24. e1–3. doi: 10.1053/j.gastro.2009.02.063. [DOI] [PubMed] [Google Scholar]
- 32.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 33.Khan Z, Vijayakumar S, de la Torre TV, Rotolo S, Bafico A. Analysis of endogenous LRP6 function reveals a novel feedback mechanism by which Wnt negatively regulates its receptor. Mol Cell Biol. 2007;27:7291–301. doi: 10.1128/MCB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bioulac-Sage P, Rebouissou S, Thomas C, Blanc JF, Saric J, Sa Cunha A, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–8. doi: 10.1002/hep.21743. [DOI] [PubMed] [Google Scholar]
- 35.Iyer R, Fetterly G, Lugade A, Thanavala Y. Sorafenib: a clinical and pharmacologic review. Expert Opin Pharmacother. 2010;11:1943–55. doi: 10.1517/14656566.2010.496453. [DOI] [PubMed] [Google Scholar]
- 36.Han L, Yang Y, Yue X, Huang K, Liu X, Pu P, et al. Inactivation of PI3K/AKT signaling inhibits glioma cell growth through modulation of beta-catenin-mediated transcription. Brain Res. 2010;1366:9–17. doi: 10.1016/j.brainres.2010.09.097. [DOI] [PubMed] [Google Scholar]
- 37.Chai H, Luo AZ, Weerasinghe P, Brown RE. Sorafenib downregulates ERK/Akt and STAT3 survival pathways and induces apoptosis in a human neuroblastoma cell line. Int J Clin Exp Pathol. 2010;3:408–15. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.