Abstract
The eukaryotic initiation factor 3 (eIF3) is an essential, highly conserved multi-protein complex that is a key component in the recruitment and assembly of the translation initiation machinery. To better understand the molecular function of eIF3, we examined its composition and phosphorylation status in Saccharomyces cerevisiae. The yeast eIF3 complex contains five core components: Rpg1, Nip1, Prt1, Tif34, and Tif35. 2-D LC-MS/MS mass spectrometry analysis of affinity purified eIF3 complexes showed that several other initiation factors (Fun12, Tif5, Sui3, Pab1, Hcr1, and Sui1) and the casein kinase 2 complex (CK2) co-purify. In vivo metabolic labeling of proteins with 32P revealed that Nip1 is phosphorylated. Using 2-D LC-MS/MS analysis of eIF3 complexes, we identified Prt1 phosphopeptides indicating phosphorylation at S22 and T707 and a Tif5 phosphopeptide with phosphorylation at T191. Additionally, we used immobilized metal affinity chromatography (IMAC) to enrich for eIF3 phosphopeptides and tandem mass spectrometry to identify phosphorylated residues. We found that three CK2 consensus sequences in Nip1 are phosphorylated: S98, S99, and S103. Using in vitro kinase assays, we showed that CK2 phophorylates Nip1 and that a synthetic Nip1 peptide containing S98, S99, and S103 competitively inhibits the reaction. Replacement of these three Nip1 serines with alanines causes a slow growth phenotype.
Keywords: phosphorylation, protein complex, protein kinase, IMAC, mass spectrometry, translation, eIF3, Nip1, yeast
1 Introduction
Translation initiation is the ribosome-mediated pairing of the methionyl initiator tRNA (met-tRNAi) anti-codon with the AUG start codon of the mRNA. This essential and highly conserved process is mediated by interactions of numerous multi-subunit eukaryotic initiation factors (eIFs) with the ribosome, mRNA, and tRNAs. In the first step, a ternary complex consisting of eIF2 bound to GTP and met-tRNAi is recruited to the 40S ribosomal subunit to form the 43S pre-initiation complex 1,2. In vitro experiments suggest that this interaction is promoted by eIF1, eIF1A, and eIF3 3–9. In the next step, eIF4F, which is composed of eIF4A (RNA helicase), eIF4E (5´ m7GTP cap binding protein), and eIF4G (scaffold protein) 10 facilitates interaction of the 43S pre-initiation complex with the 5’ cap structure of the mRNA producing the 48S pre-initiation complex. Efficient formation of the 48S pre-initiation complex is also dependent on eIF3 11. The 40S complex scans the mRNA from the 5’ end for the start codon in a process mediated by eIF1 and eIF1A 12. Once the met-tRNAi anti-codon pairs with the AUG codon, eIF2-bound GTP is hydrolyzed in a reaction stimulated by eIF5 13. The GTP hydrolysis triggers the release of the bound eIFs and the joining of the 60S subunit to create the 80S ribosome 14. At this point, the 80S complex accepts the next aminoacyl-tRNA in the A site of the ribosome and the process of translation elongation begins.
A key player in the assembly and function of the 48S pre-initiation complex is the multi-protein eIF3 complex. While the mammalian eIF3 complex contains thirteen subunits, much of what is known about the mechanisms of eIF3 function have been characterized in studies of the five-subunit eIF3 complex in S. cerevisiae 11. The yeast eIF3 subunits Rpg1, Nip1, Prt1, Tif34, and Tif35 are homologs of the human eIF3 subunits eIF3a, eIF3c, eIF3b, eIF3i, and eIF3g, respectively 15. Yeast Hcr1 is homologous to human eIF3j, and appears to associate with the core comples of eIF3 at non-stoichiometric levels 16. The eIF3 complex, as well as eIF1, eIF5, and the eIF2 ternary complex, have been identified as part of a multifactor complex (MFC) that mediates the assembly of the 48S pre-initiation complex 4,11. Within eIF3, Rpg1 and Hcr1 interact with Prt1 via an N-terminal RNA recognition motif (RRM) 16, which mediates protein-protein interactions despite the name. These interactions are crucial for the structural integrity of the eIF3 complex and its stable association with the MFC and 40S ribosome complex 16. Nip1 and Prt1 play key roles in both the assembly and maintenance of the 48S pre-initiation complex 11. Nip1 interacts directly with eIF1 and eIF5, suggesting that it coordinates both AUG start codon recognition (in conjunction with eIF1, which interacts with the met-tRNAi) and eIF2-GTP hydrolysis (in conjunction with eIF5) 6,17. The smallest of the eIF3 subunits, Tif34 and Tif35, have recently been shown to promote the linear scanning of mRNA 18.
Protein kinases are key regulators of cell function and are believed to directly affect translation efficiency 19,20. By phosphorylating a protein, kinases can alter the substrate’s activity, interactions, and stability. The functions of several eIF complexes are regulated by phosphorylation. The best characterized of these phosphorylation events involve regulation of eIF2 and eIF4E 21. eIF2 is bound to GDP in an inactive state at the end of each round of translation initiation. To reconstitute a functional ternary complex for a new round of translation initiation, eIF2B catalyzes the exchange of GDP for GTP 2. Phosphorylation of the eIF2 subunit eIF2α effectively blocks eIF2B-mediated GDP-GTP exchange, thus inhibiting protein synthesis 22. Phosphorylation also regulates association of eIF4E with the eIF4F mRNA-cap binding complex. This interaction is required for cap-dependent mRNA translation initiation and is inhibited by the eIF4E-binding protein (4E-BP) 10. Phosphorylation of 4E-BP attenuates this inhibition 23. In its hypophorphorylated state, 4E-BP binds to eIF4E and prevents its association with eIF4F 24. In its hyperphosphoryated state, 4E-BP binds poorly to eIF4E. 4E-BP phosphorylation is stimulated by many different external stimuli including growth factors, hormones, and mitogens 24. Finally, eIF4E is also phosphorylated. Phosphorylation of S209 is stimulated by different mitogen-activated protein kinase pathways and correlates with increased translation rates 23,25,26.
We have used mass spectrometry techniques to identify the components of eIF3 and specific residues that are phosphorylated in vivo. Mass spectrometry has become the method of choice for identifying the composition of a protein complex 27. However, although protein identification by mass spectrometry has become routine, protein phosphorylation analysis remains a challenging problem. Low phosphorylation stoichiometry, heterogeneous phosphorylation sites, and low protein abundance contribute to the difficulty of phosphoprotein analysis 28. In addition, phosphopeptides are generally difficult to analyze by mass spectrometry for a number of reasons 29. Reduced ionization efficiency in positive ionization mode and suppression by nonphosphorylated peptides exacerbate the problems. To improve the efficiency of phosphopeptide analysis, various strategies have been developed to enrich for phosphopeptides including strong cation exchange chromatography (SCX), immunoaffinity capture using antiphosphotyrosine antibodies, and immobilized metal affinity chromatography (IMAC) 30–32. IMAC was initially plagued by the nonspecific retention of peptides rich in acidic amino acids, but improvements have increased both the specificity and sensitivity of IMAC 33–35. The direct coupling of IMAC with high sensitivity reversed-phase liquid chromatography and tandem mass spectrometry has enabled us to identify phosphopeptides starting with small amounts of material.
Using tandem mass spectrometry analysis of tandem affinity-purified (TAP) eIF3 complexes, we detected the core yeast components Rpg1, Nip1, Prt1, Tif34, and Tif35. Several other initiation factors co-purified, including Fun12, Tif5, Sui3, Pab1, Hcr1, Sui1, and the casein kinase 2 complex (CK2). Using phosphorylation assays and IMAC enrichment prior to mass spectrometry, we found that Prt1, Nip1, and Tif5 are phosphorylated in vivo and identified three specific sites on Nip1, two on Prt1 and one on Tif5. We used an in vitro kinase assay to test individual kinases for their ability to phosphorylate eIF3 in vitro. CK2, which associates with the eIF3 complex, phosphorylates the eIF3 component Nip1. A synthetic NIP1 peptide containing the three serine residues identified as phosphorylated inhibits in vitro phosphorylation of Nip1 by CK2. Mutagenesis of these three phosphorylation sites in Nip1 indicates that they are important for a normal growth rate.
2 Materials and Methods
Plasmids
Plasmid pNip1entry (ScCD00008839) containing the wild-type Nip1 gene in a Gateway entry vector was obtained from the Harvard Institute of Proteomics (Boston, MA) and sequenced verified. It was recombined with the destination clone pAG415GPD-ccdB-TAP from Addgene (Cambridge, MA) as previously described 36 to generate pNip1a. The identified sites of phosphorylation, S98, S99 and S101, were converted to alanines by site directed mutagenesis 37 using the primers nip1mutfwd (GCTCTAACTATGATGCCGCTGATGAAGAAGCCGATGAAGAAG) and nip1mutrev (CTTCTTCATCGGCTTCTTCATCAGCGGCATCATAGTTAGAGC) to generate pNip1b. The endogenous stop codon was removed from both pNip1a and pNip1b with the primers nip1stopfwd (CCATCAAATCGTCGTGCGACCCAGC) and nip1stoprev (GCTGGGTCGCACGACGATTTGATGG) to generate pNip1wtTAP and pNip1mutTAP. All clones were sequence verified. pNip1comp, expressing Nip1 with a URA3 selectable marker 17, was obtained from Dr. Alan Hinnebusch’s laboratory.
Yeast strains
TAP-tagged yeast strains used in this study have been previously described 38. We tested pNip1a (wild-type) and pNip1b (with three phosphorylation sites replaced by alanines) by transforming the strain H2997 (MATa, ura3-52, leu2-3, 112, his4-303, (AUU)SUI1, nip1Δ [pNIP1+(B3940)] 17, obtained from Dr. Alan Hinnebusch and selecting for loss of the parent strain’s plasmid with 5-FOA. Strains containing pNip1wtTAP and pNip1mutTAP were similarly generated. pNip1comp was used to transform the pNip1b strain, generating a complemented strain.
Purification and Identification of eIF3 Components
The eIF3 complex was purified using a previously described protocol from TAP-tagged yeast strains grown to early stationary phase (O.D.600 2–4) in YPD medium 39,40. For identifying the protein composition of eIF3, 2 L cultures of strains expressing TAP-tagged alleles of RPG1, NIP1, PRT1, and TIF5 were analyzed. BY4741, the parental strain for the TAP clones, was used as a negative control since it lacks any affinity tagged gene. For analysis of the association of casein kinase 2 (CK2) with eIF3, a TAP-tagged allele of CKB2 was used for a similar purification. For validation experiments, TAP-tagged FUN12 and PAB1 strains were used. Purified proteins were identified using two different methodologies. In one approach, proteins were separated by 10% acrylamide SDS-PAGE. Stained bands were excised and in-gel trypsin digested, and purified proteins were identified using reversed-phase LC-MS/MS analysis 39,41. In the second approach, components of eIF3 were identified directly in solution with Multidimensional Protein Identification Technology (MudPIT) using an LTQ quadrupole ion trap (Thermo Electron, Inc) 39,42,43. Protein samples were reduced with 1/10 volume of 50 mM DTT at 65°C for 5 min, cysteines were alkylated with 1/10 volume of 100 mM iodoacetamide at 30°C for 30 min, and proteins were trypsinized with modified sequencing grade trypsin at ~25:1 subtrate:enzyme ratio (Promega, Madison, WI) at 37°C overnight. A fritless, microcapillary (100 µm-inner diameter) column was packed sequentially with the following: 9 cm of 5 µm C18 reverse-phase packing material (Synergi 4 µ Hydro RP80a, Phenomenex), 3 cm of 5 µm strong cation exchange packing material (Partisphere SCX, Whatman) and 2 cm of C18 reverse-phase packing material. The trypsin-digested samples were loaded directly onto the triphasic column equilibrated in 0.1% formic acid, 2% acetonitrile, which was then placed in-line with an LTQ linear ion trap mass spectrometer (Thermo, Inc.). An automated six-cycle multidimensional chromatographic separation was performed using buffer A (0.1% formic acid, 5% acetonitrile), buffer B (0.1% formic acid, 80% acetonitrile) and buffer C (0.1% formic acid, 5% acetonitrile, 500 mM ammonium acetate) at a flow rate of 300 nL/min. The first cycle was a 20-min isocratic flow of buffer B. Cycles 2–6 consisted of 3 min of buffer A, 2 min of 15–100% buffer C, 5 min of buffer A, followed by a 60-min linear gradient to 60% buffer B. In cycles 2–6, the percent of buffer C was increased incrementally (from 15, 30, 50, 70 to 100%) in each cycle. During the linear gradient, the eluting peptides were analyzed by one full MS scan (200–2000 m/z), followed by five MS/MS scans on the five most abundant ions detected in the full MS scan while operating under dynamic exclusion.
Mass Spectrometry Data Analysis
To process and analyze the mass spectrometry data, the program extractms2 was used to generate an ASCII peak list and identify +1 or multiply charged precursor ions from native *.RAW mass spectrometry data files (Jimmy Eng and John R. Yates III, unpublished). Tandem spectra were searched with no protease specificity using SEQUEST-PVM 44 against the SGD yeast_orfs database containing 6,000 entries with a static modification of +57 on C (addition of a carbamidomethyl group). For multiply charged precursor ions (z ≥ +2), an independent search was performed on both the +2 and +3 mass of the parent ion. Data were processed and organized using the BIGCAT software analysis suite45. A weighted scoring matrix was used to select the most likely charge state of multiply charged precursor ions42,45. From the database search, tryptic peptide sequences with SEQUEST cross-correlation scores (Cn) ≥ 1.5 for +1 ions, ≥ 2 for +2 ions, and ≥ 2 for +3 ions were considered significant and used to create a list of identified proteins.
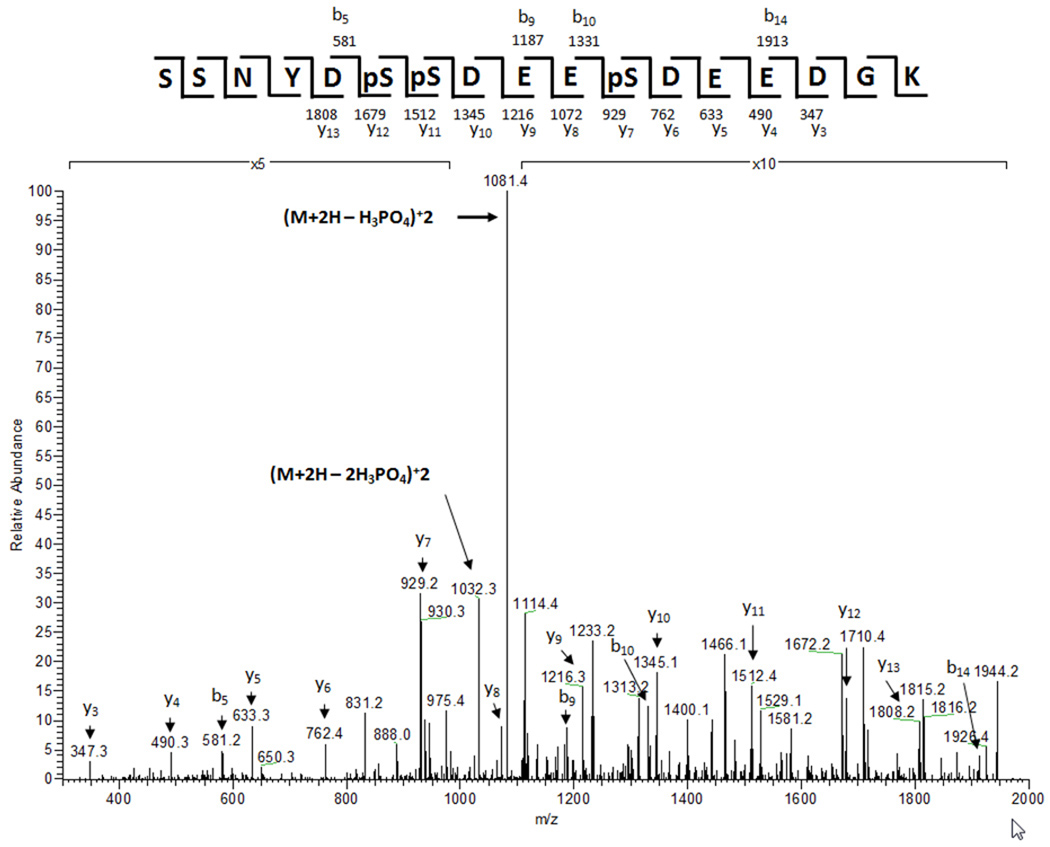
To validate candidate phosphopeptides, tandem mass spectra were manually evaluated to identify evidence of neutral loss of phosphoric acid caused by fragmentation of the precursor ion (98 (z=+1), 49 (z=+2), or 32.7 (z=+3) m/z units relative to the precursor). For all IMAC mass spectrometry data, the acquired mass spectrometry data were searched against the yeast ORF database using the Sequest algorithm assuming static modifications of +14 on D, E, and the C-termini, static modification of +57 on C, and variable modifications of +80 on S, T, and Y 46. Our criteria in identifying phosphorylations were: 1) a series of at least 3 y or b ions with an appropriate +80 Dalton shift, and 2) ions indicating a neutral loss of phosphoric acid from the precursor ion. Annotated MS/MS spectra illustrating such evidence are presented in Figure 3 and Supplemental Figure 1.
Figure 3. Nip1 is phosphorylated at S98, S99, and S103.
An MS/MS spectrum generated after IMAC-enrichment of phosphopeptides from trypsin-digested eIF3 showing in vivo phosphorylation of a Nip1 peptide at S98, S99, and S103. The spectrum significantly matched the trypsin-generated peptide of Nip1 (Sequest Xcorr = 3.1). The spectrum shows the addition of 80 Da at S98, S99, and S103. The m/z values of the most prominent ions in the MS/MS spectrum correspond to the neutral loss of one and two phosphoric acids from the precursor ion of 1131.6 (+2).
A protein abundance factor (PAF) representing relative protein abundance, was calculated for each identified protein by normalizing the total number of non-redundant spectra that correlated significantly with each cognate protein to the molecular weight of the protein and multiplying by 104 39.
In Vivo 32P Labeling of the eIF3 Complex
12 mL cultures of strains expressing TAP-Rpg1, TAP-Nip1, and TAP-Nip1S98,99,101A were grown in low-phosphate YPD at 30°C to an O.D.600 of 1.0 47. The cultures were centrifuged, and the cells were resuspended in 5 mL low-phosphate YPD containing 5 mCi carrier-free 32P-orthophosphate (ICN, Costa Mesa, CA). An unlabeled culture was grown in parallel. The cells were labeled for 30 min at 30°C. The cultures were centrifuged, and the cells were resuspended in 20 mL fresh low-phosphate YPD and incubated for 4 h at 30°C. Cells were harvested by centrifugation, washed twice with 10 mL ice-cold water and resuspended in 1 mL lysis buffer containing 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% NP-40, and 1x CompleteR protease inhibitor (Roche Diagnostics Corp., Indianapolis, IN). Cells were lysed by vortexing with 500 µL 0.5 mm zirconia/silica beads (BioSpec Products, Inc., Bartlesville, OK) for 6 min with cycles of 30 sec of active vortexing followed by 30 sec off. The lysates were centrifuged at 20,000xg for 15 min to remove particulate material. The proteins were purified using the TAP method 39,40 with the volumes reduced proportionally to the amount of cells isolated. Denatured proteins were separated by 10% acrylamide SDS-PAGE. 32P labeled-proteins were detected by autoradiography. Densitometric measurements were quantified using Scion image software 48.
Direct Mapping of Phosphorylated Sites on eIF3 Components with Tandem Mass Spectrometry
The eIF3 complex was purified from whole-cell lysates prepared from 4 L overnight cultures of the TAP-Rpg1 strain 39,40. Purified complexes were digested directly in solution with sequencing grade trypsin (Promega, Madison, WI) or chymotrypsin (Roche Diagnostics Corp., Indianapolis, IN) 43 and analyzed by MudPIT using an LTQ linear ion trap mass spectrometer (Thermo Electron, Inc).
Mapping eIF3 Phosphopeptides using IMAC Enrichment and Tandem Mass Spectrometry
The S. cerevisiae strain expressing a TAP-Rpg1 was grown overnight (O.D.600 2–4) in 16 L of YPD medium. eIF3 complexes were TAP-isolated from whole cell lysates 40. The pH of the eluted eIF3 mixture was adjusted to 8.0 using 1 M Tris pH 8.0. Cysteine residues were reduced with DTT and alkylated with iodoacetamide. The proteins were digested with 20 µg of sequencing grade trypsin 43 (Promega, Madison, WI). The tryptic peptide mixture was divided into 1 mL aliquots and lyophylized. To methyl esterify the peptides, a pre-mix of 1 mL anhydrous methanol and 50 µL thionly chloride (Sigma-Aldrich, St. Louis, MO) was added to the dried peptides. The mixture was sonicated for 15 min at room temperature and incubated at room temperature for 2 h. Esterified peptides were lyophilized to dryness.
An IMAC column was constructed using a fritted 20 cm long piece of 200 × 360 µm fused silica capillary (FSC). The column was packed with POROS MC 20 beads (Applied Biosystems, Foster City, CA) to a length of 15 cm. The column was rinsed with 100 mM EDTA (Sigma-Aldrich, St. Louis, MO), pH 8.5 and then with deionized water. The column was charged with 100 mM Iron(III) chloride solution (Sigma-Aldrich, St. Louis, MO). Finally, the IMAC column was rinsed with 0.1% acetic acid.
The dried, esterified peptides were resuspended in 180 µL of 33% methanol, 33% acetonitrile, and 0.034% acetic acid and loaded onto the IMAC column. Following loading, the column was rinsed with a solution of 25% acetonitrile, 100 mM NaCl, and 1% acetic acid.
A reversed-phase capture column was constructed from 20 cm of 100 × 360 µm FSC fritted at one end using an inline MicroFilter Assembly (Upchurch Scientific, Inc, Oak Harbor, WA). The RP-capture column was packed with POROS 10 R2 (Applied Biosystems, Foster City, CA) to a length of 10 cm. Using a 0.012 × 0.060 inch piece of Teflon tubing (Zeus, Orangeburg, SC), the RP-capture column was butt-jointed to the IMAC column. Phosphopeptides were eluted from the IMAC column onto the RP-capture column with 250 mM sodium phosphate, pH 8.0 solution.
The RP-capture column was rinsed overnight with 5% acetonitrile and 0.1% formic acid and attached to an analytical RP chromatography column (100 × 365 µm FSC with an integrated laser-pulled emitter tip packed with 10 cm of Synergi 4 µm RP80A (Phenomenex)) via the inline MicroFilter Assembly for LC-ESI-MS/MS analysis. Peptides were eluted using the following linear gradient: 0 min: 0% B, 120 min: 40% B, 140 min: 60% B, 160 min: 100% B at a flow rate of 400 nL/min (mobile phase A: 5% acetonitrile, 0.1% formic acid and mobile phase B: 80% acetonitrile, 0.1% formic acid).
Spectra were acquired with a LTQ linear ion trap mass spectrometer (Thermo Electron, Inc). During LC-MS/MS analysis, the mass spectrometer performed automated data-dependent acquisition with a full MS scan followed by three MS/MS scans on the most intense ions while operating under dynamic exclusion.
Kinase Purification and In Vitro Kinase Assays
Specific protein kinases were purified from S. cerevisiae strains expressing individual amino-terminally GST-tagged kinases 49. The GST-tagged kinase strains were kindly provided by Dr. Mike Snyder. GST-tagged strains were cultured in 25 mL synthetic complete medium lacking uracil (SC - URA) with 2% raffinose at 30°C for 16 h. GST-kinase expression was induced by incubation in SC-URA with 2% galactose for an additional 2 h at 30°C. Cells were harvested by centrifugation and resuspended in 1 mL lysis buffer containing 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% NP-40, and 1x CompleteR protease inhibitor (Roche Diagnostics Corp., Indianapolis, IN). Cells were lysed using an equal volume of 0.5 mm zirconia/silica beads and a Mini-Beadbeater-8 (BioSpec Products, Inc., Bartlesville, OK). Lysates were centrifuged at 20,000xg for 15 min to remove particulate material. Supernatants were incubated with 100 µL glutathione-coupled agarose beads (Amersham Biosciences, Piscataway, NJ) for 1 h at 4°C. The beads were washed twice with 1 mL lysis buffer then twice with 1 mL buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, and 10% glycerol. The beads were then resuspended in 100 µL kinase buffer (25 mM HEPES, 25 mM β-glycerolphosphate, 25 mM MgCl2, 2 mM DTT, 0.1 mM NaVO3).
Phosphorylation reactions with the eIF3 complex were performed by incubating each purified kinase with 10 µCi [γ-32P]ATP (ICN, Costa Mesa, CA) and 1 µg eIF3 complex (isolated by purification of TAP-Rpg1) in 30 µL kinase buffer for 1 h at 30°C. To eliminate endogenous kinase activity, eIF3 purifications were first denatured with 8 M urea, which was later diluted to 1 M prior to the kinase assays 41. Kinase reactions were terminated by the addition of Laemmli-SDS sample buffer and heating at 100 °C for 3 min. The kinase reaction products were separated by SDS-PAGE. The proteins were silver stained and phosphorylation was detected by autoradiography.
Inhibition of In Vitro Phosphorylation
A peptide inhibitor containing Nip1 phosphorylation sites (H2N-SSNYDSSDEEDSDDDGK-OH) was synthesized to mimic the tryptic peptide of Nip1 identified by mass spectrometry (New England Peptides Gardner, MA). A control peptide H2N-ISQAVAHAHAEINEAGR-OH that contains an identical number of amino acids but lacks CK2 consensus sequence sites for phosphorylation was synthesized (New England Peptides yes/no). The peptides were suspended at 1 mg/mL in 100 mM Tris, pH 8.0 to a concentration of 1 mg/mL. The CK2 inhibitor (E)-3-(2,3,4,5-Tetrabromophenyl)acrylic acid (TBCA) and the CK1 inhibitor 3-[(2,4,6-Trimethoxyphenyl)methylidenyl]-indolin-2-one (IC261) were obtained from Calbiochem (Gibbstown, NJ) and resuspended to 1 mg/mL in DMSO 50,51. The peptides and kinase inhibitors were added to kinase reactions described above to a final concentration of 33 µg/mL.
Quantifying the Doubling time of Wild-type and Mutant of Nip1
We measured the growth rate of nip1 deletion mutants carrying either pNip1a (NIP1+), pNip1b (nip1-S98,99,101A) or pNIP1b plus pNip1comp (NIP1+). Single colonies from each strain were inoculated into YPD and grown overnight at 30°C. Cell density was calculated using a Countess automated cell counter from Invitrogen (Carlsbad, CA). 10,000 cells of each strain were transferred to fresh 6 mL cultures of YPD at 30°C. Aliquots from each strain were collected periodically, and cell density was calculated in triplicate until the cultures reached stationary phase. The triplicate density measurements were averaged. The doubling time (tdub) for each strain was determined from the logarithmic growth phase of the cells using the following formula:
Where D is the cell density and Δtmin is the change in time in min from D1 to D2. The experiment was repeated in triplicate and the resulting doubling times were averaged for each strain. A Student’s T-test (95% confidence interval) was performed on the data to determine its statistical significance.
3 Results
In Vivo Interaction of eIF3 Components
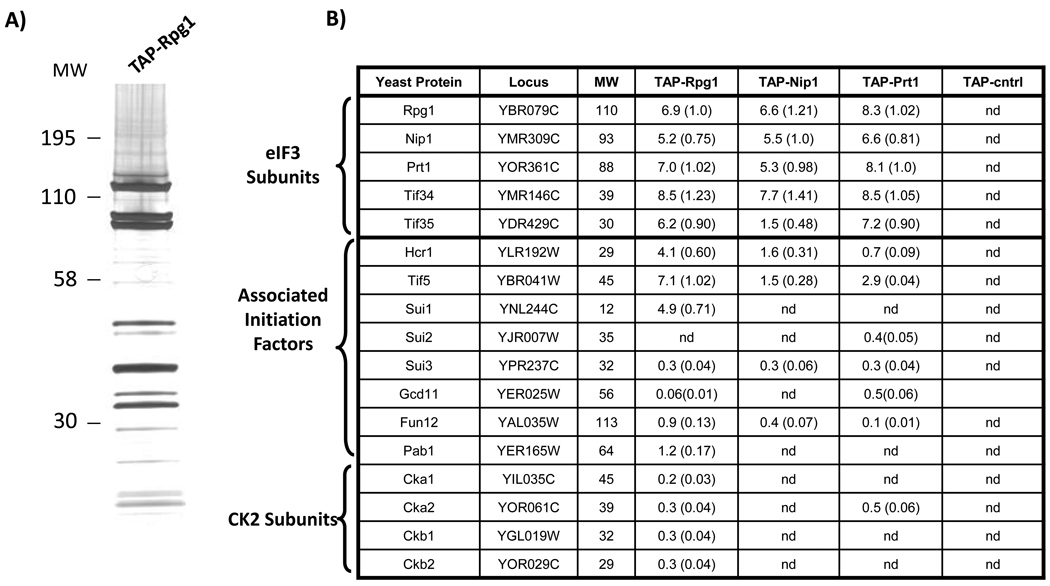
The goals of this study were to identify comprehensively the composition of the S. cerevisiae eIF3 complex and dissect the sites of in vivo phosphorylation. To this end, we first established an approach for high-quality purification of the eIF3 complex using epitope tags and tandem affinity purification (TAP). This approach enables the efficient recovery of proteins present at low cellular concentrations under native conditions 52. We used individually TAP-tagged yeast strains in which the fusion proteins are driven by native promoters, avoiding overexpression of the affinity tagged protein38,53. A representative purification is shown for TAP-Rpg1 (Fig. 1A). eIF3 complexes were purified in duplicate from four different yeast strains, each expressing a different TAP-tagged protein (Rpg1, Nip1, Prt1, or Tif5) 15. To control for purification of non-specific proteins, three independent extracts were prepared and analyzed in parallel from an isogenic untagged yeast strain (BY4741). The isolated eIF3 complexes and control purifications were trypsin-digested in solution, and the proteins were identified using the 2-D LC-MS/MS mass spectrometry approach termed Multidimensional Protein Identification Technology (MudPIT) (Fig. 1B) 42,54,55. To estimate the relative abundance of each protein, a Protein Abundance Factor (PAF) was calculated using the acquired mass spectrometry data 39. A PAF is a label-free, semi-quantitative measure of a protein’s relative abundance and is based on the fact that a protein’s abundance is directly related to the frequency at which its peptides are selected for MS/MS analysis.
Figure 1. Purification and mass spectrometry identification of eIF3 components.
The eIF3 complex was isolated from S. cerevisiae by tandem affinity purification (TAP). Complexes and control extracts were prepared in duplicate or triplicate from strains expressing affinity tags specific for three core eIF3 components (Rpg1, Nip1, and Prt1) and the translation initiation factor Tif5, as well as an isogenic, untagged yeast strain. (A) Silver-stained SDS-PAGE separation of a TAP-Rpg1 purification. (B) MudPIT mass spectrometry analysis of the purifications from the four different TAP-tagged strains and the control strain (TAP-cntrl). The results from one of the replicate experiments are shown. The predicted molecular weights (MW) of the detected yeast proteins (rows) are shown in kDa. For each tagged strain and the control, protein abundance factors (PAFs) were calculated for the identified proteins (PAF = (the number of non-redundant spectra identifying the protein) / (MW of the protein) ×10439. The relative stoichiometry of each protein (in parentheses) was determined by normalizing its PAF to the PAF of the TAP-tagged protein targeted in the purification. (nd: not detected).
We routinely detected the five primary components of yeast eIF3 (Rpg1, Nip1, Prt1, Tif34, and Tif35) at greater than 75% coverage (Fig. 1B). The PAFs suggest that these proteins have equivalent stoichiometries. We also identified other translation initiation factors co-purifying in our eIF3 preparations, including the yeast orthologs of mammalian eIF5 (Tif5), eIF3j (Hcr1), eIF1 (Sui1), eIF2β (Sui3), eIF5B (Fun12), and PABC1 (Pab1) (Fig 1B). With the exception of Fun12 and Pab1, earlier studies have reported similar eIF3 interactions 11. As expected, the PAFs values were lower for these six non-core, co-purifying translation initiation factors.
Interestingly, purifications based on different tagged targets yielded distinct subsets of co-purifying translation initiation factors, and the results were consistent in replicate purifications. We performed reciprocal purifications targeting the previously uncharacterized interactions of Fun12 and Pab1 and detected co-purifying eIF3 components but at lower abundances (see the TAP-Tif5 results in Fig. 1B, Table 1 and Supplemental Table 1). These results are consistent with a model in which these six translation initiation factors act as accessories in eIF3 function. Additionally, we consistently observed the yeast casein kinase 2 complex (CK2) copurifying with the TAP-Rpg1 isolation of eIF3 (Fig. 1B). In yeast, CK2 is a tetramer consisting of two catalytic subunits (Cka1 and Cka2) and two regulatory subunits (Ckb1 and Ckb2) 56,57. However, the PAF values were relatively low, indicating that the CK2 subunits associate with eIF3 at substoichiometric levels, consistent with the transient nature of kinase-substrate interactions. The CK2 subunits were not detected in the controls or in a large number of independent TAP/MudPIT experiments using yeast strains with different TAP-tagged proteins (Link et al, manuscript in preparation), suggesting a specific interaction of CK2 with eIF3. We further investigated the physical association of CK2 with eIF3 by targeting TAP-Ckb2 for protein purification. LC-MS/MS analysis of purified TAP-Ckb2 confirmed that the isolate contains all components of CK2 as well as all five core subunits of eIF3. Control purifications from an untagged strain contained neither CK2 nor eIF3 (Table 3).
Table 1. Mass spectrometry identification of Fun12 and Pab1 interactions with eIF3.
Fun12 protein complexes was isolated from an S. cerevisiae strain expressing TAP-Fun12 and an isogenic, untagged yeast strain as a negative control. Mudpit mass spectrometry analysis of the purified complexes were performed in triplicate. PAF values were used to approximate protein abundances as in Fig. 1.
| Locus | TAP-Fun12 | TAP-Pab1 | negative control |
|
|---|---|---|---|---|
| Targeted Proteins | ||||
| Fun12 | YAL035W | 5.1(1.0) | nda | |
| Pab1 | YER165W | 12(1.0) | nd | |
| eIF3 Core Proteins | ||||
| Rpg1 | YBR079C | 0.18(0.035) | 0.030(0.0025) | nd |
| Nip1 | YMR309C | 0.071(0.014) | 0.036(0.0030) | nd |
| Prt1 | YOR361C | 0.038(0.0074) | nd | nd |
| Tif34 | YMR146C | 0.43(0.084) | nd | nd |
| Tif35 | YDR429C | 0.33(0.064) | nd | nd |
nd = none detected
Table 3. Summary of Nip1 peptides identified by LC/MS/MS after IMAC-enrichment of phosphopeptides from trypsin-digested eIF3.
The table shows the precursor ion and the m/z of the most intense fragment ion (base peak) in the corresponding MS/MS spectrum. The observed neutral loss is given and corresponds to a loss of 98 Da from the precursor peptide. The predicted charge state of the precursor ion (Z) is listed. Sequest cross-correlation values (Cn) to the phosphopeptide sequences are shown. This value is a statistical measurement of how well the theoretical spectrum generated for the given peptide sequence corresponds to the experimental spectrum. The peptide sequences show the predicted upstream and downstream amino acids flanking the peptides.
| Parent m/z |
m/z of MS/MS base peak |
Neutral Loss | Z | Cn | Peptide Sequence |
|---|---|---|---|---|---|
| 1131.6 | 1081.4 | 50.2 | +2 | 3.1 | K.SSNYDpSpSDEEpSDEEDGK.K |
| 796.8 | 764.1 | 32.7 | +3 | 2.6 | K.SSNYDpSpSDEEpSDEEDGKK.V |
| 1090.5 | 1040.7 | 49.1 | +2 | 2.8 | K.SSNYDpSpSDEESDEEDGK.K |
| 1151.1 | 1105.6 | 49.3 | +2 | 2.2 | K.SSNYDpSpSDEESDEEDGKK.V |
| 770.1 | 737.0 | 33.1 | +3 | 2.3 | K.SSNYDpSpSDEESDEEDGKK.V |
| 1090.3 | 1041.5 | 48.8 | +2 | 2.3 | K.SSNYDSpSDEEpSDEEDGK.K |
| 1155.2 | 1105.1 | 50.1 | +2 | 2.7 | K.SSNYDSpSDEEpSDEEDGKK.V |
The eIF3 complex was effectively purified using all four TAP-tagged eIF3 components, however the mass spectrometry results indicated that the TAP-Rpg1 strain produced the highest yield of both the eIF3 core components and other associated initiation factors. Therefore, from this point on eIF3 proteins were purified and analyzed using the TAP-Rpg1 strain.
Identification of Phosphorylated Components of eIF3
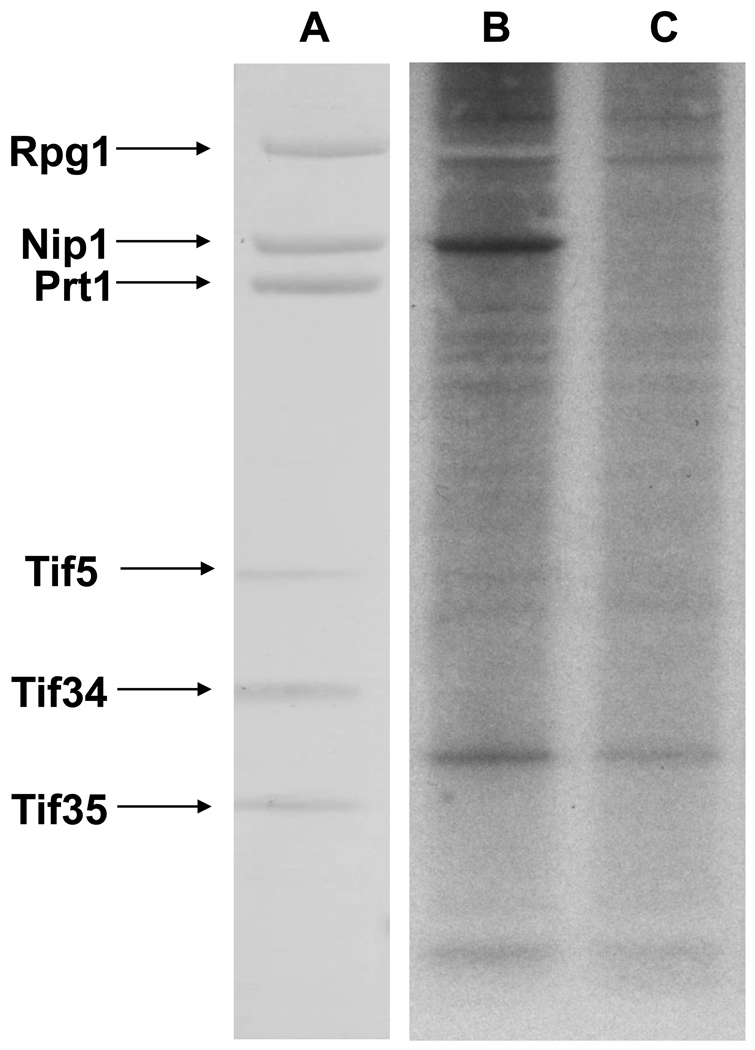
The observation that CK2 consistently copurifies with components of eIF3 and the important role of phosphorylation in a wide array of biological processes lead us to examine whether it can target phosphorylations on components of the eIF3 complex. We metabolically labeled the TAP-Rpg1 and an unlabeled strain with 32P-orthophosphate at early stationary phase in rich media. eIF3 was purified, the protein components were separated by SDS-PAGE, and phosphorylation was detected by autoradiography (Fig. 2). One prominent band was observed that corresponded to the molecular weight of Nip1. The identity of the phosphorylated protein was confirmed to be Nip1 by mass spectrometry analysis of the corresponding band excised from the unlabeled eIF3 complex analyzed in parallel.
Figure 2. In vivo phosphorylation of the yeast eIF3 complex.
(A) SDS-PAGE separation and silver staining of eIF3 components isolated from a TAP-Rpg1 strain following in vivo 32P metabolic labeling. In-gel trypsin digestion and LC/MS/MS were used to identify the bands (B) The corresponding autoradiograph indicates that Nip1 is phosphorylated in vivo. (C) Negative control lane using a TAP purification from an untagged strain metabolically labeled with 32P.
Mass spectrometry Identification of In Vivo Prt1 and Tif5 Phosphorylation Sites
Mass spectrometry techniques can be used to pinpoint exact sites of protein modification, including phosphorylation. Our initial approach used MudPIT to identify eIF3 phosphopeptides directly from complexes purified from the TAP-Rpg1 strain. This direct approach avoided sample loss associated with strategies to enrich for phosphopepetides. TAP-isolated eIF3 samples were digested separately with either trypsin or chymotrypsin and analyzed using MudPIT. We used two different proteolytic enzymes to maximize peptide coverage and to facilitate identification of candidate phosphorylation sites on multiple overlapping peptides 58.
Phosphorylations present several hallmarks in mass spectrometry analysis. First, the mass of a phosphorylated residue is increased by 80 Da. Second, when peptides containing phosphorylated serine or threonine residues are fragmented by resonance excitation in an ion trap they commonly undergo a gas-phase β-elimination reaction, resulting in the neutral loss of phosphoric acid (−H3PO4 or 98 Da) 29. Depending on the charge of the precursor ion, neutral loss of phosphoric acid appears as a decrease of 98 (z=+1), 49 (z=+2), or 32.7 (z=+3) m/z units from the precursor. Finally, because a phosphorylated residue increases the net negativity of the peptide in solution, it is expected to have a shorter retention time during SCX chromatography compared to the unmodified peptide 30. Our criteria in identifying phosphorylations were: 1) a series of at least 3 y or b ions with an appropriate +80 dalton shift for the addition of a phosphate group, and 2) ions indicating a neutral loss of 98 Da from the precursor ion
To identify candidate phosphorylation sites we used the Sequest algorithm to search for additions of 80 Da at serine, threonine, and tyrosine residues 59. We anticipated that the in vivo phosphorylation would be incomplete (< 50%), and therefore we expected to identify the corresponding unmodified peptide as well. Once candidate sites were identified, we manually inspected the data for evidence of neutral loss events indicating phosphorylation. In this way, we identified two phosphorylation sites in Prt1 (Supplemental Figure 1). Phosphorylation of a high stringency CK2 consensus site at S22 (CK2 consensus sequence: PVDDIDFpSDLEEQYK) was identified in two independent trypsin and chymotrypsin experiments (http://scansite.mit.edu) (Supplemental Data). Phosphorylation of a low stringency CK2 consensus site at T707 (CK2 consensus sequence: DASSDDFTpTIEEIVEE) was identified following trypsin digestion, but not using chymotrypsin (Supplemental Data). The corresponding unmodified versions of these peptides were also identified (data not shown). As predicted, analysis of the SCX fractions in the MudPIT runs showed the phosphorylated peptides eluted first followed by the unmodified peptides (data not shown). Additionally, a single Tif5 phosphopeptide was detected that contained a phosphorylated T191 (SQNAPSDGTGSSpTPQHHDEDEDELSR). However, the site does not fit the CK2 consensus sequence. To our surprise, we did not detect phosphopeptides from Nip1.
Our results using metabolic labeling and mass spectrometry illustrate the need for a multifaceted approach to comprehensively identify phosphorylations. 32P-metabolic labeling revealed that Nip1 is phosphorylated. Mass spectrometry analysis of eIF3 complexes identified specific sites of phosphorylation in Prt1 and Tif5, but not Nip1.
Mass spectrometry Identification of In Vivo Phosphorylation of Nip1 at S98, S99, and S103 using IMAC phosphopeptide enrichment
In considering our failure to identify Nip1 phosphopeptides in our direct mass spectrometry analysis, we speculated that either Nip1 phosphopeptides were not retained in either the 2-D LC’s SCX or RP chromatography steps or that the phosphopeptide ion signals were being repressed by co-eluting nonphosphorylated peptides. Therefore, we enriched for phosphorylated peptides from digested eIF3 complexes using an optimized immobilized metal affinity chromatography (IMAC) protocol 35. Purified eIF3 complexes were digested with trypsin, and the peptides were converted to methyl esters. Phosphorylated peptides were enriched by Fe(III)-charged IMAC prior to LC-MS/MS analysis. Using this approach and the criteria described earlier for validating the MS/MS spectra, we detectedmultiple Nip1 peptides that were phoshorylated at S98, S99, and S103 (Fig. 3 and Table 3). All three sites are predicted to be high stringency substrates for CK2 (CK2 consensus sequence: LKSSNYDpSpSDEEpSDEEDGKK) 60. S99 was phosphorylated in all the Nip1 phosphopeptides that we detected, suggesting constitutive phosphorylation, while the phosphorylation of S98 and S103 was variable (Table 1). The corresponding unmodified peptides were not detected in our LC-MS/MS analysis of unenriched samples. Thus the IMAC enrichment enabled us to analyze these particular peptides. However, the IMAC approach did not allow us to detect the singly modified peptides previously identified in this study for Prt1 and Tif5. This is a commonly cited limitation of the IMAC protocol in that it preferentially binds multiply phosphorylated peptides 61.
Casein Kinase 2 Phosphorylates Nip1, Prt1, and Tif5 In Vitro
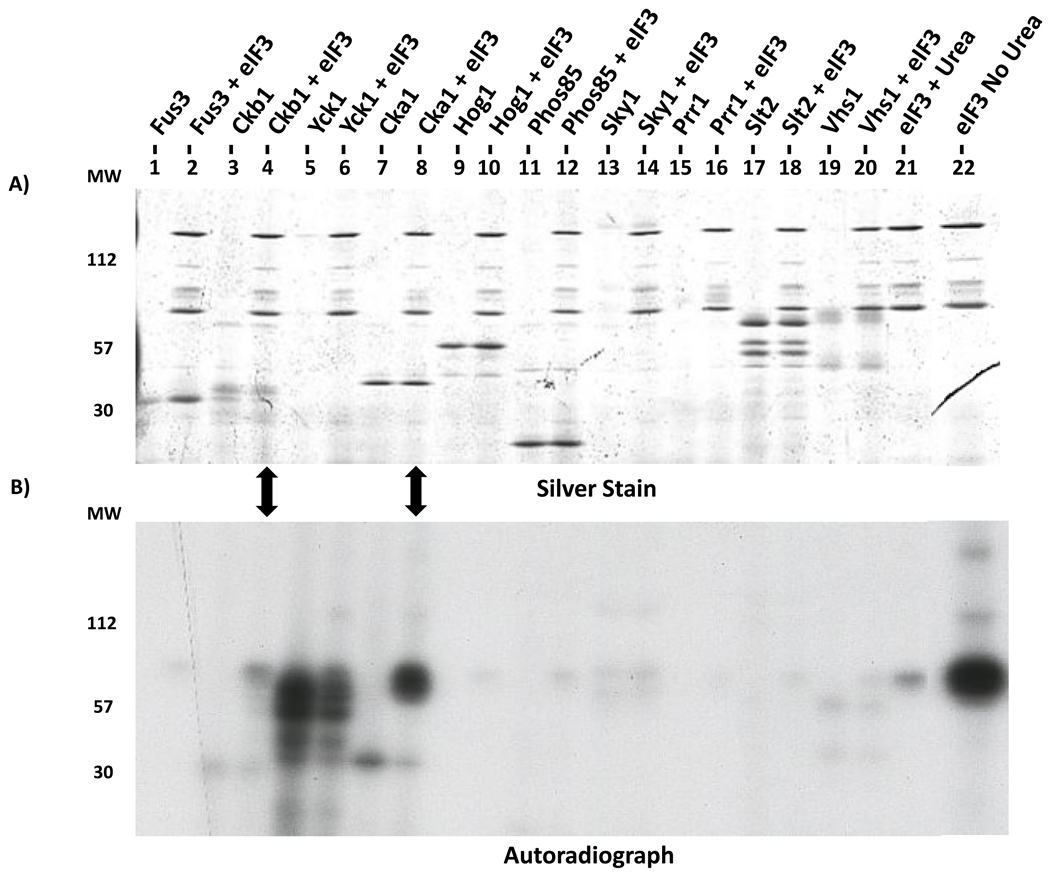
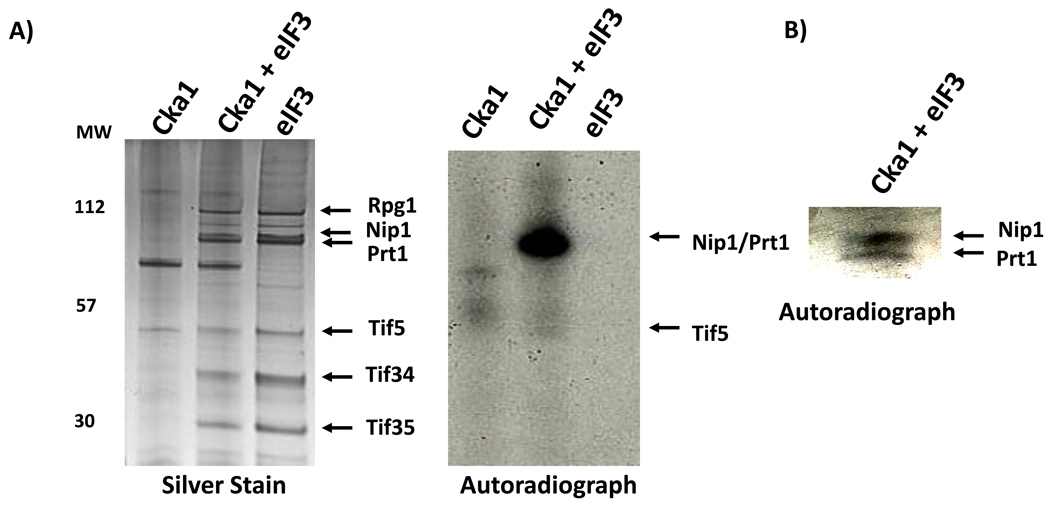
To test whether CK2 directly phosphorylates eIF3 components, we used an in vitro kinase assay. We purified GST-tagged Cka1 and Ckb1 as well as eight randomly selected serine/threonine yeast protein kinases with GST tags as controls. Isolation of the kinases was inferred from observation of SDS-PAGE bands with appropriate molecular sizes. To identify autophosphorylation, control reactions with no eIF3 substrate were performed for each kinase. The Cka1 catalytic subunit significantly phosphorylated eIF3 components above background (Fig. 4). The CK2 regulatory subunit Ckb1 phosphorylated eIF3 to a lesser extent. The kinase activity of the Ckb1 preparation suggests that catalytic components of the CK2 complex co-purified with this regulatory subunit. The results of the Yck1 assays are ambiguous due to apparent Yck1 auto-phosphorylation. None of the other kinases had any effect, although we cannot say for certain they are active as no autophosphorylation was detected. Yet it is clear that Cka1 has a striking ability to phosphorylate eIF3. Following this positive result, we confirmed the identity of the purified Cka1 using LC-MS/MS analysis (data not shown).
Figure 4. In vitro kinase screen identifies CK2 as an eIF3 kinase.
GST-tagged Cka1, Ckb1, and eight control kinases were purified and incubated with [γ-32P]ATP plus purified eIF3 treated with urea to reduce background kinase activity. Reaction products were separated on a 10% SDS-polyacrylamide gel and subjected to autoradiography. In the far right lane, purified eIF3 without urea treatment shows significant endogenous kinase activity. Arrows highlight phosphorylation of eIF3 components above background levels by the Cka1 catalytic subunit and to a lesser extent the Ckb1 regulatory subunit.
Analysis of the eIF3-Cka1 reaction products allowed us to determine which eIF3 components are phosphorylated by Cka1. SDS-PAGE showed the major phosphorylated product is in the size range of Nip1 and Prt1 (Fig. 5A). A gel with a lower percentage of acrylamide allowed separation of Nip1 and Prt1 and showed that both proteins are Cka1 substrates, with Nip1 the predominant target (Fig. 5B). Mass spectrometry analysis of these two bands confirmed that only Nip1 and Prt1 were present (data not shown). Finally, we also tested inhibitors of specific kinases. The addition of IC261, a CK1 inhibitor, had no affect on phosphorylation of Nip1. Conversely, addition of TBCA, a CK2 inhibitor, abolished the Nip1 signal (Fig. 6).
Figure 5. CK2 phosphorylates Nip1 and Prt1 in vitro.
SDS-PAGE separation was used to identify eIF3 components phosphorylated in in vitro kinase reactions with GST-tagged Cka1. (A) Separation on a 10% gel indicates that CK2 phosphorylates Nip1 and/or Prt1. (B) Separation on a 7.5 % gel resolves the most prominent phosphorylated band in (A) into a doublet and shows both Nip1 and Prt1 are phosphorylated by Cka1. Nip1 appears to be more prominently 32P-labeled compared to Prt1.
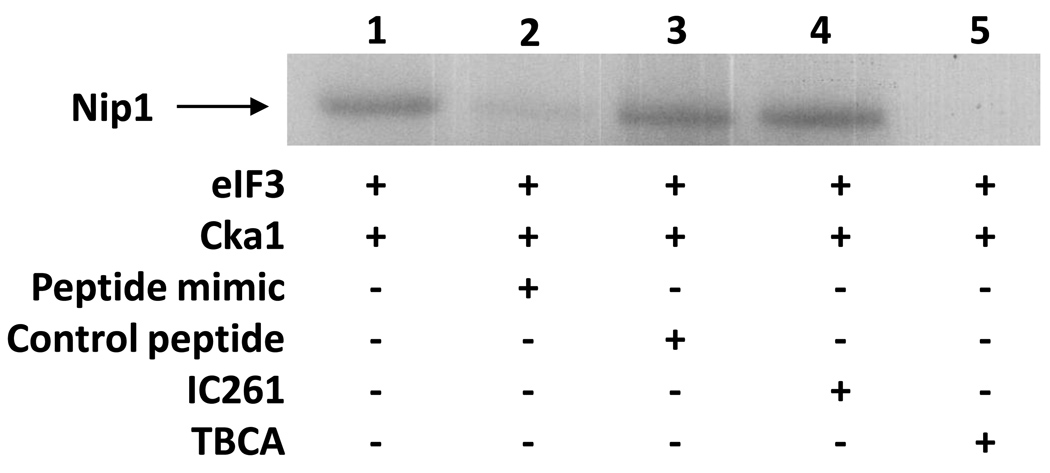
Figure 6. Synthetic Nip1 peptide inhibits the in vitro phosphorylation of Nip1.
A peptide was synthesized that mimics the unmodified tryptic peptide of Nip1 identified as phosphorylated by mass spectrometry. Separation on a 4–12% gradient gel for an extended time allows for better resolution of the Nip1-Prt1 region of the gel. The ability of this peptide mimic to inhibit in vitro phosphorylation is shown (lane 2). A control peptide added to the kinase assay indicates that this inhibition is specific to the Nip1 peptide (lane 3). Addition of a CK2 inhibitor, TBCA, abolishes the in vitro phosphorylation of Nip1 (lane 5). An unrelated kinase inhibitor, IC261, does not affect the in vitro phosphorylation of Nip1 (lane 4).
Phosphorylation of Nip1 at Specific Sites
To test whether CK2 targets Nip1 specifically at S98, S99, and S103, the sites identified by mass spectrometry, a synthetic peptide with a sequence identical to the tryptic fragment was generated and added to the in vitro kinase assays. Addition of the competitor peptide reduced phosphorylation of Nip1 (Fig. 6, compare lanes 1 and 2), indicating that these specific sites identified on Nip1 are phosphorylated by CK2 in vitro. The residual phosphorylation of Nip1 in the lane with the peptide mimic may result from either incomplete competition or CK2 phosphorylation of Nip1 at other site(s). The addition of a control peptide of unrelated sequence had no effect on Nip1 phosphorylation (Fig 6, lane 3).
Phosphorylation of Nip1 at Additional Sites
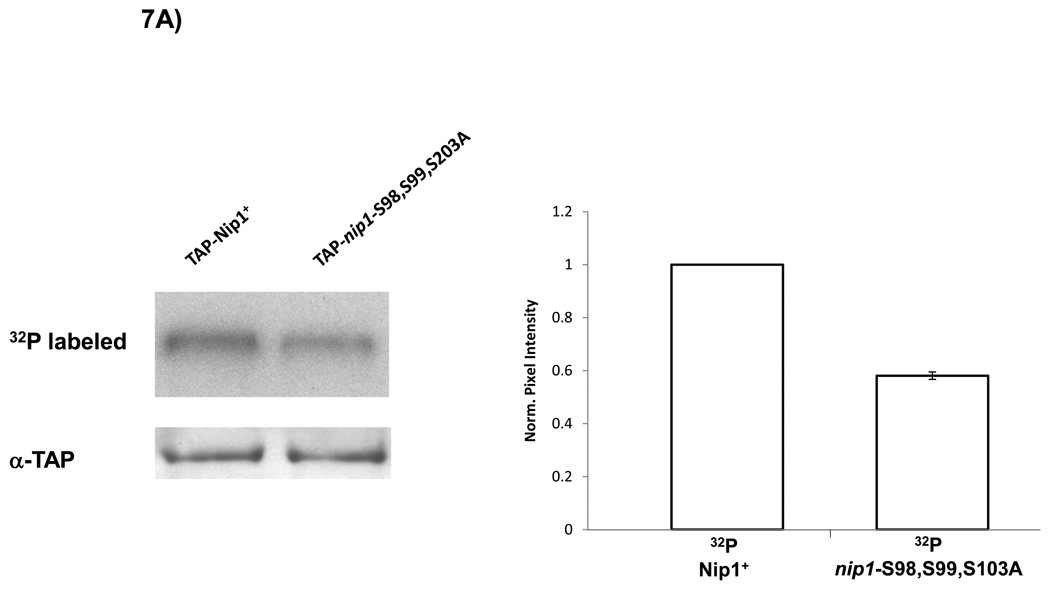
The incomplete inhibition of in vitro Nip1 phosphorylation by the peptide containing S98, S99, and S103 suggested that there may be additional phosphorylation sites on Nip1. To address this possibility, we examined in vivo phosphorylation of a mutant version of Nip1 lacking these three CK2 sites. Using a TAP-tagged Nip1 plasmid, we replaced S98, S99, and S103 with alanines. Strains with the wild-type and Nip1S98,99,101A mutant plasmids in a nip1 null background were metabolically labeled with 32P-orthophosphate. Following TAP purification of Nip1, autoradiography showed a considerable level of phosphorylation of Nip1S98,99,101A (Fig. 7A). The signal for the mutant strain represents a 41% reduction compared to the wild-type as determined by densitometry (Fig. 7A), consistent with the phosphorylation of other Nip1 sites.
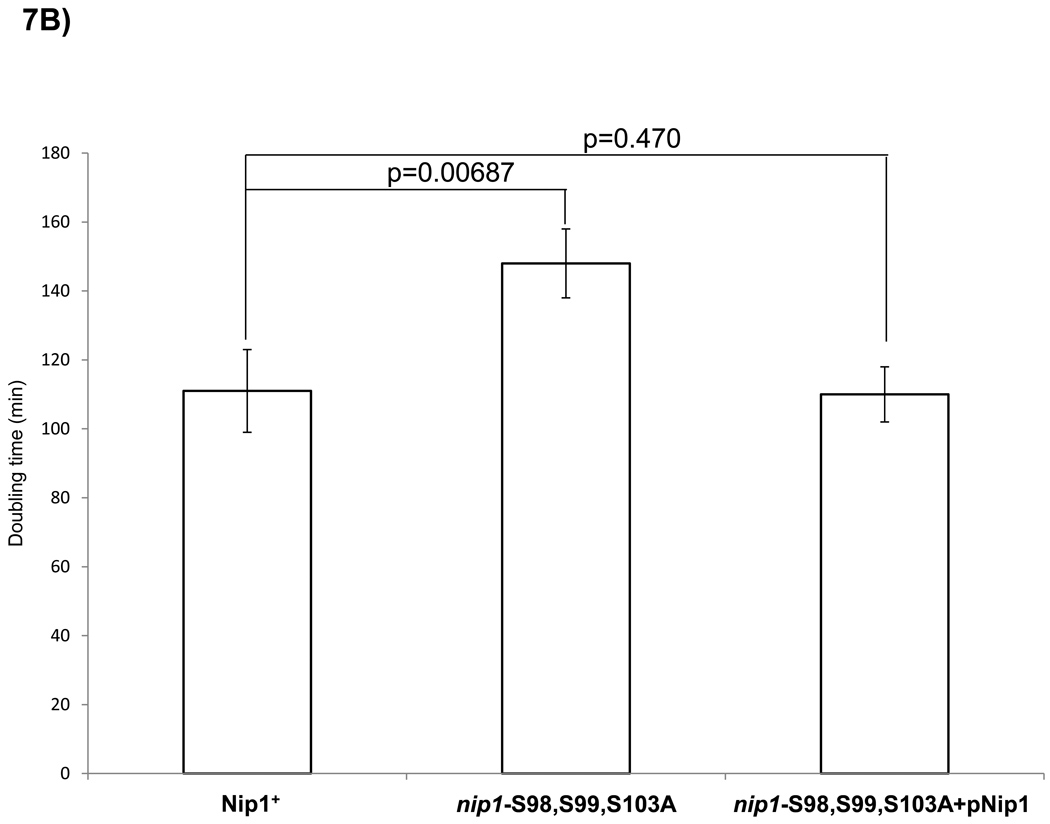
Figure 7. Mutation of S98, S99 and S103 in Nip1 reduces in vivo phosphorylation and growth rate.
nip1 null mutants containing plasmids expressing either wild-type Nip1 or a Nip1 mutant protein in which S98, S99 and S103 were replaced by alanines were examined. (A) SDS-PAGE separation of TAP-Nip1 following in vivo 32P metabolic labeling. The S98,S99, S103A mutation results in a 41% reduction of the mutant signal relative to wild-type based on densitometry using values normalized for the α-TAP loading control. As a control, western blotting with an α-TAP antibody shows equivalent amounts of TAP-Nip1 in each lane. (B) Doubling time for each strain (wild-type and mutant) was determined from the logarithmic growth phase of the cells in triplicate. Comparison of doubling times reveals that the strains harboring the mutation have a doubling time increased by 33%. A one-tailed t-test reveals a p-value between the wild-type and mutant doubling times of 0.00687. This defect is rescued with a wild-type copy of Nip1 in the mutant strain.
The Biological Significance of the Nip1 Phosphorylation at S98, S99, and S103
To test the functional importance of the S98, S99, and S103 phosphorylation sites, we closely examined a Nip1S98,99,101A mutant strain. We compared yeast strains with either wild-type Nip1 or Nip1S98,99,101A expressed from low-copy-number plasmids in a nip1 null background. Following a multitude of assays including drug selection, temperature sensitivity, polysome profiling, 35S incorporation, and selective growth conditions, the most striking difference observed between the mutant and wild-type strains was a marked increase in doubling time for the Nip1S98,99,101A mutant (Fig. 7B). The wild-type strain doubled every 111 ± 12 min, within the normal range for S. cerevisiae. However, the Nip1S98,99,101A mutant strain doubled every 148 min ± 10 min, a 33% increase (p-value = 0.00687). Complementing the mutant strain with a wild-type Nip1 plasmid results in a restoration of the doubling time to normal levels. (112 min ±8 min).
4 Discussion
In this study, we have used variouis mass spectrometry approaches to study the molecular composition of the highly conserved translation initiation factor eIF3 in S. cerevisiae. We used tandem affinity purification to isolate eIF3 complexes from tagged yeast strains and direct LC-MS/MS analysis of the complexes to identify core components of eIF3 as well as more loosely associated factors. To optimize our purification strategy, we affinity purified eIF3 based on three different tagged core components as well as the associated factor Tif5. The TAP-Rpg1 and TAP-Prt1 purifications resulted in the highest yield of eIF3 subunits. This finding is consistent with the previously described roles of Rpg1 and Prt1 in scaffolding and structural maintenance of the core eIF3 complex and in mediating interactions with Hcr1, eIF1, and the eIF2 ternary complex 3,4,16.
We confirmed that the yeast eIF3 complex is composed of five conserved core components: Rpg1 (eIF3a), Nip1 (eIF3c), Prt1 (eIF3b), Tif34 (eIF3i), and Tif35 (eIF3g). We also confirmed that other translation initiation factors co-purifying with eIF3. Similar to previous reports, we found Tif5 (eIF5), Sui1 (eIF1), Hcr1 (eIF3j), Sui3 (eIF2β) associated with eIF3 at substoichiometric levels 3,11,16. Importantly, we also identified novel interactions of eIF3 with Fun12 (eIF5B) and Pab1 (PabC). Like the other eIF3-associated factors, Fun12 and Pab1 were found at low levels and were absent from some eIF3 preparations. Fun12 has been shown to facilitate the joining of the 40S and 60S ribosomal subunits in the terminal step of translation inititation 62. Δfun12 null strains have a slow growth phenotype and altered polysome profiles suggesting a defect in translation initiation 63. Our finding of a possible interaction between Fun12 and eIF3 is consistent with the previous findings that both complexes play an active role in translation initiation. We postulate that the interaction of eIF3 with eIF5B links the formation of the pre-initiation complex with the binding of the 60S ribosome and the termination of translation initiation. Pab1 has also been shown to stimulate translation initiation 64,65. Pab1 facilitates the joining of the 5’ and 3’ ends of mRNA by binding the initiation scaffold protein eIF4G 66,67. Similar to Fun12, Δpab1 deleted strains have decreased levels of translation initiation 68. Our finding that PABP interacts with the eIF3 complex complements the previous findings and suggests a role in this interaction in the efficient formation of the 48S pre-initiation complex. Taken together, we speculate that the novel interactions Fun12 and Pab1with the eIF3 complex facilitate their optimal biological function. We believe that our ability to identified novel interactions of a previously well characterized complex stems from our approach combining gentle affinity purification based on multiple eIF3 components with a sensitive and comprehensive protein identification using mass spectrometry.
As part of our characterization of eIF3, we have also identified several sites of phosphorylation. LC/MS/MS analysis can yield very specific and detailed information about protein modification, but we found it necessary to use a variety of approaches. Using 32P metabolic labeling followed by TAP-Rpg1 purification of eIF3, we demonstrated in vivo phosphorylation of Nip1 and Prt1, two core eIF3 components. The findings are consistent with previous studies investigating the global phosphoproteome of S. cerevisiae 33,69,70. To identify specific sites of phosphorylation, we analyzed TAP-isolated eIF3 complexes directly by LC/MS/MS. In this way, we identified two phosphorylation sites in Prt1 (S22 and T707) and one in Tif5 (T191). To our surprise, we were unable to identify any phosphorylation sites in Nip1 using this method. We enriched for eIF3 phosphopeptides using IMAC and then analyzed the peptides using LC/MS/MS. This approach enabled us to identify three phosphorylation sites in Nip1 (S98, S99, and S103). Using the Scansite algorithm (http://scansite.mit.edu), Nip1 is predicted to contain 5 sites that are high stringency CK2 targets. Of these five, S98, S99, and S103 were identified in this study. There was no evidence found to support the remaining two sites S56 and S62. However, our mass spectrometry data showed evidence of additional phosphorylations. In a number of tandem spectra, the most intense fragment ion was due to a neutral loss of 98, 49, or 32.6 Da, indicating phosphorylation. However, insufficient fragmentation prevented us from confidently identifying the sequences. Additionally, Rpg1 is the only other member of the eIF3 complex predicted to contain a high stringency CK2 site at T513. No evidence was found to support this and the lack of a signal in the Rpg1 region of the autoradiography (Fig. 2, lane C) supports the conclusion that this particular CK2 site is not targeted for phosphorylation.
To comprehensively identify the eIF3 phosphorylation sites, complete sequence coverage of proteins by mass spectrometry will be required. However, current mass spectrometry protocols seldom achieve complete coverage. A fraction of the proteolytic peptide fragments are either too small or too large to be confidently analyzed by mass spectrometry. In addition, the physiochemical properties of phosphopeptides compound the problems of obtaining comprehensive coverage. Because of the negative charge on phosphate groups, the ionization efficiency of phosphopeptides in positive ion mode is reduced. Furthermore, because a phosphopeptide is typically at a lower abundance relative to the equivalent nonphosphorylated peptide, the more prevalent mass spectrometry signal of nonphosphorylated peptide can suppress the signal of the phosphopeptides 71–73.
We have found that casein kinase 2 (CK2) plays an important role in phosphorylating eIF3 components. Most of the phosphorylation sites we have identified fit the CK2 consensus sequence with high stringency: S98, S99, and S103 on Nip1 and S22 on Prt1. T707 on Prt1 fits the consensus with low stringency, while T191 on Tif 5 does not match. We found that CK2 subunits co-purify with eIF3 and that, in a reciprocal purification of TAP-Ckb1, eIF3 components co-purify with the CK2 complex. Our data are consistent with a previously described interaction between the CK2 kinase and eIF3 53. Using in vitro kinase assays, we found that CK2 phosphorylates both Nip1 and Prt1. Interestingly, the phosphorylated regions of Nip1 and Prt1 are predicted to stabilize both protein interactions within the eIF3 complex and interactions of eIF3 with other components of the multifactor pre-initiation complex 11.
Casein Kinase 2 is a conserved serine/threonine protein kinase that is ubiquitous in eukaryotes. Despite numerous studies, its regulation and physiological role is still poorly understood 74. Yeast strains with null alleles of either one of the two catalytic subunits (CKA1 or CKA2) are viable. However, deletion of both genes is lethal, showing that CK2 activity is essential for S. cerevisiae viability 75. Yeast strains with null alleles of either or both regulatory subunits (CKB1 or CKB2) are viable but are salt sensitive 76. A number of CK2 substrates have been reported, leading to the hypothesis that this kinase plays an essential role in several biological processes 57.
Several other components of the yeast translation initiation machinery are CK2 targets. Phosphorylation of the eIF2-α subunit of the eIF2 translation initiation complex is required for its optimal function but is not essential for viability 77. The yeast eIF4E cap binding protein and the 4E-BP protein Caf20/p20 are also phosphorylated in vitro by CK278.
To further our understanding of how phosphorylation might regulate translation initiation, we used mutational analysis to investigate the functional importance of the phosphorylated CK2 consensus sites in Nip1. Mutation of S98, S99 and S103 causes a 33% increase in the doubling times for the mutant strain. This observation suggests that the phosphorylation events occurring in the region of these sites S98, S99 and S103 are necessary for normal cell growth or division. In the absence of these three sites, in vivo phosphorylation of Nip1 was reduced by only 41%. This is consistent with our mass spectrometry analysis in which we were tantalized by several spectra that showed hallmarks of phosphorylation but did not contain enough fragment ions to provide sequence information. Therefore, we believe there are additional phosphorylation sites in the eIF3 complex yet to be identified. Additional genetic and biochemical experiments will be required to dissect the functional and mechanistic consequences of these phosphorylation events.
Collectively, our results provide a comprehensive characterization of the phosphorylation of eIF3 and its targeting by CK2. The work can serve as a blueprint for the discovery of kinases that phosphorylate components of other protein complexes in vivo. In addition, this approach can be employed in the discovery of enzymes responsible for other types of post-translational modification including ubiquination, acetylation, and methylation.
Supplementary Material
Table 2. Mass spectrometry identification of CK2 interactions with eIF3.
CK2 complexes were isolated from an S. cerevisiae strain expressing TAP-Ckb2 and an isogenic, untagged yeast strain as a negative control. MudPIT mass spectrometry analyses of the purified complexes were performed in triplicate. PAF values were used to approximate protein abundances as in Fig. 1.
| Yeast Protein |
Locus | TAP-Ckb2 | negative control |
|---|---|---|---|
| Cka1 | YIL035C | 6.3(1.2) | nda |
| Cka2 | YOR061C | 5.6(1.0) | nd |
| Ckb2 | YOR029C | 5.5(1.0) | nd |
| Ckb1 | YGL019W | 3.6(0.66) | nd |
| Tif34 | YMR146C | 2.1(0.38) | nd |
| Rpg1 | YBR079C | 2.1(0.39) | nd |
| Tif35 | YDR429C | 1.9(0.34) | nd |
| Nip1 | YMR309C | 1.5(0.27) | nd |
| Prt1 | YOR361C | 1.5(0.28) | nd |
| Tif5 | YBR041W | nd | nd |
nd = none detected
Acknowledgements
We are grateful to Tracey Fleischer and Vince Gerbasi for experimental advice throughout this work and for discussions and comments during the preparation of the manuscript. We thank Elizabeth M. Link for comments during the preparation of this manuscript. The project was supported by NIH grant GM64779. D.W.P. was supported by a NRSA GM073549-02. A.R.F is supported by NIH training grant T32 CA009385. C.M.W. and J.L.J. were supported by NIH grants GM64779 and HL68744. A.J.L. was supported by NIH grants GM64779, HL68744, ES11993, and CA098131. In addition, A.J.L. was supported in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400079C/N01-AI-40079.
Abbreviations
- met-tRNAi
methionyl initiator tRNA
- eIF3
eukaryotic Initiation Factor 3
- eIFs
eukaryotic Initiation Factors
- TAP
Tandem Affinity Purification
- FSC
fused silica capillary
- MudPIT
Multidimensional Protein Identification Technology
- CK2
Casein Kinase 2
- IMAC
Immobilized Metal Affinity Chromatography
References
- 1.Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 2.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano K, Phan L, Valasek L, Schoenfeld LW, Shalev A, Clayton J, Nielsen K, Donahue TF, Hinnebusch AG. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harb Symp Quant Biol. 2001;66:403–415. doi: 10.1101/sqb.2001.66.403. [DOI] [PubMed] [Google Scholar]
- 5.Valasek L, Nielsen KH, Hinnebusch AG. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. Embo J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. Embo J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. Embo J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh CR, Curtis C, Yamamoto Y, Hall NS, Kruse DS, He H, Hannig EM, Asano K. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol Cell Biol. 2005;25:5480–5491. doi: 10.1128/MCB.25.13.5480-5491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev. 2005;122:865–876. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch AG, Asano K, Olsen DS, Phan L, Nielsen KH, Valasek L. Study of translational control of eukaryotic gene expression using yeast. Ann N Y Acad Sci. 2004;1038:60–74. doi: 10.1196/annals.1315.012. [DOI] [PubMed] [Google Scholar]
- 12.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 13.Das S, Maitra U. Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the beta subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol Cell Biol. 2000;20:3942–3950. doi: 10.1128/mcb.20.11.3942-3950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Maitra U. Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol. 2001;70:207–231. doi: 10.1016/s0079-6603(01)70018-9. [DOI] [PubMed] [Google Scholar]
- 15.Browning KS, Gallie DR, Hershey JW, Hinnebusch AG, Maitra U, Merrick WC, Norbury C. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci. 2001;26:284. doi: 10.1016/s0968-0004(01)01825-4. [DOI] [PubMed] [Google Scholar]
- 16.Valasek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. Embo J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuchalova L, Kouba T, Herrmannova A, Danyi I, Chiu WL, Valasek L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol. 2010;30:4671–4686. doi: 10.1128/MCB.00430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 20.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 21.Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 22.Sudhakar A, Ramachandran A, Ghosh S, Hasnain SE, Kaufman RJ, Ramaiah KV. Phosphorylation of serine 51 in initiation factor 2 alpha (eIF2 alpha) promotes complex formation between eIF2 alpha(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry. 2000;39:12929–12938. doi: 10.1021/bi0008682. [DOI] [PubMed] [Google Scholar]
- 23.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 25.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. Embo J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheper GC, Morrice NA, Kleijn M, Proud CG. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol Cell Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 28.Farley AR, Link AJ. Identification and quantification of protein posttranslational modifications. Methods Enzymol. 2009;463:725–763. doi: 10.1016/S0076-6879(09)63040-8. [DOI] [PubMed] [Google Scholar]
- 29.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 30.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 32.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 33.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 34.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brock A, Peters EC. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 35.Moser K, White FM. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. J Proteome Res. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- 36.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishii TM, Zerr P, Xia XM, Bond CT, Maylie J, Adelman JP. Site-directed mutagenesis. Methods Enzymol. 1998;293:53–71. doi: 10.1016/s0076-6879(98)93007-5. [DOI] [PubMed] [Google Scholar]
- 38.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 39.Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Link AJ, Fleischer TC, Weaver CM, Gerbasi VR, Jennings JL. Purifying protein complexes for mass spectrometry: applications to protein translation. Methods. 2005;35:274–290. doi: 10.1016/j.ymeth.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Powell DW, Rane MJ, Joughin BA, Kalmukova R, Hong JH, Tidor B, Dean WL, Pierce WM, Klein JB, Yaffe MB, McLeish KR. Proteomic identification of 14-3-3zeta as a mitogen-activated protein kinase-activated protein kinase 2 substrate: role in dimer formation and ligand binding. Mol Cell Biol. 2003;23:5376–5387. doi: 10.1128/MCB.23.15.5376-5387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 43.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the Eukaryotic Transcription Machinery: Identification of Proteins Associated with Components of Yeast TFIID by Multidimensional Mass Spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 45.McAfee KJ, Duncan DT, Assink M, Link AJ. Analyzing proteomes and protein function using graphical comparative analysis of tandem mass spectrometry results. Mol Cell Proteomics. 2006;5:1497–1513. doi: 10.1074/mcp.T500027-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 47.Maiti T, Bandyopadhyay A, Maitra U. Casein kinase II phosphorylates translation initiation factor 5 (eIF5) in Saccharomyces cerevisiae. Yeast. 2003;20:97–108. doi: 10.1002/yea.937. [DOI] [PubMed] [Google Scholar]
- 48.Sotanaphun U, Phattanawasin P, Sriphong L. Application of Scion image software to the simultaneous determination of curcuminoids in turmeric (Curcuma longa) Phytochem Anal. 2009;20:19–23. doi: 10.1002/pca.1086. [DOI] [PubMed] [Google Scholar]
- 49.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 50.Knippschild U, Milne DM, Campbell LE, DeMaggio AJ, Christenson E, Hoekstra MF, Meek DW. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene. 1997;15:1727–1736. doi: 10.1038/sj.onc.1201541. [DOI] [PubMed] [Google Scholar]
- 51.Pagano MA, Poletto G, Di Maira G, Cozza G, Ruzzene M, Sarno S, Bain J, Elliott M, Moro S, Zagotto G, Meggio F, Pinna LA. Tetrabromocinnamic acid (TBCA) and related compounds represent a new class of specific protein kinase CK2 inhibitors. Chembiochem. 2007;8:129–139. doi: 10.1002/cbic.200600293. [DOI] [PubMed] [Google Scholar]
- 52.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 53.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 54.Sanders SL, Garbett KA, Weil PA. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol. 2002;22:6000–6013. doi: 10.1128/MCB.22.16.6000-6013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ackermann K, Waxmann A, Glover CV, Pyerin W. Genes targeted by protein kinase CK2: a genome-wide expression array analysis in yeast. Mol Cell Biochem. 2001;227:59–66. [PubMed] [Google Scholar]
- 57.Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 58.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR., 3rd Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates JR, 3rd, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 60.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 62.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 63.Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 64.Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. Embo J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell. 2002;94:205–216. doi: 10.1016/s0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 67.Tarun SZ, Jr., Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. Embo J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 68.Sachs A, Davis R. The poly(A)-binding protein is required for translation initiation and poly(A) tail shortening. Mol Biol Rep. 1990;14:73. doi: 10.1007/BF00360421. [DOI] [PubMed] [Google Scholar]
- 69.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang EJ, Archambault V, McLachlin DT, Krutchinsky AN, Chait BT. Analysis of protein phosphorylation by hypothesis-driven multiple-stage mass spectrometry. Anal Chem. 2004;76:4472–4483. doi: 10.1021/ac049637h. [DOI] [PubMed] [Google Scholar]
- 72.Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J Am Soc Mass Spectrom. 2003;14:971–979. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- 73.Zhen Y, Xu N, Richardson B, Becklin R, Savage JR, Blake K, Peltier JM. Development of an LC-MALDI method for the analysis of protein complexes. J Am Soc Mass Spectrom. 2004;15:803–822. doi: 10.1016/j.jasms.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Glover CV., 3rd On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1998;59:95–133. doi: 10.1016/s0079-6603(08)61030-2. [DOI] [PubMed] [Google Scholar]
- 75.Padmanabha R, Chen-Wu JL, Hanna DE, Glover CV. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bidwai AP, Reed JC, Glover CV. Cloning and disruption of CKB1, the gene encoding the 38-kDa beta subunit of Saccharomyces cerevisiae casein kinase II (CKII). Deletion of CKII regulatory subunits elicits a salt-sensitive phenotype. J Biol Chem. 1995;270:10395–10404. doi: 10.1074/jbc.270.18.10395. [DOI] [PubMed] [Google Scholar]
- 77.Feng L, Yoon H, Donahue TF. Casein kinase II mediates multiple phosphorylation of Saccharomyces cerevisiae eIF-2 alpha (encoded by SUI2), which is required for optimal eIF-2 function in S. cerevisiae. Mol Cell Biol. 1994;14:5139–5153. doi: 10.1128/mcb.14.8.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zanchin NI, McCarthy JE. Characterization of the in vivo phosphorylation sites of the mRNA.cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J Biol Chem. 1995;270:26505–26510. doi: 10.1074/jbc.270.44.26505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.