Abstract
Background and methods
A new amphiphilic comb-shaped copolymer (SP) was synthesized by conjugating poly(styrene-co-maleic anhydride) with low molecular weight polyethyleneimine for gene delivery. Fourier transform infrared spectrum, 1H nuclear magnetic resonance, and gel permeation chromatography were used to characterize the graft copolymer.
Results
The buffering capability of SP was similar to that of polyethyleneimine within the endosomal pH range. The copolymer could condense DNA effectively to form complexes with a positive charge (13–30 mV) and a small particle size (130–200 nm) at N/P ratios between 5 and 20, and protect DNA from degradation by DNase I. In addition, SP showed much lower cytotoxicity than polyethyleneimine 25,000. Importantly, the gene transfection activity and cellular uptake of SP-DNA complexes were all markedly higher than that of complexes of polyethyleneimine 25,000 and DNA in MCF-7 and MCF-7/ADR cell lines.
Conclusion
This work highlights the promise of SP as a safe and efficient synthetic vector for DNA delivery.
Keywords: poly(styrene-co-maleic anhydride), polyethylenimine, DNA, gene delivery
Introduction
Gene therapy has enormous clinical potential for treating human disease, particularly cancer, but still lacks safe and efficient delivery systems. Viral vectors are very effective for achieving highly efficient gene delivery and expression,1 but the drawbacks associated with viral-based carriers in terms of safety, immunogenicity, and high cost have encouraged researchers to focus on nonviral gene delivery systems, eg, cationic polymers and cationic lipids, with attractive biological traits such as low immunogenicity, simplicity of preparation, high gene encapsulation capability, and excellent biocompatibility.2–4 Among the nonviral vectors, cationic polymers have shown particular promise for gene delivery due to their unique properties, including stability, a size range suitable for passive tumor targeting, high water solubility, and easy modification to increase specificity.5–8
Polyethyleneimine (PEI) has been shown to be effective in gene delivery due to its excellent ability to condense DNA, which facilitates endocytosis, as well as its “proton sponge effect”, which can improve escape of DNA from endosomes. Many studies have shown that the molecular weight of PEI is the most important determinant of gene transfection efficiency and cytotoxicity.9,10 High molecular weight PEI (eg, 25 kDa) shows high transfection efficiency, but also has significant cytotoxicity. In contrast, low molecular weight PEI (eg, ≤2 kDa) has been shown to have low toxicity, but cannot condense DNA effectively and has very poor transfection activity. To overcome this problem, one rational strategy is to combine low molecular weight PEI using stimuli-responsive or biodegradable linkages to form combined PEI of a suitable molecular weight. The transfection efficiency of these crosslinked PEI is considerably high, with cytotoxicity being much lower than that of PEI 25,000.11–14 Other strategies have included using linear chains to link PEI together, such as grafting PEI to a linear polymer backbone to give comb-shaped copolymers,15–17 conjugating low molecular weight PEI to a biocompatible polymer core to form star-shaped copolymers,18,19 and constructing supramolecular polymers with grafting of low molecular weight PEI.20,21
Poly(styrene-co-maleic anhydride) (SMA), an amphiphilic macromolecule, is an intermediate polymer that can be conveniently converted to a functional polymer by reacting with a bioactive agent containing amino or hydroxyl groups via a ring-opening reaction between the active agent and the succinic anhydride unit. SMA has been shown to be biologically safe and to confer immunopotentiation in host animals by activation of macrophages, T cells, and natural killer cells, and induction of interferons.22,23 SMA has been used for the preparation of polymeric prodrugs with controlled chemical and diffusion properties, which can enhance the solubility and stability of drugs, prolong their half-life in plasma, and improve their pharmacokinetic and pharmacodynamic profiles, thereby enhancing the therapeutic effects.24 The most successful example is attachment of the protein drug, neocarzinostatin, to SMA. The half-life of neocarzinostatin when attached to SMA was prolonged by 10-fold in blood, which led to tumor-selective drug delivery and retention (tumor/blood ratio ≥ 2500); antitumor activity became more pronounced, but the toxicity was reduced to one quarter of that of the parent compound. Clinical testing of this compound in the anticancer field has been extremely promising, with a success rate of 70%–90% in treated patients.25–27 SMA-conjugated YIGSR (a core sequence located in the β1 chain of laminin) also showed a strong antimetastatic effect (approximately 50-fold greater than that of native YIGSR).28 SMA-doxorubicin29 and SMA-pirarubicin30 micelles, formed by hydrophobic interaction between the styrene portion of SMA and the drug, showed much higher tumor accumulation after intravenous administration than that of the free drug. All these studies highlight the potential of SMA in drug delivery, but the use of SMA in gene delivery has not been well investigated.
In the present work, we synthesized a new amphiphilic comb-shaped copolymer (SP) by conjugating SMA with low molecular weight PEI (PEI 800) for gene delivery. Grafted PEI residues could form stable complexes with DNA due to their strong positive charges, while SMA could reduce the cytotoxicity of the compound by controlling the charge density. The DNA condensation ability, DNA protection ability, and cytotoxicity were determined. The in vitro transfection efficiency, cellular uptake, and subcellular location were investigated in MCF-7 and MCF-7/ADR cell lines in the presence of serum. It was expected that this new amphiphilic copolymer would show high transfection efficiency with low cytotoxicity.
Materials and methods
Materials
Branched PEI with average molecular weights of 800 Da and 25 kDa were purchased from Sigma-Aldrich (St Louis, MO). SMA with a molar styrene to maleic anhydride ratio of 1:1 in the backbone (molecular weight 5500 Da) was gifted by Sartomer Company Inc (Guangzhou, China). DNase I was supplied by Beyotime Biotechnology (Jiangsu, China), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), ethidium bromide, and trypan blue were purchased from Sigma-Aldrich. Trypsin-ethylenediamine tetra-acetic acid and agarose were obtained from Gibco-BRL (Burlington, ON, Canada). YOYO-1, Hoechst 33342, and a LysoTracker® red kit were purchased from Molecular Probes (Eugene, OR). Heparin was obtained from Aladdin (Shanghai, China). All other reagents were of analytical grade.
The reporter plasmid, enhanced green fluorescent protein (pEGFP-N1, 4.7 kb), purchased from Clontech (Palo Alto, CA), was amplified in a DH5α strain of Escherichia coli and purified using an EndFree plasmid mega kit (Qiagen GmbH, Hilden, Germany). Purity was confirmed by ultraviolet spectrophotometry (A260/A280), and the concentration was determined by measurement of ultraviolet absorbance at 260 nm using an ultraviolet-visible spectrophotometer (UV-2450, Shimadzu, Tokyo, Japan).
Cell culture
MCF-7 (human breast cancer) and MCF-7/ADR (adriamycin- resistant human breast cancer) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 (Gibco) containing 10% fetal bovine serum (Gibco), 100 U/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate at 37°C and 5% CO2 in a humidified incubator. Drug resistance of the MCF-7/ADRcells was maintained by addition of adriamycin 1 μg/mL to the medium.
Synthesis and characterization of SMA-PEI 800 (SP) copolymer
SP was synthesized by reacting the primary amine in PEI with the anhydride groups in the SMA backbone according to a method previously described,31 with minor modification. Briefly, PEI 800 (792 mg, 0.99 mmol) dissolved in 20 mL of dimethyl sulfoxide was put into a round-bottomed flask equipped with a mechanical stirrer, followed by dropwise addition of SMA (100 mg, 0.495 mmol of maleic anhydride) in 20 mL of dimethyl sulfoxide. The reaction was carried out at room temperature for 12 hours with stirring. The reaction mixture was then dialyzed using cellulose dialysis membranes (molecular weight cutoff 10 kDa, Spectrum Co) against deionized water for 3 days to remove the dimethyl sulfoxide and unreacted PEI 800. Water was then removed by rotary evaporation, and the polymer was dried under vacuum overnight.
The Fourier transform infrared spectra of the polymers, recorded in pressed KBr pellets on a Fourier transform infrared spectrometer (Equinox 55, Bruker Optics, Billerica, MA), were done to demonstrate successful synthesis of SP. The structure of the polymers was confirmed by 1H nuclear magnetic resonance (NMR) spectra recorded on a Varian Mercury Plus-400 NMR spectrometer (Varian, Palo Alto, CA) operated at 400 MHz. The molecular weight and polydispersity of SP were measured using a gel permeation chromatography system equipped with a separation module (Waters 2695, Milford, MA), ultrahydrogel columns (250 PKGD and 1000 PKGD, 30°C) and a refractive index detector (Waters 2414).
The buffer capacities of SP, PEI 800, and PEI 25,000 were evaluated by acid-base titration assay over pH values ranging from 10.0 to 2.0. An NaCl (150 mM) solvent was used as the control.32 Polymers, equivalent to 0.25 mmol of protonable amine groups, were dissolved in 5 mL of 150 mM NaCl solution. The pH of the polymer solution was adjusted to 10.0 by adding 0.1 M NaOH. The solution was then titrated against 0.1 M HCl, and the pH of the solution was measured using an acidimeter (Sartorius, Goettingen, Germany).
Preparation and characterization of polymer-DNA complexes
All polymer-DNA complexes were freshly prepared before use by adding different concentrations of polymer solutions into equal volumes of pEGFP-N1 to obtain the desired polymer nitrogen to DNA phosphate (N/P) ratio, followed by vortexing for 30 seconds and incubating at room temperature for 30 minutes.
An agarose gel retardation assay was used to determine the DNA binding abilities of the polymers. SP-DNA complexes, PEI 800-DNA complexes, and PEI 25,000-DNA complexes containing 1 μg of DNA at various N/P ratios ranging from 0.5 to 20 were prepared by the same process as described above. The complexes, premixed with appropriate amounts of 6 × gel-loading buffer (Generay, Shanghai, China), were loaded on 0.8% (w/v) agarose gel containing ethidium bromide (0.5 μg/mL of gel) in Tris-acetate-ethylenediamine tetra- acetic acid and subjected to electrophoresis for 45 minutes at 80 V. The location of DNA in the gel was visualized using an ultraviolet illuminator and photographed using a bioimage system (UVP, Upland, CA).
The particle size and zeta potential of the SP-DNA complexes were determined by dynamic light scattering measurement using a Zetasizer (Malvern Nano-ZS90, Worcestershire, UK). As control, PEI 800-DNA complexes and PEI 25,000-DNA complexes were also prepared and analyzed using the same procedure as for the SP-DNA complexes.
DNase I protection assay
Protection of the loaded DNA molecules against enzyme degradation by the polymers was determined by treating the complexes with DNase I (1 U/μg of DNA) for 30 minutes at 37°C. DNase I was inactivated by adding ethylenediamine tetra-acetic acid to a final concentration of 2.5 mM, followed by heating at 65°C for 10 minutes. The DNA molecules were then released from the complexes using heparin (100 U/μg of DNA) at 37°C for 30 minutes, and analyzed using 0.8% agarose gel electrophoresis.
In vitro cytotoxicity assay
The MTT method was used to evaluate the cytotoxicity of the polymers. The cells were seeded in 96-well plates at a density of 5 × 104 cells per well in 200 μL of RPMI 1640 medium and incubated overnight. The growth medium was then replaced with 200 μL fresh RPMI 1640 medium containing different concentrations of the polymers. After 48 hours of incubation, 20 μL of MTT solution (5 mg/mL) was added to each well and incubated for an additional 4 hours. After that, the medium was removed and 150 μL of dimethyl sulfoxide was added to dissolve the crystals formed by living cells. Absorbance was measured at 570 nm using a microplate reader (Infinite 2000, Tecan, Durham, NC). Cell viability was expressed as a percentage of the absorbance to that of the control experiment without polymers. The results were presented as the average values of three runs. The cytotoxicity of the complexes at different N/P ratios was also investigated as described above, except that the medium was replaced with 200 μL of fresh RPMI 1640 medium containing complexes with 0.8 μg/well of DNA at different N/P ratios.
In vitro transfection experiment
The transfection efficiency of the SP-DNA complexes was evaluated on MCF-7 and MCF-7/ADR cells using pEGFP-N1 as the model DNA. The cells were seeded into 24-well plates at a density of 1 × 105 cells per well in 500 μL of complete medium and incubated for 24 hours, yielding a cell density at about 80% confluence. The medium was then replaced with fresh growth medium containing SP-DNA complexes with 2 μg per well of DNA at different N/P ratios. The medium was changed again after 4 hours, and the cells were incubated for a further 48 hours. Untransfected cells were used as the negative control, and cells transfected with PEI 25,000-DNA complexes at an N/P ratio of 10 were used as the positive control. The EGFP-expressing cells were visualized using a fluorescence inversion microscope system (Olympus, Tokyo, Japan) and quantified using a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ). All transfection experiments were performed in triplicate.
Cellular uptake
MCF-7 and MCF-7/ADR cells were seeded into 24-well plates (1 × 105 cells/well) with 500 μL of complete medium and incubated further for 24 hours. pEGFP was fluorescently labeled with YOYO-1, with a ratio of one dye molecule to 300 bp and incubated for 30 minutes at room temperature in the dark. The polymers-YOYO-1-labeled DNA complexes were prepared as described above at their optimal N/P ratios and added to the cells at a DNA concentration of 2 μg/well. After 2 hours of incubation, the medium was removed, and the cells were washed twice with phosphate-buffered solution, collected, and resuspended in phosphate-buffered solution. The fluorescence was measured using the FACSCalibur system.
Subcellular localization
For the subcellular localization experiment, cells were seeded onto 10 mm2 glass coverslips placed in 24-well plates (5 × 104 cells/well) and incubated for 24 hours. pEGFP was fluorescently labeled with YOYO-1 as described above. SP-DNA complexes were added to the cells with 2 μg DNA per well and incubated for 1.5 hours, followed by addition of Hoechst 33342 (6 μg/mL, Invitrogen, Carlsbad, CA) and LysoTracker red (10 nmol/mL, Invitrogen) to the cells, and incubation at 37°C for a further 30 minutes. Extracellular fluorescence was quenched using 0.4% trypan blue for 2 minutes. After the medium was removed, the cells were washed twice with phosphate-buffered solution and fixed with 4% paraformaldehyde for 30 minutes in the dark. The cells were then mounted onto glass slides with 3 μL of MobiGlow (MoBiTec, Goettingen, Germany) and visualized using confocal microscopy (FluoView TM FV1000, Olympus).
Statistical analysis
Statistical analysis was performed using the Student’s t-test. The differences were considered to be statistically significant at P < 0.05, and highly significant at P < 0.01.
Results and discussion
Synthesis and characterization of SP
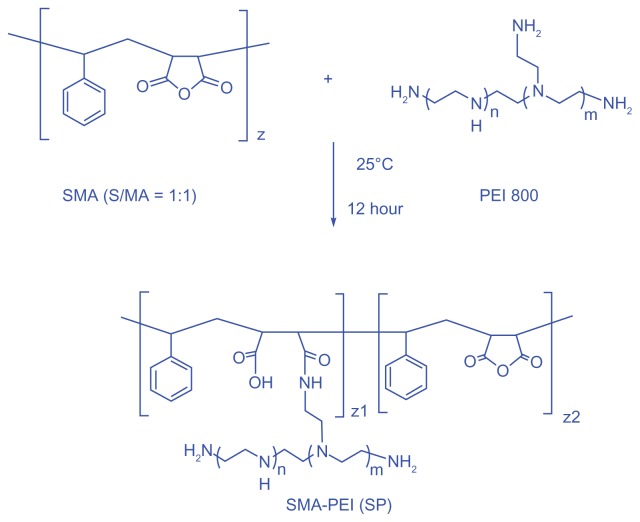
A new comb-shaped copolymer was synthesized by grafting PEI 800 onto SMA (Scheme 1), which involved the reaction of cyclic anhydride and primary amine to generate amido acid functionalities in the linking sites. An excess equivalent amount of PEI and dropwise addition of SMA was undertaken to avoid a possible cross-linking side reaction as a result of steric hindrance. Excess PEI was then removed by dialysis against deionized water for 3 days. The reaction mixture became cloudy upon addition of SMA and turned clear again during dialysis, which indicated that the reaction had indeed occurred, with no or negligible cross-linking.
Scheme 1.
Synthesis of comb-shaped copolymer SP. A two-fold excess of PEI 800 to SMA was used to avoid a possible cross-linking side reaction.
Abbreviations: SMA, poly(styrene-co-maleic anhydride); SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
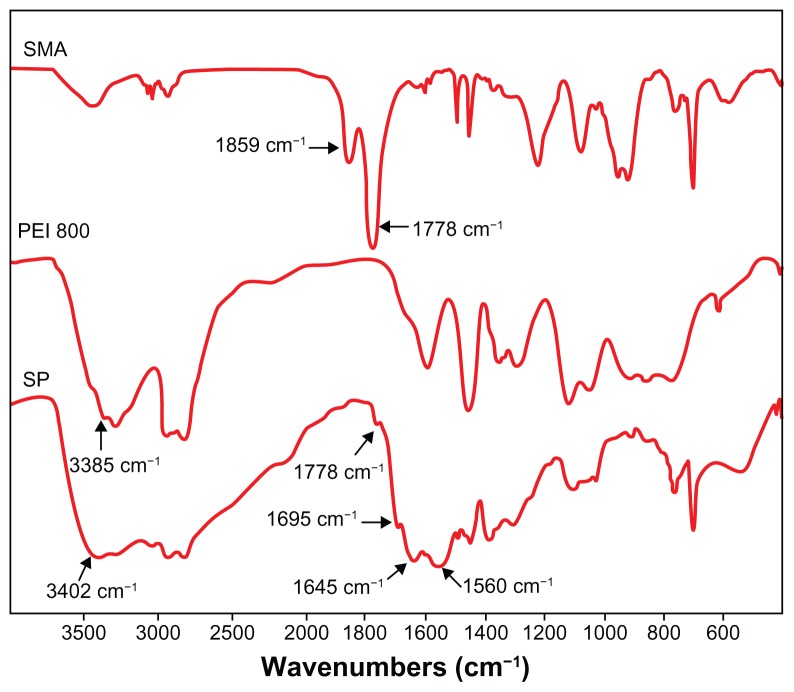
Fourier transform infrared spectra showed that the absorption peaks of anhydride carbonyl groups at 1859 and 1778 cm−1 disappeared almost completely (Figure 1). Instead, new peaks emerged in the spectrum of SP. A peak of 1695 cm−1 was attributed to the carboxyl group and the absorption of 1645 cm−1 (amide I) and 1560 cm−1 (amide II) corresponded to amide groups, indicating that an aminolysis reaction had taken place. A new strong peak at 3402 cm−1 assigned to the N-H stretch vibration of PEI demonstrated that PEI was successfully grafted to SMA.
Figure 1.
Fourier transform infrared spectra of SMA, PEI 800, and SP (KBr).
Abbreviations: SMA, poly(styrene-co-maleic anhydride); SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
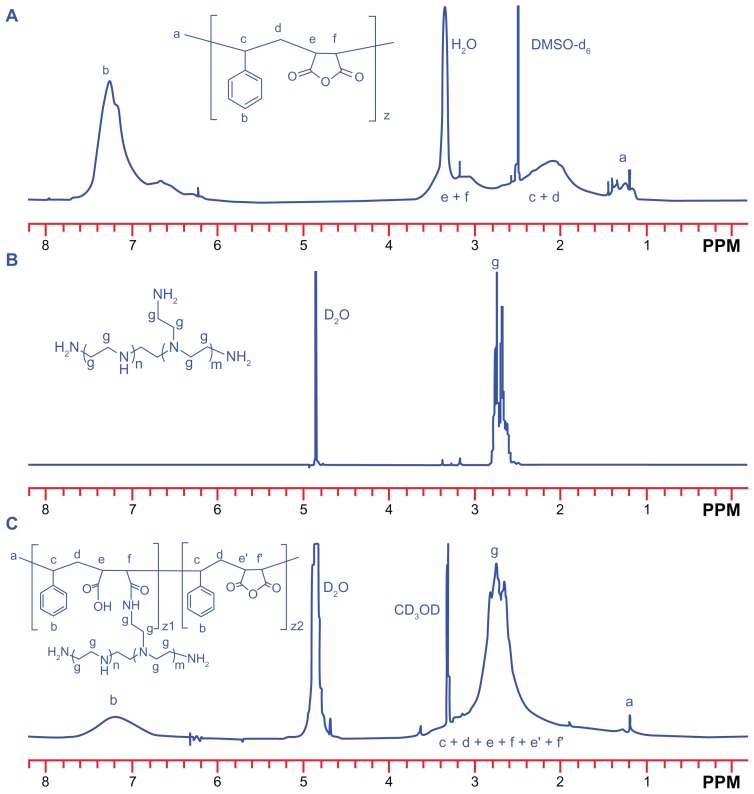
The structure of the SP copolymer was also confirmed by 1H NMR spectra (Figure 2). A broad bimodal peak at 1.8–3.6 ppm in SMA was attributable to the methylene and tertiary carbon of the main chain. The peak near 1.25 ppm was the signal from the methyl group from the terminal cumene residue in SMA. The signal at 7.0–7.6 ppm was the proton signal from the styrene residue. In the case of the SP conjugate, although the peak areas were inconsistent, their peak shifts were similar to those of SMA, except that the peaks of the methylene and tertiary carbons were partly overlapped with the characteristic resonances of the PEI block (2.4–3.1 ppm, –CH2CH2 NH–). These results were consistent with the above Fourier transform infrared measurements, and suggested successful synthesis of the SP copolymer.
Figure 2.
1H nuclear magnetic resonance (400 MHz) spectra of (A) SMA (DMSO-d6), (B) PEI 800 (D2O) and (C) SP (D2O–CD3OD, volume ratio of 1:1).
Abbreviations: SMA, poly(styrene-co-maleic anhydride); SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
The molecular weight and polydispersity of SP measured by gel permeation chromatography were 23.6 kDa and 2.17, respectively, which meant that about 83% of maleic anhydride was reacted with PEI according to the molecular weight of SMA (5500 Da) and PEI (800 Da). In addition, gel permeation chromatography of SP showed only one peak, which further verified the successful synthesis of copolymer and the absence of any cross-linked product.
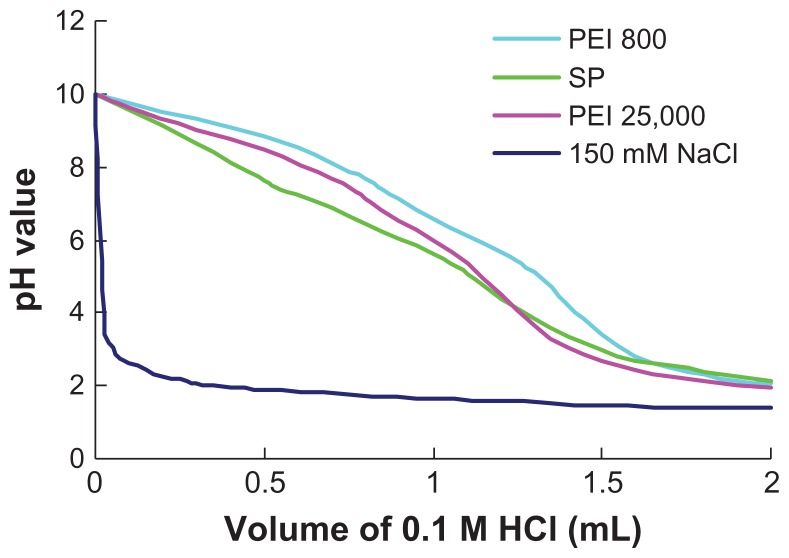
The buffer capacity of a cationic polymer gene vector is an important factor influencing the escape of nucleic acid from endosomes according to the “proton sponge” hypothesis. 33 In this work, the buffer capacities of polymers were determined by acid-base titration assay. As shown in Figure 3, the acid-base titration curve of SP was almost identical to that of PEI 25,000 and PEI 800. The high buffer capacity of SP could result from the high amino density of PEI 800 grafted to SMA. A slower decrease in pH was observed from the titration curve of SP when the volume of HCl was over 1 mL, which suggested that carboxylate ions on the backbone could also be protonated during titration and endow the polymer with high buffer capacity, except for amine protonation from PEI 800.
Figure 3.
Titration curves of PEI 800, SP, and PEI 25,000 with 0.1 M HCl, NaCl (150 mM) solvent was used as a control.
Abbreviations: SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
Characterization of SP-DNA complexes
DNA must be compacted into stable nanosized complexes for efficient delivery. A gel retardation assay was performed to investigate the binding behavior of SP with DNA at different N/P ratios from 0.5 to 20, PEI 800 and PEI 25,000 were used as controls. The intensity of migrating free DNA bands decreased gradually for all polymers, with an increasing charge ratio until DNA was completely retarded (Figure 4). At that or higher N/P ratios, the DNA was completely condensed into nanoparticles with a positive surface charge and could not migrate during agarose gel electrophoresis, and the ethidium bromide could not gain access to the DNA, so the DNA band could not be observed. The capacity of SP to condense DNA with complete retardation at an N/P ratio of 2 was comparable with that of PEI 25,000. However, PEI 800 showed a much weaker ability to condense DNA with complete retardation at an N/P ratio of 5. Further, PEI 800 formed rather loose complexes with DNA. These results indicated that modification of SMA by grafting PEI 800 to the main chain to form a PEI combined copolymer could ameliorate the DNA condensation ability, which could result from the extended conformation of SP and the high molecular weight PEI block of SP. Furthermore, phenyl groups, carboxyl groups, and amide linkages in the polymer might interact with DNA base pairs via hydrogen bonding and π-π stacking interactions, and improved the compacting capability of SP, except for the typical electrostatic interactions between the cationic amine and anionic phosphate groups of DNA.34
Figure 4.
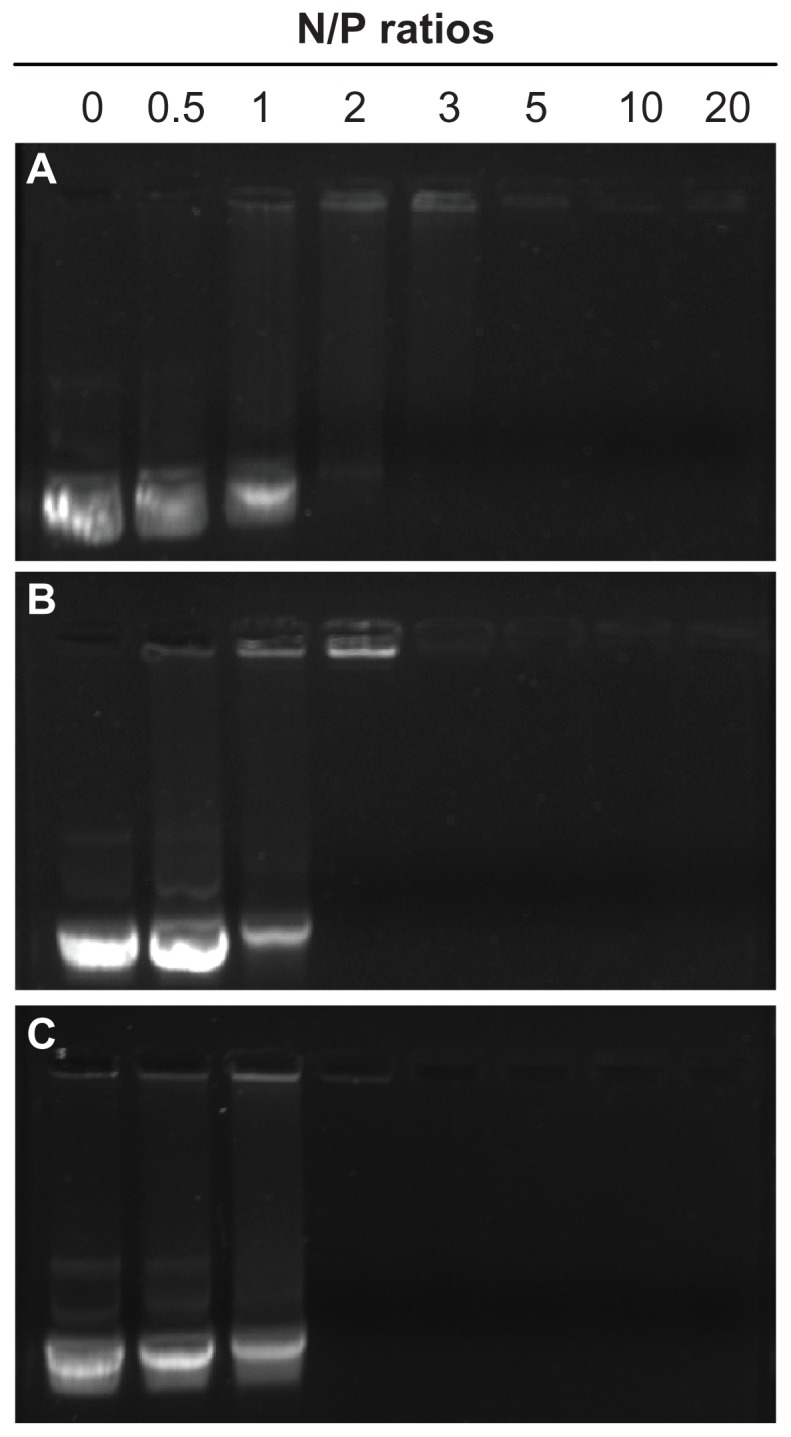
Agarose gel electrophoresis of SP-DNA complexes (B) in comparison with PEI 800-DNA complexes (A) and PEI 25,000-DNA complexes (C) at varying N/P ratios.
Abbreviations: SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
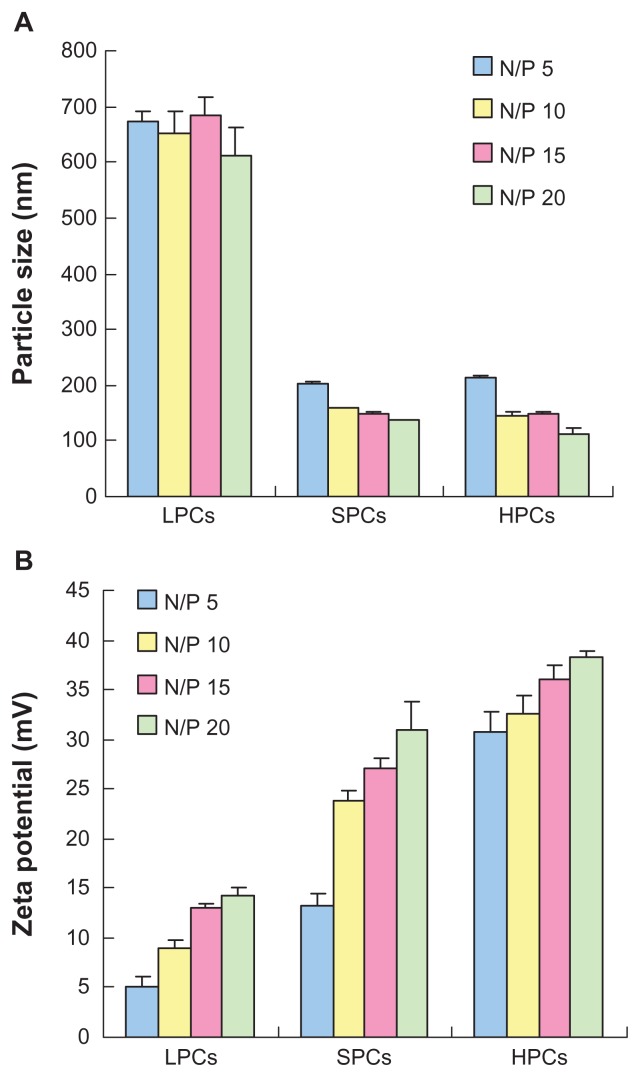
Figure 5A showed the particle sizes of complexes at various N/P ratios. PEI 800 formed loose complexes with a size of approximately 700 nm from an N/P ratio of 5 to an N/P ratio of 20. The trend of the sizes was in accordance with the results of the gel retardation experiments, indicating that PEI 800 could not compact DNA efficiently. In contrast, SP could effectively compact DNA into small nanoparticles with a particle size in the range of 135–200 nm, which was similar to that of complexes prepared using commercially available PEI 25,000. The zeta potentials of the polymer-DNA complexes were measured by dynamic light scattering. As shown in Figure 5B, even at an N/P ratio of 20, the zeta potential of PEI 800-DNA complexes remained below 15 mV. As expected, complexes of PEI 25,000 with DNA showed a high zeta potential of 30 mV to 40 mV at an N/P ratio of 5 to 20. In particular, SP-DNA complexes showed a lower surface charge (13–30 mV) compared with PEI 25,000-DNA complexes, which might be due to the presence of carboxyl groups in the backbone which could partly neutralize the positive charge. The moderate zeta potential of the SP-DNA complexes might be more beneficial for obtaining a better balance between cellular uptake and cytotoxicity because a positive surface charge of untargeted complexes is necessary for attachment to a negatively charged cell surface, which could lead to efficient intracellular trafficking.36 However, an excessively high charge density was related to cytotoxicity and resulted in aggregation of the complexes in a physiologic environment.
Figure 5.
Particle size (A) and zeta potential (B) of complexes at various N/P ratios as determined by dynamic light scattering.
Abbreviations: LPCs, polyethyleneimine 800-DNA complexes; SPCs, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride)-DNA complexes; HPCs, polyethyleneimine 25,000-DNA complexes.
DNase I protection assay
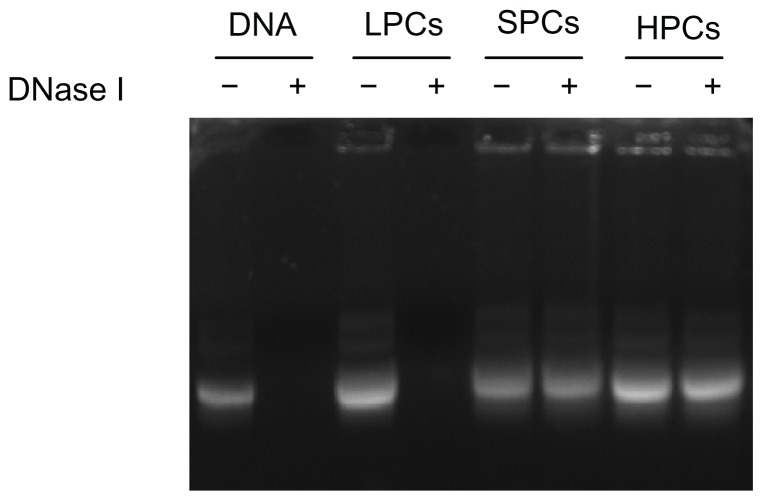
An efficient gene delivery system must be able to protect DNA from degradation by nucleases in serum and in the extracellular matrix.35 As shown in Figure 6, after incubation with DNase I, the naked DNA and the DNA in PEI 800-DNA complexes were all completely degraded, while the DNA in SP-DNA complexes or PEI 25,000-DNA complexes remained intact, indicating that both SP-DNA complexes and PEI 25,000-DNA complexes can effectively protect DNA from degradation by DNase I. Quantification of the intact DNA (ImageJ, NIH) revealed that 93.0% and 94.4% of the loaded DNA was recovered from the SP-DNA complexes and PEI 25,000-DNA complexes, respectively, indicating that the ability of SP to protect DNA from enzymatic cleavage is comparable with that of PEI 25,000.
Figure 6.
DNase I protection assay.
Note: DNA was released by adding heparin (100 U/μg DNA) to the complexes after treatment with DNase I (1 U/μg DNA).
Abbreviations: LPCs, polyethyleneimine 800-DNA complexes; SPCs, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride)-DNA complexes; HPCs, polyethyleneimine 25,000-DNA complexes.
In vitro cytotoxicity assay
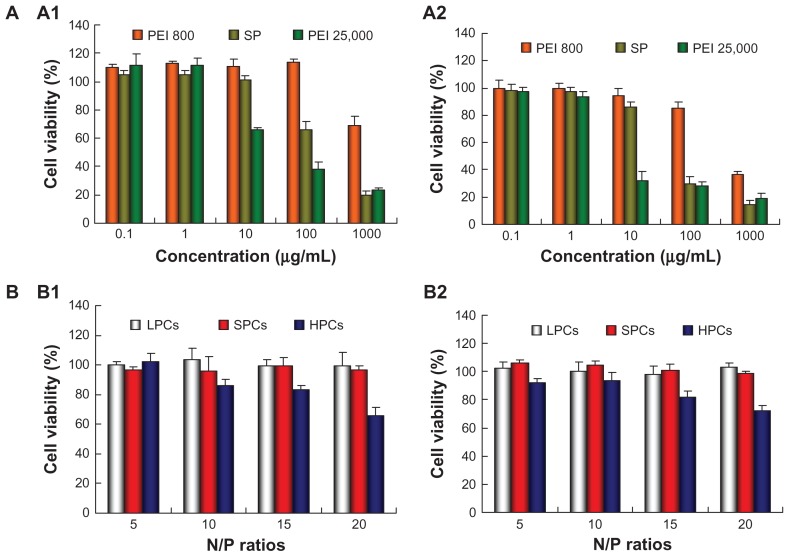
Cationic polymers are known to be potentially cytotoxic because they damage cell membranes via electrostatic interactions. Figure 7A showed the viability of MCF-7 and MCF-7/ADR cells after incubation for 48 hours with SP, PEI 25,000, and PEI 800 at different concentrations by the MTT assay. PEI 800 showed very low cytotoxicity with over 90% cell viability at concentrations ranging from 0.1 to 100 μg/mL. SP also showed good biocompatibility with 100% and 85% cell viability for MCF-7 and MCF-7/ADR cells, respectively, at 10 μg/mL. In contrast, PEI 25,000 showed higher cytotoxicity (65% and 30% cell viability for MCF-7 and MCF-7/ADR cells, respectively) than SP. With increasing polymer concentrations, cell viability decreased gradually, but the viability of cells exposed to SP was obviously higher than that of cells exposed to PEI 25,000 at the same concentration.
Figure 7.
Cell viability determined by MTT for (A) polymers at various concentrations and (B) polymer-DNA complexes at different N/P ratios against MCF-7 (A1 and B1) cells and MCF-7/ADR (A2 and B2) cells.
Abbreviations: LPCs, polyethyleneimine 800-DNA complexes; SPCs, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride)-DNA complexes; HPCs, polyethyleneimine 25,000-DNA complexes; PEI, polyethyleneimine; SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride).
Given that the cytotoxicity of DNA complexes is more relevant and important to transfection, the cytotoxicity of complexes at different N/P ratios was also evaluated in the MCF-7 and MCF-7/ADR cells (Figure 7B). PEI 800-DNA complexes and SP-DNA complexes did not show any cytotoxicity in cells at any N/P ratios. In contrast, the cytotoxicity of PEI 25,000-DNA complexes increased with the N/P ratio, showing about 70% cell viability at an N/P ratio of 20.
In vitro transfection experiment
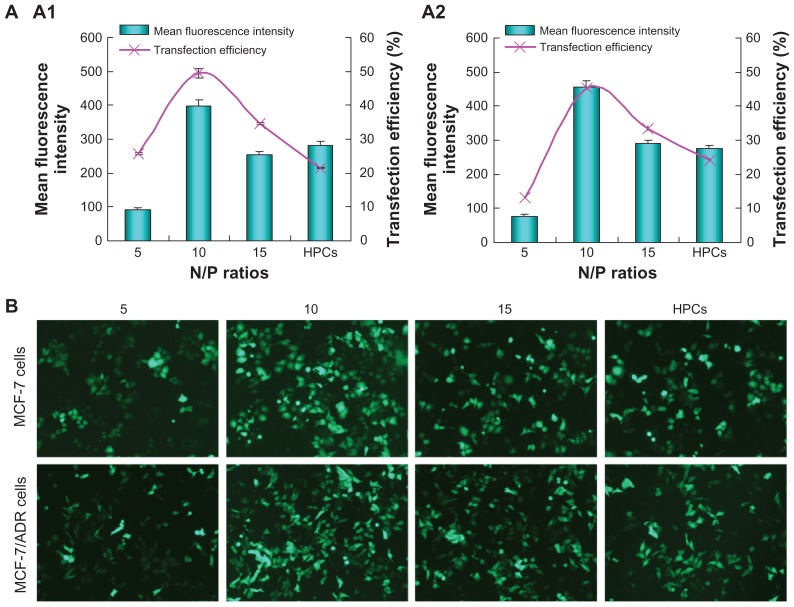
The in vitro transfection of SP-DNA complexes at various N/P ratios in MCF-7 and MCF-7/ADR cells was compared with that of PEI 25,000-DNA complexes in the presence of serum. As shown in Figure 8A, SP-DNA complexes showed maximum transfection efficiency at an N/P ratio of 10, with up to 50% and 45% transfection achieved by SP-DNA complexes in MCF-7 and MCF-7/ADR cells, respectively, which was higher than that from PEI 25,000-DNA complexes (22% for MCF-7 cells and 25% for MCF-7/ADR cells) at the optimal N/P ratio of 10. In addition, the mean fluorescent intensity of cells treated with SP-DNA complexes was also higher than that of cells treated with PEI 25,000-DNA complexes, indicating that cells incubated with SP-DNA complexes expressed more fluorescent proteins than those incubated with PEI 25,000-DNA complexes.
Figure 8.
(A) In vitro gene transfection efficiency of SP-pEGFP-N1 complexes at various N/P ratios in the presence of 10% fetal bovine serum in MCF-7 (A1) cells and MCF-7/ADR (A2) cells, in comparison with that of PEI 25,000-pEGFP-N1 complexes (at its optimal N/P ratio of 10). (B) Fluorescence images of MCF-7 cells and MCF-7/ADR cells transfected with SP-pEGFP-N1 complexes at various N/P ratios.
Note: PEI 25,000-pEGFP-N1 complexes at an N/P ratio of 10 were used as control.
Abbreviations: PEI, polyethyleneimine; SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); HPCs, polyethyleneimine 25,000-DNA complexes; pEGFP, plasmid-enhanced green fluorescent protein.
Furthermore, MCF-7 and MCF-7/ADR cells transfected with the complexes were observed under an inverted fluorescent microscope. In the microscopic images (Figure 8B), cells treated with SP-DNA complexes at an N/P ratio of 10 showed the most numerous and brightest green fluorescent spots. In contrast, the number of green cells transfected with PEI 25,000-DNA complexes was less, and the green fluorescent spots were also much dimmer. These results were in accordance with those from flow cytometry, suggesting that SP could be more effective than PEI 25,000 for gene delivery.
Cellular uptake
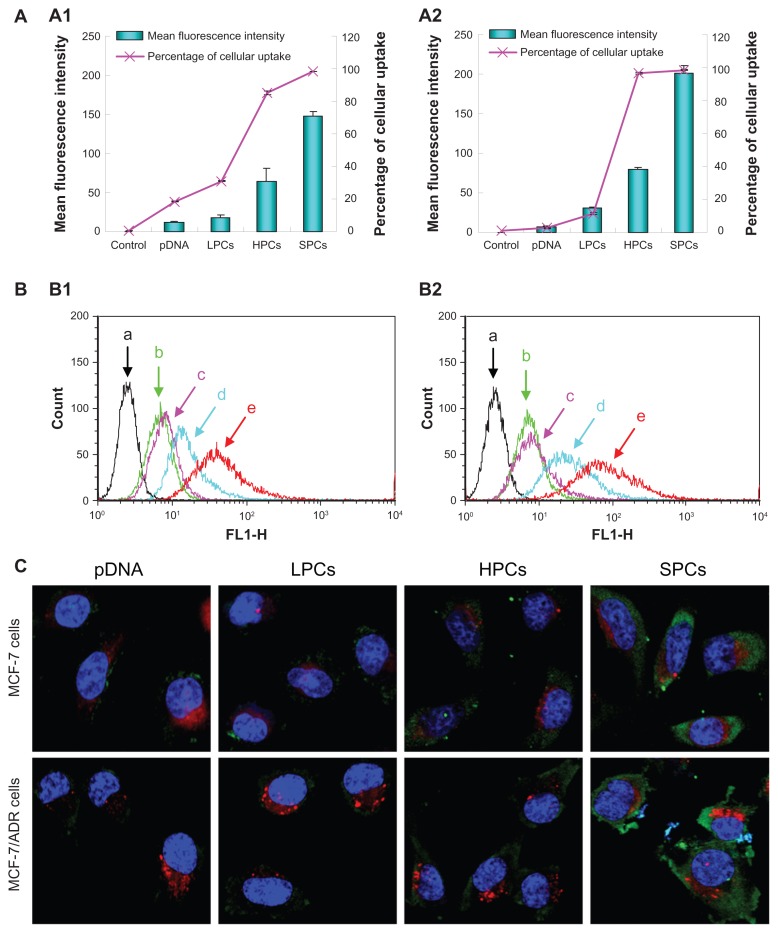
Cellular uptake is one of the crucial steps during the gene transfer process. The capacity of SP-DNA complexes to internalize plasmid DNA into cells was performed using pEGFP-N1 labeled with YOYO-1 on MCF-7 and MCF-7/ADR cells (Figure 9A and B). The cellular uptake of YOYO-1-labeled naked DNA was negligible, and complexing with PEI 800 achieved no obvious improvement in internalization. However, when DNA was complexed with SP, cellular uptake increased significantly in both cell lines, as indicated by the mean fluorescence intensity and percentage of cellular uptake. Although the number of green cells treated with PEI 25,000-DNA complexes was similar to that treated with SP-DNA complexes, the mean fluorescence intensity of the PEI 25,000-DNA complexes was much lower than that of the SP-DNA complexes. These results showed that SP-DNA complexes could internalize plasmid DNA into cells more effectively than PEI 25,000-DNA complexes. For vectors with a positive surface charge, uptake into cells may occur predominantly via adsorptive endocytosis.37 In addition, the amphiphilic properties of SP could be very beneficial for cellular uptake of the complexes.
Figure 9.
(A) Quantitative analysis of cellular uptake of pDNA, PEI 800-DNA complexes, PEI 25,000-DNA complexes, and SP-DNA complexes in MCF-7 cells (A1) and MCF-7/ADR cells (A2). (B) Flow cytometry images of control cells (a, black), and cells treated with pDNA (b, green), PEI 800-DNA complexes (c, pink), PEI 25,000-DNA complexes (d, turquoise), SP-DNA complexes (e, red) in MCF-7 cells (B1), and MCF-7/ADR cells (B2). (C) Confocal microscopic images of MCF-7 cells and MCF-7/ADR cells after treatment with pDNA, PEI 800-DNA complexes, PEI 25,000-DNA complexes, and SP-DNA complexes for 2 hours.
Note: Nuclei (blue), lysosomes (red), and pDNA (green) were stained with Hoechst, LysoTracker red, and YOYO-1, respectively.
Abbreviations: LPCs, polyethyleneimine 800-DNA complexes; SPCs, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride)-DNA complexes; HPCs, polyethyleneimine 25,000-DNA complexes; SP, polyethyleneimine 800 conjugated poly(styrene-co-maleic anhydride); PEI, polyethyleneimine.
Subcellular localization
Localization of the complexes inside the cells was observed using confocal laser scanning microscopy. The uptake of plasmid DNA into cells was observed by the presence of green dots from YOYO-1-labeled DNA in the cytoplasm around the nucleus (blue structures from Hoechst staining). The plasmid DNA (pEGFP-N1) was not translated within 2 hours, so the green fluorescence in the cells represented the complexes, not the expressed EGFP. As shown in Figure 9C, naked DNA and PEI 800-DNA complexes showed little sign of cellular uptake, while the fluorescence intensity of cells treated with PEI 25,000-DNA complexes increased significantly, but was still much lower than that of cells incubated with SP-DNA complexes, indicating that more SP-DNA complexes than PEI 25,000-DNA complexes were taken up by the cells. Given that the capture and degradation of DNA by lysosomes is one of the obstacles to efficient gene delivery, we also focused on colocalization of endocytosed plasmid DNA and lysosomes (red dots from LysoTracker red staining). As expected, both SP-DNA complexes and PEI 25,000-DNA complexes showed little colocalization with lysosomes due to their high buffering capacity. Generally, the results were in good agreement with the qualitative results of the transfection and cellular uptake assays.
There are several reasons for the superior transfection efficiency of SP-DNA complexes. First, the phenyl groups, carboxyl groups, and amide linkages in SP might interact with DNA base pairs via hydrogen bonding and π-π stacking interactions, except for the typical electrostatic interactions between the cationic amine and anionic phosphate groups of DNA, thereby improving the stability of SP-DNA complexes. In addition, cellular uptake of SP-DNA complexes was higher than for PEI 25,000-DNA complexes because of the amphiphilic properties of SP and its moderate positive charge. Furthermore, the cytotoxicity of SP-DNA complexes was lower than that of PEI 25,000-DNA complexes, which could be very beneficial for transfection of the complexes.
Conclusion
A novel amphipathic comb-shaped copolymer was synthesized by conjugating PEI 800 with SMA and evaluated as a gene vector. SP showed good ability to condense DNA and protect it against degradation by DNase I. The buffering ability of SP was similar with that of PEI. The nanosized diameter and moderate zeta potential of SP-DNA complexes were beneficial for obtaining a better balance between cellular uptake and cytotoxicity. As expected, in vitro experiments demonstrated that SP had significantly higher transfection efficiency with lower cytotoxicity compared with PEI 25,000 in MCF-7 and MCF-7/ADR cells in the presence of 10% fetal bovine serum. SP could be a promising new nonviral gene delivery vector due to its high transfection efficiency with low cytotoxicity.
Acknowledgments
The National Basic Research Program of China (2012CB932502, 2010CB934000), National Natural Science Foundation of China (30901866, 30925041), Shanghai Nanomedicine Program (11NM0505900), Shanghai Rising Star Program (11QA1407900), and Shanghai Outstanding Academic Leaders Program (11XD1406200) are gratefully acknowledged for their financial support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wu TL, Zhou DM. Viral delivery for gene therapy against cell movement in cancer. Adv Drug Deliv Rev. 2011;63(8):671–677. doi: 10.1016/j.addr.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in devheloping cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29(24–25):3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 3.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13(18):1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 4.El-Aneed A. An overview of current delivery systems in cancer gene therapy. J Control Release. 2004;94(1):1–14. doi: 10.1016/j.jconrel.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58(4):467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Chen XA, Zhang LJ, He ZJ, et al. Plasmid-encapsulated polyethylene glycol-grafted polyethylenimine nanoparticles for gene delivery into rat mesenchymal stem cells. Int J Nanomedicine. 2011;6:843–853. doi: 10.2147/IJN.S17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong X, Tian HY, Chen L, Chen J, Chen XS. Biodegradable mPEG-b-P(MCC-g-OEI) copolymers for efficient gene delivery. J Control Release. 2011;152(1):135–142. doi: 10.1016/j.jconrel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Liu KH, Wang XY, Fan W, et al. Degradable polyethylenimine derivate coupled to a bifunctional peptide R13 as a new gene-delivery vector. Int J Nanomedicine. 2012;7:1149–1162. doi: 10.2147/IJN.S28819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliv Rev. 2001;53(3):341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 10.Neu M, Fischer D, Kissel TJ. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. Gene Med. 2005;7(8):992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 11.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003;14(5):934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, Park JH, Lee M, Park TG, Kim SW. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J Control Release. 2005;103(1):209–219. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Jiang XL, Xu L, Wang XM, Hennink WE, Zhuo RX. Novel reduction-responsive cross-linked polyethylenimine derivatives by click chemistry for nonviral gene delivery. Bioconjug Chem. 2010;21(10):1827–1835. doi: 10.1021/bc100191r. [DOI] [PubMed] [Google Scholar]
- 14.Wang YQ, Su J, Wu F, et al. Biscarbamate cross-linked polyethylenimine derivative with low molecular weight, low cytotoxicity, and high efficiency for gene delivery. Int J Nanomedicine. 2012;7:693–704. doi: 10.2147/IJN.S27849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong K, Sun GB, Zhang XQ, et al. PEI-g-chitosan, a novel gene delivery system with transfection efficiency comparable to polyethylenimine in vitro and after liver administration in vivo. Bioconjug Chem. 2006;17(1):152–158. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 16.Wen YT, Pan SR, Luo X, Zhang X, Zhang W, Feng M. A biodegradable low molecular weight polyethylenimine derivative as low toxicity and efficient gene vector. Bioconjug Chem. 2009;20(2):322–332. doi: 10.1021/bc800428y. [DOI] [PubMed] [Google Scholar]
- 17.Wang CF, Lin YX, Jiang T, He F, Zhuo RX. Polyethylenimine-grafted polycarbonates as biodegradable polycations for gene delivery. Biomaterials. 2009;30(27):4824–4832. doi: 10.1016/j.biomaterials.2009.05.053. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Li H, Goh SH, Li J. Cationic star polymers consisting of α-cyclodextrin core and oligoethylenimine arms as nonviral gene delivery vectors. Biomaterials. 2007;28(21):3245–3254. doi: 10.1016/j.biomaterials.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Ping Y, Xu FJ, et al. Construction of a star-shaped copolymer as a vector for FGF receptor-mediated gene delivery in vitro and in vivo. Biomacromolecules. 2010;11(9):2221–2229. doi: 10.1021/bm100141y. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Yang C, Li HZ, et al. Cationic supramolecules composed of multiple oligoethylenimine-grafted β-cyclodextrins threaded on a polymer chain for efficient gene delivery. Adv Mater. 2006;18(22):2969–2974. [Google Scholar]
- 21.Hu QD, Fan H, Ping Y, Liang WQ, Tang GP, Li J. Cationic supramolecular nanoparticles for co-delivery of gene and anticancer drug. Chem Commun. 2011;47(19):5572–5574. doi: 10.1039/c1cc10721f. [DOI] [PubMed] [Google Scholar]
- 22.Winek CL, Burgun JJ. Acute and subacute toxicology and safety evaluation of SMA 1440-H resin. Clin Toxicol. 1977;10(2):255–260. doi: 10.3109/15563657708987970. [DOI] [PubMed] [Google Scholar]
- 23.Maeda H. SMANCS and polymer-conjugated macromolecular drugs: advantages in cancer chemotherapy. Adv Drug Deliv Rev. 2001;46(1–3):169–185. doi: 10.1016/s0169-409x(00)00134-4. [DOI] [PubMed] [Google Scholar]
- 24.Zovko M, Barbarić M, Zorc B, Hafner A, Filipović-Grčić J. Synthesis of fenoprofen and gemfibrozil styrene-maleic acid copolymer conjugates. Acta Pharm. 2005;55(2):169–176. [PubMed] [Google Scholar]
- 25.Iwai K, Maeda H, Konno T. Use of oily contrast medium for selective drug targeting to tumor: enhanced therapeutic effect and x-ray image. Cancer Res. 1984;44(5):2115–2121. [PubMed] [Google Scholar]
- 26.Maeda H, Ueda M, Morinaga T, Matsumoto T. Conjugation of poly(styrene-co-maleic acid) derivatives to the antitumor protein neocarzinostatin: pronounced improvements in pharmacological properties. J Med Chem. 1985;28(4):455–461. doi: 10.1021/jm00382a012. [DOI] [PubMed] [Google Scholar]
- 27.Konno T. Targeting chemotherapy for hepatoma: arterial administration of anticancer drugs dissolved in lipiodol. Eur J Cancer. 1992;28(2–3):403–409. doi: 10.1016/s0959-8049(05)80063-2. [DOI] [PubMed] [Google Scholar]
- 28.Mu Y, Kamada H, Kaneda Y, et al. Bioconjugation of laminin peptide YIGSR with poly(styrene co-maleic acid) increases its antimetastatic effect on lung metastasis of B16-BL6 melanoma cells. Biochem Biophys Res Commun. 1999;255(1):75–79. doi: 10.1006/bbrc.1999.9930. [DOI] [PubMed] [Google Scholar]
- 29.Greish K, Sawa T, Fang J, Akaike T, Maeda H. SMA-doxorubicin, a new polymeric micellar drug for effective targeting to solid tumours. J Control Release. 2003;97(2):219–230. doi: 10.1016/j.jconrel.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Greish K, Nagamitsu A, Fang J, Maeda H. Copoly(styrene-maleic acid)-pirarubicin micelles: high tumor-targeting efficiency with little toxicity. Bioconjug Chem. 2005;16(1):230–236. doi: 10.1021/bc040297g. [DOI] [PubMed] [Google Scholar]
- 31.Lin JJ, Hsu YC. Temperature and pH-responsive properties of poly(styrene-co-maleic anhydride)-grafting poly(oxypropylene)-amines. J Colloid Interface Sci. 2009;336(1):82–89. doi: 10.1016/j.jcis.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 32.Yin Q, Gao Y, Zhang ZW, Zhang PC, Li YP. Bioreducible poly (β-amino esters)/shRNA complex nanoparticles for efficient RNA delivery. J Control Release. 2011;151(1):35–44. doi: 10.1016/j.jconrel.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Boussif O, Lezoualćh F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chabaud P, Camplo M, Payet D, et al. Cationic nucleoside lipids for gene delivery. Bioconjug Chem. 2006;17(2):466–472. doi: 10.1021/bc050162q. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Tang A, Law LP, et al. Amphiphilic core-shell nanoparticles with poly(ethylenimine) shells as potential gene delivery carriers. Bioconjug Chem. 2005;16(1):139–146. doi: 10.1021/bc049895l. [DOI] [PubMed] [Google Scholar]
- 36.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 37.Remy-Kristensen A, Clamme JP, Vuilleumier C, Kuhry JG, Mely Y. Role of endocytosis in the transfection of L929 fibroblasts by polyethylenimine/DNA complexes. Biochim Biophys Acta. 2001;1514(1):21–32. doi: 10.1016/s0005-2736(01)00359-5. [DOI] [PubMed] [Google Scholar]