Abstract
The invasion of Squamous Cell Carcinoma (SCC) is a significant cause of morbidity and mortality. Here we identify an E3 ligase, Traf6, and a de-ubiquitinating enzyme, Cezanne/ZA20D1, as important regulators of this process in organotypic models. Traf6 can promote the formation of Cdc42-dependent F-actin microspikes. Furthermore, Traf6 plays a key role in autocrine Interleukin-1β signalling in SCC cells which in turn is required to drive the expression of Tumour Necrosis Factor α (TNFα). TNFα acts in a paracrine manner to increase the invasion promoting potential of carcinoma-associated fibroblasts. Exogenous TNFα signalling can restore invasion in cells depleted of Traf6. In conclusion, Traf6 plays two important roles in SCC invasion: it promotes cell intrinsic Cdc42-dependent regulation of the actin cytoskeleton and enables production of the paracrine signal, TNFα, that enhances the activity of carcinoma-associated fibroblasts.
Keywords: Traf6, Interleukin-1, TNFα, invasion, carcinoma-associated fibroblast
Introduction
Squamous cell carcinoma is an epithelial derived cancer of variable clinical presentation and histological appearance. Important tumour-related factors which govern patient prognosis include the tumour size and grade, the depth of invasion, the degree of infiltration of neighbouring tissue, and the presence or absence of metastases. In particular, the extent of local invasion has significant consequences when planning surgery with overall management being largely determined by the nature and pattern of metastatic disease (1-3). A greater understanding of the basic biology of squamous cell carcinoma invasion is therefore likely to lead to significant benefits in the treatment of the disease and thus its long-term prognosis.
Carcinoma invasion is greatly influenced by non-tumour cells within the tumour micro-environment. Fibroblasts within tumours are able to promote cancer cell invasion through the production of soluble factors and also by remodelling the extra-cellular matrix (ECM) as well as the generation of permissive tracks for invasion (4, 5). It is clear that Carcinoma-Associated Fibroblasts (CAFs) have distinct properties from normal fibroblasts (6). However, the mechanisms by which cancer cells co-opt fibroblasts to promote invasion are varied and incompletely understood. Paracrine TGFβ and Hedgehog signalling can promote the activation of fibroblasts (4, 7), but genetic blockade of TGFβ signalling in fibroblasts can also be pro-tumourigenic (Bhowmick et al., 2004). More recently cytokine signalling has been implicated in the invasion promoting properties of CAFs (8-10). The inflammatory cytokines TNFα, IL-1α/β and IL-6 can all modulate tumourigenesis by signalling to fibroblasts.
Regulators of ubiquitin and ubiquitin-like modification can affect cancer invasion in many ways, including modulation of cytokine signalling. Both TNFα and IL-1α/β bind to membrane spanning receptors which then trigger a range of intracellular signalling pathways. Canonical NFκB signalling is activated by both cytokines. Once the receptors have engaged their ligand a multi-protein complex is assembled that ultimately leads to the activation of IκB Kinases (IKK’s). Traf2 is a critical adaptor molecule in the activation of IKK following TNFα stimulation (11); while the K63 ubiquitin linkage E3 ligase, Traf6, performs a similar function after IL-1 stimulation (12). The action of Traf6 can be antagonised by the De-UBiquitinating enzyme (DUB), A20 (13). Active IKKs then phosphorylate IκB leading to its ubiquitin-dependent degradation. The degradation of IκB enables NFκB transcription factors to accumulate in the nucleus and drive a broad transcriptional programme. Besides NFκB activation, TNFα and IL-1α/β also trigger many other changes within the cell, including alteration of the actin cytoskeleton (14, 15). The ubiquitination and SUMOylation machinery can also target Rho-family proteins that regulate actin organisation and cell migration (16). Both the HECT domain E3 ligase, Smurf1, (17) and CRL3, a component of Cullin-RING E3 ligase complexes, target RhoA for degradation (18). In contrast, SUMOylation of Rac1 can promote its activity, the formation of actin protusions and cell migration in response to HGF (19).
We hypothesised that other regulators of ubiquitin and SUMO modifications would also regulate F-actin organisation and thus cell migration in squamous cell carcinoma. Therefore, we undertook a systematic siRNA screen for the effect of depletion of E2 ligases, E3 ligases and De-UBiquitinating enzymes (DUBs) on F-actin. Following the analysis of several hundred genes we focused on two that had effects on F-actin organisation. We find that the E3 ligase Traf6 and the DUB Cezanne/ZA20D1 have contrasting effects on actin architecture, though they do not simply antagonise one another. Divergent pathways downstream of Traf6 regulate SCC invasion. Cdc42 function is required for Traf6 to promote the formation of F-actin microspikes, while the regulation of NFκB signalling by Traf6 sustains an autocrine IL-1β loop that facilitates cell invasion through the expression of TNFα. Moreover, TNFα participates in paracrine signalling that promotes the activity of carcinoma-associated fibroblasts and thereby enhances cancer cell invasion.
Results
Identification of Traf6 and Cezanne/ZA20D1 as regulators of SCC morphology
To investigate the role of the ubiquitin regulatory machinery in the control of cancer cell invasion we hypothesised that key regulators of invasion would affect the F-actin organisation of cells. We therefore investigated F-actin architecture in A431 squamous cell carcinoma cells grown on thick collagen-rich deformable matrices following siRNA-mediated depletion of 426 E2 ligases, E3 ligases and DUBs. Cells were fixed 72 hours after transfection and stained with phalloidin to label F-actin structures. Control cells grew in small well packed groups with F-actin protrusions extending at the edges of the clusters. We observed a diverse range of phenotypes following depletion of E3 ligases and DUBs using siRNA smartpools: these ranged from elongated cells to contracted cells with membrane blebs. To identify genes that consistently affected F-actin in an unbiased manner the screen was performed twice and each time the F-actin organisation was scored by three ‘blinded’ observers using standardised nomenclature: this gave a total of six scores per gene. Forty-five genes scored 5/6 or 6/6 and these were chosen for further investigation (Supplementary Figure 1a).
To exclude the possibility of ‘off-target’ effects sometimes associated with siRNA reagents, we tested multiple siRNA oligonucleotides targeting the 45 genes that we identified in our initial screen. Twenty-nine genes showed consistent phenotypes (Supplementary Figure 1b). We noted that within this set of genes there was an E3 ligase, Traf6, and a DUB, ZA20D1/Cezanne (hereafter referred to as ZA20D1) which had been reported to interact with each other (20, 21). We therefore chose these genes for further study.
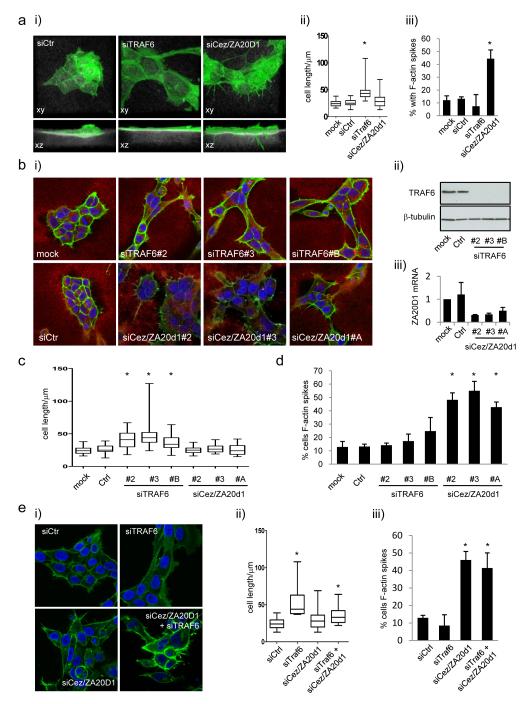
Figure 1a shows that depletion of Traf6 generates cells with an elongated and spread morphology. 3D confocal analysis also revealed that these cells were flatter than controls (Figure 1a). In contrast, depletion of ZA20D1 leads to an increase in the number of F-actin microspikes which frequently protrude into the underlying collagen-rich matrix (Figure 1a). We confirmed that these phenotypes could be reproduced with three independent siRNA duplexes targeting either Traf6 or ZA20D1 (Figure 1b-d) and that these siRNA effectively depleted Traf6 and ZA20D1 (Figure 1b). Traf6 depletion also led to more elongated cells in two other SCC cell lines, Detroit 562 and SCC12 (Supplementary Figure 1b). The effects of ZA20D1 depletion were less pronounced in these cell lines (Supplementary Figure 1b).
Figure 1.
Traf6 and Cezanne/ZA20D1 regulate SCC morphology. A) i) (upper panels) or ‘the side’ (lower panels) depleted for either Traf6 or Cezanne/ZA20D1 using siRNA smart-pools. F-actin is shown in green and reflectance imaging of the thick collagen-rich deformable matrix is shown in white. Panels are 121μm wide. ii) Quantification of the long axis of A431 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. iii) Quantification of the percentage of cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. (B) i) Single confocal sections (146×146μm) of A431 cells shown depleted for either Traf6 or Cezanne/ZA20D1 using three independent siRNA sequences. F-actin is shown in green, reflectance imaging of matrix is shown in red and nuclei are shown in blue. ii) Western blot showing Traf6 protein levels in A431 cells following transfection of three independent siRNA sequences. iii) QRT-PCR showing Cezanne/ZA20D1 mRNA levels in A431 cells following transfection of three independent siRNA sequences. C) Quantification of the long axis of A431 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. D) Quantification of the percentage of cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. E) i) Single confocal sections (146×146μm) of A431 cells shown depleted for either Traf6 or Cezanne/ZA20D1 or both using siRNA smartpools. F-actin is shown in green and and nuclei are shown in blue. ii) Quantification of the long axis of A431 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. iii) Quantification of the percentage of cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 one way t-test.
In view of the reported antagonistic relationship of these two proteins (21), we next tested whether the microspikes resulting from ZA20D1 depletion were dependent on Traf6 function. When both ZA20D1 and Traf6 were simultaneously knocked down the number of F-actin microspikes were similar to that in ZA20D1 depleted cells (Figure 1e). Although we could not confirm that every cell received equal amounts of both siRNAs, western blot and immune-fluorescence analysis indicated that Traf6 was depleted by at least 90% when co-transfected with ZA20D1 siRNA (data not shown). Indeed, the phenotype of cells depleted for both Traf6 and ZA20D1 was more of a mixture of the two individual phenotypes. These data suggest that de-regulated Traf6 activity is not responsible for the F-actin microspikes observed in ZA20D1 depleted cells.
Cdc42 is required for Traf6-induced microspikes
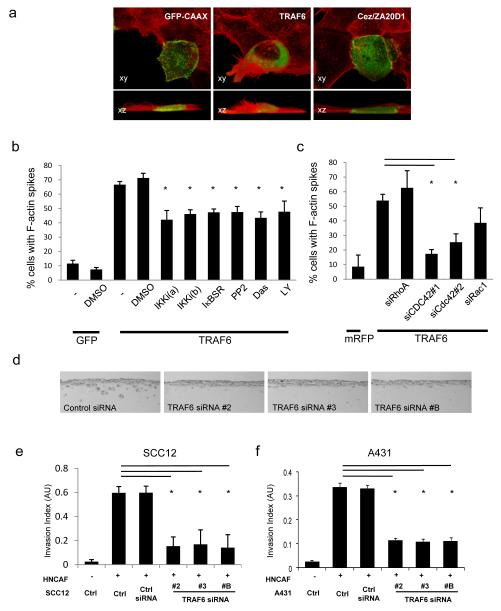
Having established that Traf6 and ZA20D1 had effects on the actin cytoskeleton we then sought to investigate the molecular mechanism. cDNA encoding Traf6 and ZA20D1 were transiently transfected into A431 cells and the F-actin organisation examined 48 hours later. The majority of Traf6 over-expressing cells showed pronounced F-actin microspikes and frequently increased levels of F-actin (Figure 2a). ZA20D1 over-expressing cells did not show striking changes in morphology (Figure 2a). Consequently, we focused our attention on Traf6.
Figure 2.
Traf6 is required for SCC invasion. A) Membrane-GFP (control), HA-tagged Traf6 or GFP-Cezanne/ZA20D1 were transfected into A431 cells. 3D reconstructions of the morphology of A431 cells viewed from ‘above’ and ‘the side’ are shown: transfected protein is shown in green and F-actin in red. Panels are 50 μm wide. B) Membrane-GFP (control), HA-tagged Traf6 and IκB-super-repressor were transfected into A431 cells as indicated. Six hours prior to fixation the indicated inhibitors were added at 10μM final concentration (except Dasatinib at 250nM). The number of transfected cells with F-actin microspikes >7.5μm is shown. Average of at least three experiments (standard error is shown). C) Membrane-RFP (control) or HA-tagged Traf6 were transfected into A431 cells together with 80nM siRNA against RhoAsi#smartpool, Cdc42si#1, Cdc42si#2 or Rac1si#smartpool as indicated. The number of transfected cells with F-actin microspikes >7.5μm is shown. Average of three experiments (standard error is shown). D) Representative images (600×280μm) showing the extent of SCC12 invasion following Traf6 depletion in organotypic cultures containing carcinoma-associated fibroblasts. E) Quantification of SCC12 invasion in organotypic assays. At least ten images from at least two independent assays are quantified for each data point. F) Quantification of A431 invasion in organotypic assays. At least ten images from at least two independent assays are quantified for each data point.
Traf6 was over-expressed in A431 cells and 6 hours before fixation cells were treated with inhibitors of pathways previously linked to Traf6 and Rho-family regulators of F-actin organisation. Specifically, IKK, PI-3K, Src-family kinases, RhoA, Rac1 and Cdc42 were inhibited (22, 23). Co-transfection of Traf6 with a ‘dominant negative’ non-degradable IκB mutant was also used to block canonical NFκB signalling. This analysis indicated that the ability of Traf6 to regulate the formation of actin microspikes was partly dependent on IKK-NFκB, PI3K and Src-family kinase signalling (Figure 2b. Supplementary Figure 2 demonstrates that effective IKK inhibition was achieved). More impressively, we consistently observed that depletion of Cdc42, but not Rac1 or RhoA, prevented Traf6 from inducing F-actin microspikes (Figure 2c).
Traf6, IKK and Cdc42 function are required for SCC invasion
We next tested the role of Traf6 depletion on squamous cell carcinoma invasion in an organotypic invasion assay. Squamous cell carcinoma cells are cultured on a collagen-rich matrix containing carcinoma-associated fibroblasts at an air-liquid interface (24). This model reproduces several features of the epidermal-dermal architecture of a carcinoma and utilises the same collagen-rich deformable matrix that was used for the cell morphology analysis. Whilst control cells invade efficiently in the presence of carcinoma-associated fibroblasts, depletion of Traf6 (using three different siRNA duplexes) significantly reduces the invasion of SCC12 and A431 squamous cell carcinoma (Figures 2d, e and f). Conventional transwell-matrigel assays also demonstrated that Traf6 is required for A431 cell invasion (Supplementary Figure 1d). Depletion of ZA20D1 also reduced SCC invasion, which further suggests that it has additional functions besides antagonising Traf6 (Supplementary Figure 1d&e).
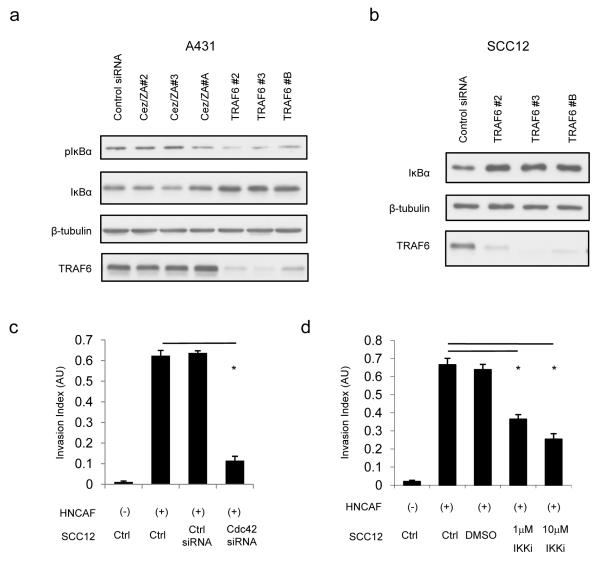
The data outlined above suggests that Traf6 mediated regulation of Cdc42 and F-actin microspikes may contribute to SCC invasion. Additionally, Traf6 is known to be a key regulator of downstream events in canonical NFκB signalling which itself has been shown to influence cancer invasion (12, 25). We therefore sought to determine whether Traf6 regulates IKK functions in SCC cells. IKK phosphorylates IκB leading to its degradation. Hence, reduced IκB phosphorylation along with increased IκB levels indicate lower levels of IKK function. Depletion of Traf6 decreased the phosphorylation of IκB and increased the levels of IκB in both A431 and SCC12 cells (Figures 3a&b and data not shown). Depletion of ZA20D1 had no effect.
Figure 3.
Role of IKK and Cdc42 in SCC invasion. A) A431 cells were depleted for either Cezanne/ZA20D1 or Traf6 using multiple independent siRNA. 48 hours after transfection the levels and phosphorylation of IκB was determined by western blot analysis. β-tubulin and Traf6 levels are shown as controls for protein loading and transfection efficiency, respectively. B) SCC12 cells were depleted for Traf6 using multiple independent siRNA. 48 hours after transfection the levels of IκB were determined by western blot analysis. β-tubulin and Traf6 levels are shown as controls for protein loading and transfection efficiency, respectively. C) The ability of SCC12 cells to invade into carcinoma-associated fibroblast conditioned matrix following the depletion of Cdc42 is shown. At least ten images from at least two independent assays are quantified for each data point. D) The ability of SCC12 cells to invade into carcinoma-associated fibroblast conditioned matrix in the presence of IKK inhibitor. At least ten images from at least two independent assays are quantified for each data point. . Mean and SEM are shown. * indicates p<0.01 relative to control + HNCAF.
We next tested whether the function of IKK or Cdc42 is required for the invasion of squamous cell carcinoma. To exclude any indirect effects of IKK inhibitors on fibroblasts, we used organotypic invasion assays with CAF-conditioned matrices from which the CAF had been removed. The ability of SCC cells treated with either siRNA or inhibitors to invade into these matrices was tested. Similar to previous findings (5), siRNA-mediated depletion of Cdc42 in SCC12 cells caused a pronounced reduction in invasion (Figure 3c). Figure 3d shows that IKK inhibition reduced SCC invasion in a dose-dependent manner (Supplementary Figure 2 confirms the efficacy of the IKK inhibitors). Together, this suggests that IKK function is required for SCC invasion, even though its inhibition only slightly reduces the formation of Traf6-driven F-actin microspikes.
Traf6 is over-expressed in SCC
Our analyses indicate that the alteration of Traf6 expression leads to changes in SCC cell morphology and that reduced Traf6 levels inhibit SCC invasion. Consequently, we evaluated TRAF6 expression in human SCC samples. Supplementary Figure 3a confirms that Traf6 can be specifically detected in formalin-fixed paraffin embedded material. In normal oral mucosa Traf6 was largely restricted to the basal layer of cells within the epidermis. However, in numerous oral SCC samples Traf6 was expressed to a higher level and in a much greater proportion of cells (Supplementary Figure 3b). Quantification of Traf6 levels in 50 oral SCC biopsies compared to 9 normal oral mucosa samples is shown in Supplementary Figure 3c. We also analysed a limited number of patient matched oral mucosa and SCC samples, examples of which are shown in Supplementary Figure 3d. Elevated levels of Traf6 in carcinoma tissue were observed compared to that of normal tissue (controls for antibody specificity on frozen sections are shown in Supplementary Figure 3e). Analysis of 10 vulval SCC biopsies also revealed elevated levels of Traf6 expression in some samples (Supplementary Figure 3f&g).
Traf6 participates in autocrine IL-1 signalling to promote cancer cell invasion
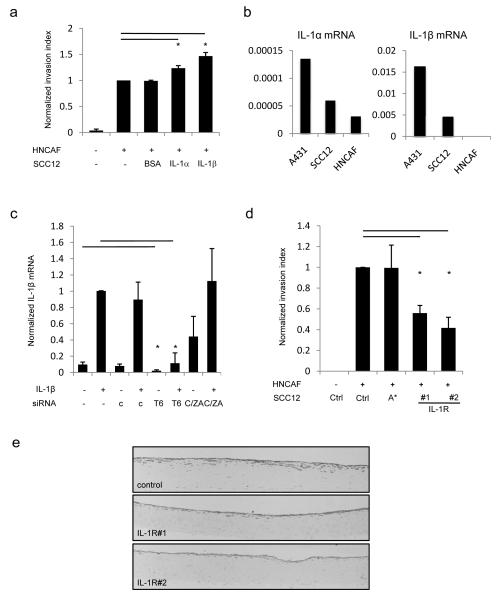
Traf6 mediates signals from a range of extra-cellular cues including IL-1, LPS and dsRNA. The effect of Traf6 depletion in the absence of exogenous ligands suggests a basal level of Traf6 signalling in standard culture conditions. Figure 4a shows that both IL-1α and IL-1β were able to promote SCC invasion in the presence of CAFs. IL-1α and IL-1β also efficiently induced IκB degradation in SCC cells but had no effect on CAFs (Supplementary Figure 4 and data not shown). In contrast, neither LPS or dI:C (a dsRNA mimetic) had any effect on invasion or NFκB signalling (Supplementary Figure 5 and data not shown). This indicates that only the carcinoma cells in our model system are able to respond to IL-1 ligands and led us to exclude a role for TLR2 and TLR4. Given the ability of exogenous IL-1α and IL-1β to promote carcinoma invasion, we investigated if they were endogenously expressed in SCC cells and CAFs. Figure 4b shows that both IL-1α and β are predominantly expressed by squamous cell carcinoma cells, suggesting that autocrine IL-1 to Traf6 signalling may be occuring. The significantly greater efficiency of IL-1β PCR (compare y-axis scales in Figure 4b) suggests that it is the predominant isoform expressed in A431 and SCC12 cells.
Figure 4.
Traf6 is required to maintain autocrine IL-1 signalling and TNFα expression. A) Quantification of SCC12 invasion in organotypic assays in the presence of either 1ngml−1 IL-1α or IL-1β. At least ten images from at least two independent assays are quantified for each data point. B) Analysis of IL-1α and IL-1 β mRNA levels in A431, SCC12 and Head and Neck Carcinoma-Associated Fibroblasts. IL-1 mRNA is normalised to GAPDH levels: one representative experiment out of three is shown. C) Analysis of IL-1β levels in A431 cells following combinations of IL-1β treatment for 60 minutes and depletion of either Traf6 or Cezanne/ZA20D1 using siRNA smart-pools. Average of two experiments performed in triplicate – error bars represent half the range of values. D) Quantification of SCC12 invasion following depletion of IL-1R using two independent sets of siRNA in organotypic assays. At least ten images from at least two independent assays are quantified for each data point. E) Representative images (3000×750μm) showing the extent of SCC12 invasion following IL-1R depletion in organotypic cultures containing carcinoma-associated fibroblasts.
Positive feedback is a common feature of autocrine loops. We tested whether IL-1β treatment increased the expression levels of either IL1α or β. IL-1β was robustly induced following IL-1β stimulation in A431 cells, while IL-1α levels remained unchanged (Figure 4c and data not shown). Similar results were obtained in SCC12 cells (Supplementary Figure 6a and data not shown). This indicates that IL-1β is involved in a positive feedback loop. We next examined whether Traf6 or ZA20D1 were involved in the maintenance of this positive feedback loop. Depletion of Traf6 reduced the levels of IL-1β mRNA in both basal conditions and following IL-1β stimulation (Figure 4c and Supplementary Figure 6b). These argue that Traf6 contributes to autocrine IL-1 signalling, even in unstimulated cells. We directly tested the importance of IL-1 autocrine signalling by depleting the IL-1 receptor in SCC12 cells in organotypic invasion assays. Figures 4d&e show that depletion of IL-1R leads to a significant reduction in cancer cell invasion thereby confirming the involvement of an IL-1 autocrine loop in this setting.
TNFα promotes Carcinoma-Associated Fibroblast function
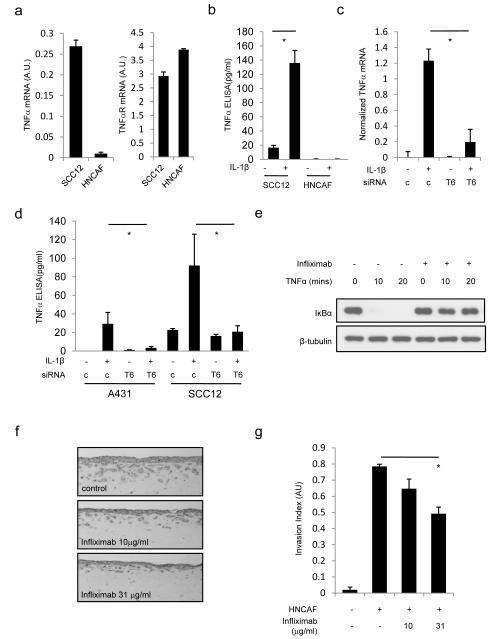
NFκB signalling regulates the expression of a broad range of cytokines. We therefore hypothesised that autocrine IL-1β>Traf6 signalling might contribute to the invasion of squamous cell carcinoma by regulating the expression of other cytokines. We found that IL-1β could increase TNFα mRNA, but not IL-6 mRNA (Figure 5a and data not shown). Thus, we investigated the role of TNFα in our organotypic co-culture invasion system. Both cancer cells and HNCAF express TNFα receptor mRNA, however only the cancer cells expressed significant levels of TNF α mRNA (Figure 5a). These data were confirmed by ELISA analysis of TNFα protein levels (Figure 5b). Furthermore, IL-1β efficiently induces TNFα expression in SCC cells, but not CAFs (Figure 4d). Both the induced and basal levels of TNFα were reduced in Traf6 depleted cells (Figure 5c&d and Supplementary Figure 6c); these data demonstrate that autocrine IL-1>Traf6 signalling is required for the expression of TNFα. Further, the data suggest that Traf6 may promote invasion by regulating the production of TNFα by SCC cells. Figure 5e demonstrates that TNFα can signal to carcinoma-associated fibroblasts and that a TNFα blocking antibody, infliximab (Remicade®), is able to inhibit TNFα function. Crucially, treatment of organotypic cultures with infliximab reduced SCC invasion in a dose-dependent manner (Figures 5f&g), confirming a role for TNFα and raising the possibility that it may act as a paracrine signal from cancer cells to CAFs.
Figure 5.
TNFα is required for SCC invasion. A) QRT-PCR analysis of TNFα and TNFα receptor mRNA levels in SCC12 and HNCAF. B) ELISA analysis showing TNF α secreted by SCC12 or HNCAF cultured in the presence or absence of 1ng/ml IL-1 β. Mean and SEM are shown. * indicates p<0.01 relative to control + IL-1β. C) Analysis of TNFα mRNA in A431 cells following combinations of IL-1β treatment for 60 minutes and depletion of Traf6 or using siRNA. Average of two experiments performed in triplicate – error bars represent half the range of values. D) ELISA analysis showing TNFα secreted by SCC12 or A431 following combinations of IL-1β treatment for 60 minutes and depletion of Traf6 or using siRNA. Mean and SEM are shown. * indicates p<0.01 relative to control + IL-1β. E) Western blots showing the modulation of IκB levels following TNFα treatment of HNCAF cells in the absence or presence of Infliximab. F) Representative images (600×240μm) of organotypic invasion assays using SCC12 cells and HNCAF with increasing doses of infliximab (Remicade®). G) Quantification of organotypic invasion assays using SCC12 cells and HNCAF with increasing doses of Infliximab. At least ten images from at least two independent assays are quantified for each data point. Mean and SEM are shown. * indicates p<0.01 relative to control + HNCAF.
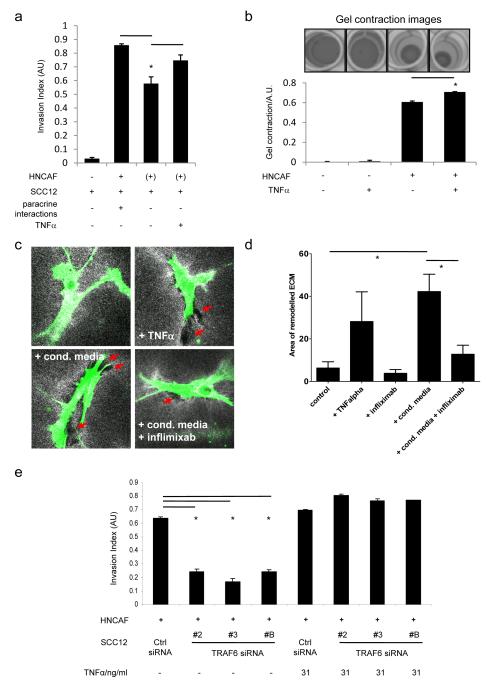
Previous work has shown that CAFs can remodel the ECM and facilitate cancer cell invasion even when they are not co-cultured with cancer cells (5). In this context soluble paracrine signals produced by cancer cells, such as TNFα, which may promote the invasion-stimulating activity of CAFs are absent. Figure 6a shows that the ability of CAFs to facilitate SCC invasion when they are not ‘co-cultured’ with cancer cells is reduced (compare columns 2 and 3). This supports the argument that soluble factors produced by SCC12 cells influence the ability of CAFs to drive invasion. We therefore tested whether TNFα could specifically affect the invasion promoting ability of CAFs by adding TNFα to cultures only during the period of fibroblast-dependent matrix remodelling. Significantly, the deficit in invasion observed in the absence of paracrine signals from SCC12 cells could be rescued by supplying additional TNFα to the CAFs.
Figure 6.
TNFα mediates paracrine signalling from cancer cells to CAFs. A) Quantification of organotypic assays using SCC12 and HNCAF: ‘+’ indicates the presence of HNCAF as the same time as SCC12 cells ‘(+)’ indicates the presence of HNCAF prior to the addition of SCC12 cells. HNCAF and SCC12 were co-cultured to allow paracrine signalling by soluble factors - 2nd column, or HNCAF and SCC12 were cultured sequentially to prevent paracrine signalling by soluble factors - 3rd column. Finally, HNCAF and SCC12 were cultured sequentially and exogenous TNF α was added during the period of HNCAF culture. B) Upper images show ECM contraction by HNCAF in the absence or presence of 31ng/ml TNFα. Lower histogram shows quantification of multiple contraction assays. Error bars are standard deviation. C) Images (146×146μm) show HNCAF (green) in collagen-rich deformable matrices (white) in the presence or absence of TNFα (31ng/ml), SCC12 conditioned media and/or infliximab (Remicade®) (31μg/ml). Red arrows indicate area of degraded matrix. D) Quantification of the average area of matrix degradation surrounding CAFs in the conditions indicated. Average of >50 cells from three independent experiments. Error bars indicate SEM and * indicates p<0.01 One-way non-parametric ANOVA. E). Quantification of SCC12 invasion following depletion of Traf6 using three independent siRNA in the presence or absence of 31ng/ml TNFα organotypic assays. At least ten images from at least two independent assays are quantified for each data point. Mean and SEM are shown. * indicates p<0.01 relative to control + HNCAF.
A key invasion-promoting function of CAFs is remodelling of the ECM. In view of this, we examined whether TNFα modulates the ability of CAFs to remodel the ECM. This can be measured by the ability of CAFs to contract collagen-rich gels (Figure 6b) and gel contraction was significantly increased in the presence of 31ng/ml TNFα (compare columns 3 & 4). Microscopic analysis also confirmed that CAFs treated with TNFα had a greater ability to remodel the ECM; with more prominent areas of matrix degradation being observed around the CAFs (indicated by the red arrows in Figure 6c). A similar pattern of matrix remodelling and degradation was induced by the addition of SCC conditioned media. The effect of conditioned media was reduced by the addition of infliximab (Figure 6c panels 3 & 4 and Figure 6d). Thus TNFα acts as a paracrine signal from SCC cells to CAFs to promote matrix remodelling.
This data demonstrates a role for TNFα in promoting cancer cell invasion and confirms that Traf6 is required for TNFα expression by SCC cells (Figure 4d). Considered together this predicts that the exogenous provision of TNFα should restore carcinoma invasion in organotypic co-cultures containing SCC cells depleted of Traf6. Figure 6e shows the results of invasion assays containing SCC12 cells depleted of Traf6 with and without daily stimulation with TNFα. In the control assays, TNFα leads to a marginal increase in invasion. In the setting of Traf6-depleted SCC cells however, exogenous TNFα caused a striking ‘rescue’ of invasion. Collectively these results indicate that Traf6 is a critical player in autocrine IL-1 signalling within SCC cells. This in turn helps to maintain a co-ordinated pro-invasive programme which involves the expression of TNFα and enhanced fibroblast-dependent remodelling of the ECM.
Discussion
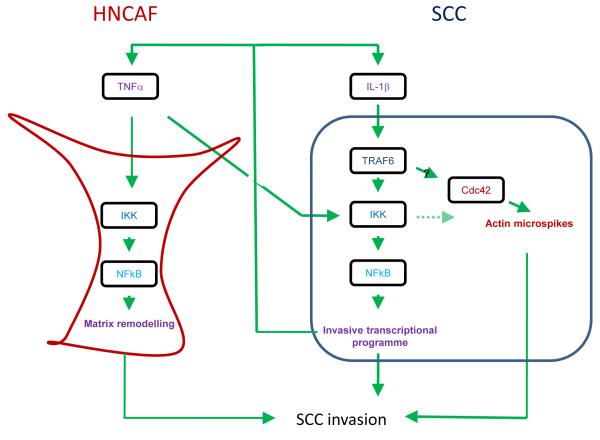
The invasion of squamous cell carcinoma and its propensity for metastasis determines patient prognosis, morbidity and ultimately mortality. To learn more about this process we screened for ubiquitin-linked regulators of SCC invasion. This work shows that Traf6 is critical for two reasons: firstly it enables canonical NFκB signalling to drive paracrine signalling to CAFs and secondly it can regulate the formation of Cdc42-dependent F-actin microspikes (Figure 7). Blockade of either activity reduces SCC invasion suggesting that regulation of F-actin and NFκB co-operate to promote invasion. Both TNFα and IL-1β are transcriptionally controlled by Traf6 as a result of NFκB signalling. This probably occurs through the well documented K63 ubiquitin ligase function of Traf6 (12, 26). The ability of exogenous TNFα to rescue invasion in Traf6 depleted cultures suggests that regulation of TNFα transcription is the predominant invasion-promoting function of Traf6. However, we noted that TNFα could induce changes in F-actin organisation in both control and Traf6 depleted cells (data not shown). These changes might provide a mechanism to compensate for the lack of F-actin microspikes in Traf6 depleted cells. The mechanism by which Traf6 might modulate Cdc42 function is unclear. Traf6 can form oligomers and has been reported to be a pleiotropic scaffold molecule, influencing a variety of signalling pathways including Akt, Src-family kinases and p38 (22, 23, 27). Our analysis confirms some role for PI3 kinase, Src-family kinases, and IKK downstream of Traf6; although no single pathway was absolutely required for Traf6 to regulate the cytoskeleton.
Figure 7.
Model summarising the roles of TRAF6, IL-1β and TNFα in SCC invasion. TRAF6 enables canonical NFκB signalling to drive an invasive transcriptional programme including TNFα and IL-1β and can also regulate the formation of F-actin microspikes in SCC cells (outlined in dark blue). Blockade of either activity reduces SCC invasion. TNFα produced by SCC cells acts in a paracrine manner to increase the invasion promoting potential of carcinoma-associated fibroblasts (outlined in red).
We also found that the DUB ZA20D1 is required for SCC invasion (Supplementary Figure 1). We believe that the reduced invasion of cells lacking ZA20D1 is not caused by their increased number of F-actin microspikes, which are usually pro-invasive structures, but by other unidentified defects. ZA20D1 can associate with Traf6 and is possibly an antagonist to Traf6 (21). However, Traf6 depletion has a minimal effect on the formation of F-actin microspikes induced by ZA20D1 depletion (Figure 1) and ZA20D1 depletion has no effect on the levels of phosphorylation of IKKα/β and IκB (Figure 4 and data not shown). We did observe a slight increase in IL-1β mRNA levels in ZA20D1 depleted cells which is consistent with the findings of Enesa et al., (28). This may reflect the greater sensitivity of the mRNA analysis compared to the IKKα/β and IκB analysis or alternatively that IL-1β mRNA can be induced in an IKKα/β/IκB-independent manner. Considered collectively, ZA20D1 most likely has additional functions besides antagonising Traf6 (29, 30).
Previous studies have identified an important role for both TNFα and IL-1α/β in the development and progression of SCC. However, these studies have primarily focused on signalling by these factors to either leukocytes or cancer cells (31-34). Here we describe interlinked but distinct roles for IL-1β and TNFα in an organotypic model of squamous cell carcinoma. Initially, we demonstrated a requirement for Traf6 in the SCC (epithelial) compartment of our system. In the absence of Traf6 the expression of both IL-1β and TNFα by SCC cells is reduced. The effect of IL-1β is limited to the SCC cells in our system and serves to maintain and possibly amplify the expression of itself and TNFα. It is also probable that Traf6 supports the expression of other cytokines that have been reported to be IL-1 regulated, including IL-8 (35). The critical role of Traf6 in driving TNFα expression is illustrated by the ability of exogenous TNFα to rescue carcinoma invasion in Traf6 depleted cells. Significantly, TNFα produced by cancer cells acts in a paracrine manner to enhance the function of carcinoma-associated fibroblasts. The matrix remodelling abilities of CAFs are promoted by the addition of exogenous TNFα. This is possibly the result of both an increase in the production of MMPs and an increase in the contractile function the CAF cytoskeleton (10, 36-38). Increased matrix remodelling by CAFs in turn leads to an increase in cancer cell invasion.
The mechanisms involved in the transformation of normal fibroblasts to invasion-promoting CAFs are incompletely understood. TGFβ has been widely implicated in this process but its inhibition does not affect the HNCAF used in this study (Hooper unpublished). Erez et al. proposed that IL-1β is a critical player in the NFκB-dependent activation of CAFs (8) but the CAFs used in this study are not responsive to either IL-1α or IL-1β. Interestingly, our work demonstrates that the expression of TNFα by cancer cells is an important strategy to co-opt stromal fibroblasts to undertake tumour-promoting functions. Inflammation associated with wound healing is usually self-limiting; however, in chronic inflammation the response is sustained. Similarly, although paracrine signalling by TNFα promotes CAF function, the removal of paracrine cues does not completely abolish the invasion promoting ability of CAFs (compare column 2&3 Figure 6a). Interestingly, we find that continued invasion promoting abilities of CAF are dependent on IKK (data not shown), but TNFα independent. We do not know the upstream activator in this context: our data exclude IL-1 and LPS. More broadly, anti-inflammatory agents such as COX-2 inhibitors have shown promise as chemo-prevention agents in certain clinical contexts and are well documented to reduce cancer cell invasion (39, 40).
Our data also suggests that blocking either IL-1 or TNFα signalling may reduce carcinoma invasion, in part through targeting stromal fibroblasts. This is of particular interest since both IL-1R antagonists such as anakinra (Kineret®) and TNFα blockers such as infliximab (Remicade®) are in routine clinical use for the treatment of several chronic inflammatory conditions including rheumatoid arthritis, psoriasis and Crohn’s disease. Indeed, we find that infliximab reduces SCC invasion in our system indicating that it could potentially be exploited as an anti-invasive cancer drug. This would be of considerable benefit to patients with SCC of the head and neck (SCCHN) in which metastatic spread along the cervical chain of lymph nodes is associated with a markedly worse clinical prognosis. In the future, it would be interesting to validate these findings in in vivo models of metastasis. The future role of IL-1 and TNFα antagonists in cancer chemotherapy will depend on a number of factors. Firstly, a greater understanding of the nature of IL-1 and TNFα-associated epithelial-stromal interactions in early and late-stage malignancy is required. Secondly, criteria for identifying the patients and the tumours most likely to respond to such therapy are needed. Thirdly, anti-TNFα agents have complex side-effects (41) and their efficacy and safety in the setting of the medically compromised cancer patient must be evaluated further.
In conclusion, siRNA-based screening of ubiquitin and ubiquitin-like regulators has identified a key role for Traf6 in SCC invasion. Furthermore, we have revealed the interconnected but distinct roles of autocrine IL-1 signalling in carcinoma cells and paracrine TNFα signalling from carcinoma cells to CAFs in promoting invasion. This work suggests new strategies for reducing cancer cell invasion using anti-inflammatory agents.
Materials and Methods
Cell lines
A431 cells (human vulval SCC) were grown in DMEM with 10% FCS in 10% CO2 at 37°C. SCC12 cells (cutaneous SCC cell line) were cultured in FAD 85 medium supplemented with 10% FCS, 5μg/ml insulin (Invitrogen, UK), 0.5μg/ml hydrocortisone and 10ng/ml EGF. Human HNCAF (head and neck carcinoma-associated fibroblasts) were cultured in DMEM with 10% FCS and insulin-transferrin-selenium in 10% CO2 at 37°C.
Transient DNA transfections
A431 cells were transfected with FuGENE® 6 transfection reagent (Roche, UK). Briefly, sub-confluent cells were washed and the media replaced with Opti-MEM®. The previously prepared DNA-liposome mix (6μl FuGENE® 6 + 1.5μg DNA for 6 well plates or 2.25μl FuGENE® 6 + 0.75μg DNA for 24 well plates) was then pipetted onto the cells and swirled briefly. After 5 hours of incubation in 10% CO2 at 37°C the transfection mix was removed, the cells washed with PBS and fresh DMEM with 10% FCS added. pUNO-hTRAF6-HA was purchased from InvivoGen™ (Autogen Bioclear UK Ltd.). Cezanne/ZA20D1 plasmids were a gift from Paul Evans (Hammersmith Hospital, UK) and IkB super-repressor plasmid was a gift from Krishna Rajalingam (Frankfurt, Germany).
siRNA transfections
Cells were cultured in standard conditions (5, 42). A431 and SCC12 cells were transfected using DharmaFECT™ 2 (Dharmacon®, UK). HNCAF were transfected using DharmaFECT™ 1 (Dharmacon®, UK). The final concentration of siRNA in all transfections was 100nM for screens and organotypic assays and 75nM for all other assays. 0.5μl, 2μl and 5μl DharmaFECT™ reagent was used for transfecting 48 well, 24 well and 6 well plates respectively. TRAF6#2 GGACAAAGUUGCUGAAAUCUU; Dharmacon® D-004712-02; TRAF6#3 GUUCAUAGUUUGAGCGUUAUU; Dharmacon® D-004712-03 TRAF6#B UACGCGAACGUGGAAGUCAAU; Qiagen® S103050145; ZA20d1#2 GAAUCUAUCUGCCUUUGGAUU; Dharmacon® D-008670-02 ZA20d1#3 GAAGGAGAAUACCAAGGAAUU; Dharmacon® D-008670-03 ZA20d1#A UUCCUCCGAUUUGUUUCAAUU; Qiagen® S100764869.
siRNA libraries and screening
Dharmacon® E3 Ubiquitin Ligase Library
ANKIB1 M-028950-01 ANKRD11 M-027299-01 ARIH2 M-020104-01 MARCH7 M-007055-01 BAZ1A M-006941-00 BBAP M-007143-01 BIRC2 M-004390-02 BIRC3 M-004099-02 BIRC4 M-004098-01 C17ORF27 M-023324-02 CBL M-003003-02 CBLB M-003004-02 CBLC M-006962-00 CBLL1 M-007069-00 CCNB1IP1 M-019251-01 CDC26 M-017290-00 CHD5 M-009878-00 RFWD2 M-007049-01CUL7 M-017673-00 DCST1 M-016740-01 DET1 M-019393-00 DKFZP547C195 M-026520-01 DKFZP564A022 M-007078-01 DKFZP761H1710 M-007082-00 DPF1 M-031779-01 DTX3 M-007156-01 DTX4 M-026663-01 DZIP3 M-006910-00 EGFL9 M-014254-00 FANCL M-021486-00 FBXL14 M-015718-00 FBXL17 M-024385-00 FBXO17 M-013128-00 FBX018 M-017404-00 FBXO27 M-018558-00 C1ORF164 M-021044-01 FLJ12875 M-007062-02 FLJ20186 M-020685-01 FLJ20315 M-007004-02 FLJ23749 M-007108-01 FLJ30092 M-027115-00 FLJ31951 M-007146-01 FLJ35757 M-007150-02 FLJ36180 M-007174-01 FLJ45273 M-027218-02 FLJ46380 M-032070-01 HACE1 M-007193-01 HECTD2 M-007198-00 FLJ21156 M-027468-03 NEDL1 M-007186-01 HECW2 M-007192-00 HERC2 M-007180-02 HOZFP M-021465-00 HRD1 M-007090-00 HUWE1 M-007185-01 IBRDC2 M-025119-00 ITCH M-007196-01 KIAA0317 M-007184-02 KIAA0804 M-023668-00 KIAA1333 M-007014-00 KIAA1542 M-026727-00 KIAA1991 M-032290-00 KIAA2025 M-021878-00 LNX2 M-007164-01 LOC285498 M-008019-00 LOC389822 M-029504-00 LOC442247 M-032292-01 LOC493829 M-032293-00 LOC51136 M-010205-01 LOC51255 M-006985-00 LRSAM1 M-007103-01 MAPKBP1 M-022225-00 FLJ20668 M-007006-02 LOC51257 M-006986-00 MARCH3 M-007116-02 MARCH9 M-015561-01 MDM2 M-003279-04 GRN1 M-022620-00 LOC142678 M-015287-00 MID2 M-006938-00 MKRN1 M-006959-00 MKRN2 M-06960-01 MKRN3 M-006581-01 MNAB M-020453-02 MYCBP2 M-006951-01 MYLIP M-006976-00 MYST3 M-019849-01 NEDD4 M-007178-02 NHLRC1 M-027323-00 NSMCE1 M-007157-01 PDZRN3 M-023265-01 PHF7 M-019179-00 PJA1 M-007045-01 PJA2 M-006916-00 PRP19 M-004668-02 RCHY1 M-006966-01 RFPL1 M-006553-01 RFPL2 M-006935-01 RFPL3 M-006934-00 RFWD3 M-017095-01 RKHD1 M-031965-00 RKHD2 M-006989-00 RKHD3 M-022292-00 RNF10 M-006918-01 RNF103 M-006594-00 RNF11 M-006971-01 RNF111 M-007002-01 RNF12 M-006982-01 RNF121 M-007011-02 RNF122 M-007068-00 RNF123 M-007041-00 RNF125 M-007005-01 RNF126 M-007015-01 RNF127 M-007067-01 RNF128 M-007061-01 TRIM60 M-007153-00 RNF13 M-006944-01 RNF130 M-007021-00 RNF133 M-007155-00 PCGF6 M-007084-01 RNF135 M-007087-01 RNF137 M-007007-01 RNF138 M-006991-01 RNF141 M-006980-00 RNF146 M-007080-00 RNF148 M-010758-00 RNF149 M-07169-00 RNF150 M-010713-01 RNF151 M-030777-00 RNF152 M-007160-00 MARCH5 M-007001-00 RNF157 M-022965-01 RNF159 M-007089-00 RNF166 M-007119-00 RNF167 M-006967-00 FLJ35794 M-007152-00 RNF17 M-007026-00 LOC285533 M-007170-00 RNF18 M-007030-00 LOC285671 M-017714-00 MGC33993 M-007162-00 MGC4734 M-007134-00 FLJ10546 M-020828-00 FLJ38628 M-007107-00 FLJ20225 M-006999-00 RNF187 M-021939-00 RNF19 M-006965-00 RNF2 M-006556-00 RNF20 M-007027-00 RNF24 M-006943-00 RNF25 M-007047-00 RNF26 M-007060-00 RNF28 M-007093-00 TRIM55 M-007092-00 RNF3 M-006926-00 TRIM54 M-007032-00 RNF34 M-007072-00 RNF36 M-007137-00 RNF39 M-007074-00 RNF40 M-006913-00 RNF41 M-006922-00 RNF44 M-006947-00 RNF5 M-006558-00 RUFY1 M-016355-00 SH3MD2 M-007037-00 SH3RF2 M-007145-00 SHPRH M-007167-00 SIAH1 M-012598-01 SIAH2 M-006561-01 SMC4L1 M-006837-00 SMURF1 M-007191-00 SMURF2 M-007194-00 STUB1 M-007201-01 SUGT1 M-012877-00 MARCH6 M-006925-00 TMEPAI M-010501-00 TRAF7 M-007086-00 TRIM10 M-006920-00 TRIM11 M-007075-00 TRIM15 M-007102-00 TRIM17 M-006981-00 TRIM2 M-006955-00TRIM25 M-006585-00 TRIM3 M-006931-00 TRIM31 M-006939-00 TRIM34 M-006997-00 TRIM35 M-006952-01 TRIM36 M-007012-00 TRIM38 M-006929-00 TRIM39 M-007028-00TRIM4 M-007101-00 TRIM40 M-007129-00 TRIM41 M-007105-00 TRIM42 M-007173-00 TRIM43 M-007127-00 TRIM45 M-007073-00 TRIM46 M-007071-00 TRIM47 M-007106-00 TRIM48 M-007059-00 TRIM5 M-007100-00 TRIM50A M-007130-00 TRIM50C M-031736-00 TRIM52 M-007095-00 TRIM56 M-007079-00 TRIM6 M-007121-00 FLJ10759 M-007010-00 LOC201292 M-018490-00 TRIM67 M-032288-00 TRIM7 M-007077-00 TRIM9 M-012974-00 TTC3 M-006570-00 UBE2L6 M-008569-00 UBE2N M-003920-01 UBQLN2 M-013566-00 VPS11 M-007022-00 VPS41 M-006972-00 KIAA1374 M-023638-00 FLJ36175 M-007203-00 WSB1 M-013015-00 WSB2 M-017223-00 WWP1 M-004251-00 WWP2 M-004252-01 ZC3HDC5 M-022950-00 ZFPL1 M-019375-00 ZFYVE19 M-017961-00 ZNF179 M-006588-00 ZNF183 M-006590-00 ZNF183L1 M-007135-00 ZNF294 M-006968-00 ZNF313 M-007024-00 ZNF364 M-006974-00 ZNF598 M-007104-00 FLJ25735 M-007148-00 ZNF650 M-016653-00ZNRF1 M-007098-00 ZNRF2 M-007165-00 FLJ12747 M-010747-00 ZNRF4 M-007141-00 ZSWIM2 M-007142-00 ARIH1 M-019984-00 HERC4 M-021426-00 HERC5 M-005174-01 HERC6 M-005175-02 UBE3C M-007183-00 UBE3B M-007197-01 UBE3A M-005137-00 MID1 M-006537-00 SSA1 M-006563-02 TRIM28 M-005046-01 TRIM32 M-006950-00 TRIM37 M-006538-01 TRIM23 M-006523-00 TRIM33 M-005392-02 TRAF6 M-004712-00 UHRF1 M-006977-00 UHRF2 M-007117-00 UBCE7IP5 M-006949-00 AMFR M-006522-00 DTX2 M-007114-01 MID1 M-006537-00 PJA2 M-006916-00 RNF139 M-006942-00 RNF144 M-006912-00 RNF14 M-006906-01 RNF7 M-006907-01 RNF8 M-006900-00 RNF6 M-006559-00 RBX1 M-004087-01 LRSAM1 M-007103-00 UBAP1 M-017474-00 UBAP2 M-013168-00 ANUBL1 M-017760-00 UBL3 M-012786-00 UBL4 M-020658-00 UBL5 M-014320-00 BARD1 M-003873-00 MDM4 M-006536-01 UBR1 M-010691-01 UBR2 M-006954-00 EDD1 M-007189-01 CHFR M-007018-01 TRIAD3 M-017305-00 PARK2 M-003603-00 NEDD4L M-007187-01 NEDD8 M-020081-00 SAE1 M-006402-00 UBA2 M-005248-00 PML M-006547-00 RFP M-006552-00.
Dharmacon® E2 Ligase and De-ubiquitinating Enzyme Library
TXN3L M-024927-00 BAP1 M-005791-00 COPS5 M-005814-01 CXORF53 M-005798-02 CYLD M-004609-00 DUB1A M-028352-00 DUB3 M-027332-00 LOC391622 M-028355-00 FBX07 M-013606-00 FBX08 M-012435-00 FLJ14981 M-018630-00 JOSD1 M-017674-00 MJD M-012013-00 MYSM1 M-005905-01 OTUB1 M-021061-00 OTUB2 M-010983-00 OTUD1 M-026487-00 OTUD4 M-009927-00 OTUD5 M-013823-00 OTUD6B M-008553-00 OTUD7A M-008841-00 PARP11 M-016115-00 PRPF8 M-012252-01 PSMD14 M-006024-00 SBBI54 M-015500-00 SENP2 M-006033-00 SHFM3P1 M-009706-01 STAMBP M-012202-00 STAMBPL1 M-005783-01 TNFAIP3 M-009919-00 UBE1 M-004509-00 UBE1C M-005249-00 UBE1DC1 M-006405-00 UBE1L M-019759-00 UBE2A M-009424-00 UBE2B M-009930-00 UBE2C M-004693-01 UBE2D1 M-009387-01 UBE2D2 M-010383-01 UBE2D3 M-008478-00 UBE2E1 M-008850-01 UBE2E2 M-031782-00 UBE2E3 M-008845-00 UBE2G1 M-010154-01 UBE2G2 M-009095-01 UBE2H M-009134-00 UBE2I M-004910-00 UBE2J1 M-007266-00 UBE2J2 M-008614-00 UBE2L3 M-010384-01 UBE2L6 M-008569-00 UBE2M M-004348-00 UBE2N M-003920-01 UBE2NL M-031625-00 UBE2Q M-008631-01 UBE2R2 M-009700-01 UBE2S M-009707-00 UBE2V2 M-008823-00 UBE3A M-005137-00 UBE3B M-007197-01 UBE4A M-007200-00 UBE4B M-007202-01 UBL3 M-012786-00 UBL4 M-020658-00 UBL5 M-014320-00 UBR1 M-010691-01 UBTD1 M-018385-00 UCHL1 M-004309-00 UCHL3 M-006059-00 UCHL5 M-006060-01 UEV3 M-008494-00 UFD1L M-017918-00 USP1 M-006061-01 USP10 M-006062-01 USP11 M-006063-00 USP12 M-027148-00 USP13 M-006064-00 USP14 M-006065-01 USP15 M-006066-00 USP16 M-006067-00 USP18 M-004236-02 USP19 M-006068-01 USP2 M-006069-00 USP20 M-006070-01 USP21 M-006071-00 USP22 M-006072-00 USP24 M-006073-01 USP25 M-006074-01 USP26 M-006075-01 USP28 M-006076-01 USP29 M-006077-00 USP3 M-006078-01 USP30 M-021294-02 USP31 M-022513-00 USP32 M-006080-01 USP33 M-006081-00 USP34 M-006082-00 USP35 M-006083-01 USP36 M-006084-01 USP37 M-006086-01 USP38 M-006086-01 USP39 M-006087-00 USP4 M-004974-01 USP40 M-006088-00 USP41 M-031434-00 USP42 M-006089-00 USP43 M-023019-00 USP44 M-006091-00 USP45 M-010054-00 USP46 M-006092-02 USP47 M-006093-00 USP48 M-006079-01 TRFP M-006094-00 USP5 M-006095-00 USP50 M-031837-00 USP51 M-032247-00 USP52 M-021192-00 USP53 M-027186-00 USP54 M-016853-00 USP6 M-006096-02 USP7 M-006097-00 USP8 M-005203-01 USP9X M-006099-01 USP9Y M-006100-01 VCPIP1 M-019137-00 YOD1 M-027369-00 ZA20D1 M-008670-00 ZRANB1 M-009270-00 SENP8 M-004071-00 SENP5 M-005946-01 SENP3 M-006034-00 SENP1 M-006357-00 PSMD7 M-009621-01 LOC220594 M-016888-01 MGC5306 M-014314-01 EIF3S3 M-003883-00 COPS8 M-015831-01.
Controls
CTRL #1 D-001210-01 CTRL #2 D-001210-02 ITGB1 #1 D-004506-01 RHOA #1 D-003860-04.
1 × 104 A431 cells were seeded per well in two 48 well plates and transfected as previously described. Forty-eight hours after transfection, the cells were trypsinised and plated in 250μl of standard media on deformable collagen-Matrigel® gels in 8 well Lab-Tek™ II chambered coverglasses (NUNC™, UK). Cells were allowed to adhere for a further 24 hours before fixing with 4% paraformaldehyde (PFA) for 10 minutes. For visualisation, cells were permeabilised with 0.25% Triton® X-100 for 10 minutes and stained with TRITC phalloidin and DAPI.
Antibody methods
Immunofluorescence: Cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes. For visualisation, cells were permeabilised with 0.25% Triton® X-100 for 10 minutes and stained with TRITC phalloidin, DAPI and the following antibodies: Traf6 (Abcam#33915 - 1:100), anti-HA epitope (12CA5 - 1:100), Cy5 Donkey anti-Rabbit (Jackson Stratech) and Alexa 555 Donkey anti-Mouse (Molecular Probes). Western blotting: SDS-PAGE and transfer to Protran nitrocellulose membrane was carried out using standard techniques. Membranes were blocked with 5% BSA before incubation with the following antibodies TRAF6 1:2000 #33915 Abcam®, IκB α 1:1000 #4814 Cell Signaling Technology®, Phospho-IκBα (Ser32) 1:1000 #2859 Cell Signaling Technology® in TBS+0.1%Tween20.
Organotypic invasion assays
The organotypic culture system was set up as previously described (5, 43). To make 1ml of organotypic matrix the following reagents were mixed: Matrigel® (11.2mg/ml) 200μl, Rat tail collagen type I (9.03mg/ml) 400μl, 5X αMEM with sodium bicarbonate 80μl, FCS 100μl FAD medium with 10% FCS 120 μl, and 5 × 105 HNCAF suspended in FAD medium with 10% FCS 100μl. 900μl of this solution was aliquoted into wells of a 24 well plate and allowed to polymerise for 30 minutes at 37°C. Once the gels were set, 1ml of complete media was added per well and the gels left to equilibrate at 37°C for 24 hours before changing the media daily thereafter. A fibroblast-free collagen gel mix was also used to coat sterile nylon discs (Millipore, UK). After incubating at 37°C for 20 minutes, the gel coated discs were fixed in 1% glutaraldehyde for 1 hour, washed four times in PBS, twice in complete media and then stored at 4°C. At 72 hours, 5 × 105 SCC12 cells were seeded on to each gel in complete media. The next day, the gels were lifted on to individual collagen coated nylon discs resting on stainless steel bridges placed in 6 well plates. Sufficient complete media was then added to feed each organotypic culture from beneath the bridge thus allowing the epithelial layer to grow at an air-liquid interface. Media was changed daily throughout the duration of the assay. One week following gel preparation, the cultures were fixed in either 4% paraformaldehyde (PFA) with 0.25% gluteraldehyde in PBS or 10% normal buffered formalin (NBF). After 4 hours, specimens were bisected, transferred to 70% EtOH and embedded in paraffin wax on their cut edge. 4μm sections taken at 50μm intervals were stained with haematoxylin and eosin for quantitative analysis. The processing, embedding and staining of samples was performed by the Cancer Research UK Experimental Histopathology Laboratory. The Invasion Index was subsequently calculated by using the formula 1 – (Non-invading Area / Invading Area) where 0 represented no invasion.
Transwell-Matrigel invasion assays
A431 cells were trypsinized one day after transfection (described above) and ~20 000 cells were plated into the upper chamber of a 24 well BD Bioscience Matrigel invasion chamber (#354480). When cells had adhered the media in the upper chamber was changed to serum free DMEM and the lower chamber was filled with DMEM containing 10% FCS and 10ng/ml EGF. Three days later the cells were fixed and stained with TRITC-phalloidin (described above). Confocal microscopy was used to observe the number of cells above and below the filter. These were then manually counted when the images were viewed using ImageJ software.
ELISA procedure
A431 or SCC12 cells were stimulated with 1ng ml IL-1β for 4hrs. The supernatant was cleared by centrifugation. Levels of TNF-α were measured by ELISA as per the protocol using commercial ELISA-based kits (R&D systems #DTA00C)
Matrix remodelling assays
The contraction of Collagen-Matrigel® matrices was measured as previously described. Briefly, 5×105 HNCAF were seeded in the same Collagen-Matrigel® mixture used for organotypic invasion assays. After 72 hours the matrices were photographed and the degree of gel contraction was then assessed by using Image J software to measure the surface area of the gel relative to its well. The Contraction Index was subsequently calculated by using the formula 1 - (Gel Area / Well Area). Microscopic analysis was carried by seeding 2.5×104 HNCAF in 50μl of the same Collagen-Matrigel® mixture used for organotypic invasion assays. These matrices were overlaid with naive FAD 85 medium supplemented with 10% FCS, 5μg/ml insulin (Invitrogen, UK), 0.5μg/ml hydrocortisone and 10ng/ml EGF or the same media conditioned by SCC12 cells over a 36 hour period. In some cases either 31ng/ml TNFα or 31μg/ml infliximab (Remicade®) was also added. Infliximab was a gift of Martin Gore, The Royal Marsden Hospital, UK.
Traf6 immunohistochemistry
Formalin-fixed paraffin embedded material: Paraffin embedded tissue oral SCC tissue arrays (#OR601) were purchased from US Biomax Inc.. Following dehydration, endogenous peroxidase activity was blocked using 1.6% hydrogen peroxide (H2O2) in PBS for 10 minutes followed by washing in distilled water. Antigen retrieval was performed by immersing samples in sodium citrate buffer and microwaving three times for 5 minutes at full power. Sections were then cooled to room temperature in running water and washed in PBS prior to blocking in goat serum diluted to 10% in 1% BSA for 30 minutes. Slides were incubated with primary antibody in 1% BSA for 1 hour at room temperature. After washing in PBS-Tween20 solution for 1 minute, biotinylated goat anti-rabbit secondary antibody (1:250 in 1% BSA) was then applied to the sections for 45 minutes, also at room temperature. Samples were washed as above and developed microscopically with VECTASTAIN® Elite ABC Reagent and Vector DAB substrate (Vector Laboratories Ltd., UK). A final wash in water was performed before the sections were counterstained lightly with haematoxylin, dehydrated, cleared and mounted. Omission of the primary antibody acted as a negative control. TRAF6 immunostained sections were viewed at 20x magnification and 3 images per sample captured. Digital images were loaded into NIS Elements software (Nikon UK Ltd) and 10 representative immunostained areas were selected for analysis per sample. TRAF6 immunostaining was cytosolic in nature and therefore cell nuclei were not included in the regions analysed. The mean absolute pixel intensity within each region of interest was measured and normalised to a ‘white balanced’ area of the image by subtracting the background pixel intensity. This normalised value was then expressed on a scale of 0 to 1, with 0 representing absolute whiteness and 1 absolute blackness. The mean and standard error of the 10 measurements was calculated to give an overall measure of mean pixel intensity for TRAF6 per sample. Frozen sections: Frozen sections of SCCHN and matched normal tissue samples were collected at St George’s Hospital, London, in accordance with NHS Research Ethics Committee (ref 06/Q0403/125) and St George’s Healthcare NHS Trust Research Office (ref 07.0003). Samples were prepared for confocal microscopy as described in ‘Antibody methods’ section. Images were acquired using an LSM510 confocal microscope.
Supplementary Material
Supplementary Figure 1: A) Schematic outline of E3 ligase and DUB screening. B) F-actin organisation in Detroit 562 and SCC12 transfected with Traf6 and ZA20D1 siRNA. C) i) Quantification of the long axis of Detroit 562 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. ii) Quantification of the percentage of Detroit 562 cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. iii) Quantification of the long axis of SCC12 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. iv) Quantification of the percentage of SCC12 cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. D) Invasion assays E) List of 29 genes that gave consistent F-actin cell morphology changes when depleted with multiple independent siRNA together with their requirement for SCC invasion (as judged by siRNA smart-pool mediated depletion in SCC12 cells).
Supplementary Figure 2: IKK inhibitors block IκB degradation. A431 cells were treated with TNFα (31ng/ml) for the times indicated in the presence of either IKK inhibitor (a) or (b). Western blot analysis shows the levels of pIκB, IκB and β-tubulin.
Supplementary Figure 3: A) Validation of Traf6 antibody on formalin-fixed paraffin embedded material. Cells pellets of either mock transfected or Traf6 siRNA#2 or #3 transfected A431 cells were formalin fixed, paraffin embedded and sectioned before staining with a Traf6 antibody or no primary antibody (control). Samples were counter-stained with haematoxylin. B) Traf6 staining of three oral SCC biopsies and three normal oral mucosa biopsies. Left-hand panels show staining in the absence of Traf6 primary antibody. Samples were counter-stained with haematoxylin. C) Quantification of Traf6 staining in multiple regions of oral SCC and normal oral mucosa biopsies. Also, included for comparison is quantification of Traf6 levels in A431 SCC cells. D) Traf6 staining in patient matched oral SCC and normal oral mucosa biopsies. β-catenin staining is also shown to emphasise epithelial and carcinoma tissue. E) Validation of Traf6 antibody. Either mock transfected or Traf6 siRNA#smartpool transfected A431 cells were fixed, paraffin embedded and stained with a Traf6 antibody (white). Samples were counter-stained with phalloidin (red). F) Traf6 staining of one vulval SCC biopsy and one normal vulval mucosa biopsy. Left-hand panels show staining in the absence of Traf6 primary antibody. Samples were counter-stained with haematoxylin. G) Quantification of Traf6 staining in multiple regions of vulval SCC and normal oral mucosa biopsies. Also, included for comparison is quantification of Traf6 levels in A431 SCC cells.
Supplementary Figure 4: A431 but not HNCAF respond to IL-1. Either A431 or HNCAF cells were treated with either IL-1α (1000pg/ml) or BSA for the times indicated. Western blot analysis shows the levels of pIκB, IκB and β-tubulin.
Supplementary Figure 5: Engagement of TLR2 or 4 does not affect SCC12 invasion. SCC12 organotypic cultures were set up in the presence of HNCAF and treated with either 100ng/ml LPS or 100μg/ml DI-C. Graph shows invasion relative to untreated control cultures.
Supplementary Figure 6: mRNA analysis in SCC12 cells. A) QRT-PCR showing IL-1β mRNA levels normalised to GAPDH in SCC12 cells treated with or without 1 hour stimulation with IL-1β (1000pg/ml). B) QRT-PCR showing IL-1β mRNA levels normalised to GAPDH in SCC12 cells transfected with Traf6 siRNA smartpool or control. C) QRT-PCR showing TNFα mRNA levels normalised to GAPDH in SCC12 cells transfected with Traf6 siRNA smartpool or control.
Acknowledgements
We thank Cancer Research UK for funding. We thank St George’s Thomas Tatum Head and Neck Unit staff involved in tissue collection, Martin Gore for providing reagents, and lab members for advice and discussions.
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- 1.Gil Z, Fliss DM. Contemporary management of head and neck cancers. Isr Med Assoc J. 2009 May;11(5):296–300. [PubMed] [Google Scholar]
- 2.Shah JP, Gil Z. Current concepts in management of oral cancer--surgery. Oral Oncol. 2009 Apr-May;45(4-5):394–401. doi: 10.1016/j.oraloncology.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao M, Epstein JB, Modi BJ, Pytynia KB, Mundt AJ, Feldman LE. Current surgical treatment of squamous cell carcinoma of the head and neck. Oral Oncol. 2007 Mar;43(3):213–223. doi: 10.1016/j.oraloncology.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 4.De Wever O, Nguyen QD, Van Hoorde L, Bracke M, Bruyneel E, Gespach C, et al. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. Faseb J. 2004 Jun;18(9):1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- 5.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007 Dec;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006 May;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 7.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009 Jun 12;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010 Feb 17;17(2):135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Sanz-Moreno V, Gaggioli C. E. ROCK and JAK1 signalling cooperate to control actomyosin contractility in tumour cells and stroma. Cancer Cell. doi: 10.1016/j.ccr.2011.06.018. al. in press. [DOI] [PubMed] [Google Scholar]
- 10.Stuelten CH, DaCosta Byfield S, Arany PR, Karpova TS, Stetler-Stevenson WG, Roberts AB. Breast cancer cells induce stromal fibroblasts to express MMP-9 via secretion of TNF-alpha and TGF-beta. J Cell Sci. 2005 May 15;118(Pt 10):2143–2153. doi: 10.1242/jcs.02334. [DOI] [PubMed] [Google Scholar]
- 11.Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001 Jun;13(6):389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 12.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000 Oct 13;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 13.Heyninck K, Beyaert R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-kappaB activation at the level of TRAF6. FEBS Lett. 1999 Jan 15;442(2-3):147–150. doi: 10.1016/s0014-5793(98)01645-7. [DOI] [PubMed] [Google Scholar]
- 14.Wojciak-Stothard B, Entwistle A, Garg R, Ridley AJ. Regulation of TNF-alpha-induced reorganization of the actin cytoskeleton and cell-cell junctions by Rho, Rac, and Cdc42 in human endothelial cells. J Cell Physiol. 1998 Jul;176(1):150–165. doi: 10.1002/(SICI)1097-4652(199807)176:1<150::AID-JCP17>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard S, Guilak F. Effects of interleukin-1 on calcium signaling and the increase of filamentous actin in isolated and in situ articular chondrocytes. Arthritis Rheum. 2006 Jul;54(7):2164–2174. doi: 10.1002/art.21941. [DOI] [PubMed] [Google Scholar]
- 16.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006 Oct;16(10):522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003 Dec 5;302(5651):1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009 Sep 24;35(6):841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, et al. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol. 2010 Nov;12(11):1078–1085. doi: 10.1038/ncb2112. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, Heyninck K, et al. A novel type of deubiquitinating enzyme. J Biol Chem. 2003 Jun 20;278(25):23180–23186. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 21.Evans PC, Taylor ER, Coadwell J, Heyninck K, Beyaert R, Kilshaw PJ. Isolation and characterization of two novel A20-like proteins. Biochem J. 2001 Aug 1;357(Pt 3):617–623. doi: 10.1042/0264-6021:3570617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang KZ, Wara-Aswapati N, Boch JA, Yoshida Y, Hu CD, Galson DL, et al. TRAF6 activation of PI 3-kinase-dependent cytoskeletal changes is cooperative with Ras and is mediated by an interaction with cytoplasmic Src. J Cell Sci. 2006 Apr 15;119(Pt 8):1579–1591. doi: 10.1242/jcs.02889. [DOI] [PubMed] [Google Scholar]
- 23.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009 Aug 28;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystrom ML, Thomas GJ, Stone M, Mackenzie IC, Hart IR, Marshall JF. Development of a quantitative method to analyse tumour cell invasion in organotypic culture. J Pathol. 2005 Mar;205(4):468–475. doi: 10.1002/path.1716. [DOI] [PubMed] [Google Scholar]
- 25.Ahn KS, Sethi G, Aggarwal BB. Nuclear factor-kappa B: from clone to clinic. Curr Mol Med. 2007 Nov;7(7):619–637. doi: 10.2174/156652407782564363. [DOI] [PubMed] [Google Scholar]
- 26.Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008 Mar;28(5):1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009 Jun;16(6):658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enesa K, Zakkar M, Chaudhury H, Luong le A, Rawlinson L, Mason JC, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008 Mar 14;283(11):7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 29.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010 Aug;17(8):939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse WK, Eisenhaber B, Ho SH, Ng Q, Eisenhaber F, Jiang YJ. Genome-wide loss-of-function analysis of deubiquitylating enzymes for zebrafish development. BMC Genomics. 2009;10:637. doi: 10.1186/1471-2164-10-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krelin Y, Voronov E, Dotan S, Elkabets M, Reich E, Fogel M, et al. Interleukin-1beta-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer Res. 2007 Feb 1;67(3):1062–1071. doi: 10.1158/0008-5472.CAN-06-2956. [DOI] [PubMed] [Google Scholar]
- 32.Szlosarek PW, Balkwill FR. Tumour necrosis factor alpha: a potential target for the therapy of solid tumours. Lancet Oncol. 2003 Sep;4(9):565–573. doi: 10.1016/s1470-2045(03)01196-3. [DOI] [PubMed] [Google Scholar]
- 33.Scott KA, Moore RJ, Arnott CH, East N, Thompson RG, Scallon BJ, et al. An anti-tumor necrosis factor-alpha antibody inhibits the development of experimental skin tumors. Mol Cancer Ther. 2003 May;2(5):445–451. [PubMed] [Google Scholar]
- 34.Moore RJ, Owens DM, Stamp G, Arnott C, Burke F, East N, et al. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nat Med. 1999 Jul;5(7):828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 35.Jeong JG, Kim JM, Cho H, Hahn W, Yu SS, Kim S. Effects of IL-1beta on gene expression in human rheumatoid synovial fibroblasts. Biochem Biophys Res Commun. 2004 Nov 5;324(1):3–7. doi: 10.1016/j.bbrc.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Hanna AN, Berthiaume LG, Kikuchi Y, Begg D, Bourgoin S, Brindley DN. Tumor necrosis factor-alpha induces stress fiber formation through ceramide production: role of sphingosine kinase. Mol Biol Cell. 2001 Nov;12(11):3618–3630. doi: 10.1091/mbc.12.11.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gronowicz G, Hadjimichael J, Richards D, Cerami A, Rossomando EF. Correlation between tumor necrosis factor-alpha (TNF-alpha)-induced cytoskeletal changes and human collagenase gene induction. J Periodontal Res. 1992 Nov;27(6):562–568. doi: 10.1111/j.1600-0765.1992.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith PC, Guerrero J, Tobar N, Caceres M, Gonzalez MJ, Martinez J. Tumor necrosis factor-alpha-stimulated membrane type 1-matrix metalloproteinase production is modulated by epidermal growth factor receptor signaling in human gingival fibroblasts. J Periodontal Res. 2009 Feb;44(1):73–80. doi: 10.1111/j.1600-0765.2007.01081.x. [DOI] [PubMed] [Google Scholar]
- 39.Elmets CA, Viner JL, Pentland AP, Cantrell W, Lin HY, Bailey H, et al. Chemoprevention of nonmelanoma skin cancer with celecoxib: a randomized, double-blind, placebo-controlled trial. J Natl Cancer Inst. 2010 Dec 15;102(24):1835–1844. doi: 10.1093/jnci/djq442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurihara Y, Hatori M, Ando Y, Ito D, Toyoshima T, Tanaka M, et al. Inhibition of cyclooxygenase-2 suppresses the invasiveness of oral squamous cell carcinoma cell lines via down-regulation of matrix metalloproteinase-2 production and activation. Clin Exp Metastasis. 2009;26(5):425–432. doi: 10.1007/s10585-009-9241-3. [DOI] [PubMed] [Google Scholar]
- 41.Rosenblum H, Amital H. Anti-TNF therapy: safety aspects of taking the risk. Autoimmun Rev. 2011 Jul;10(9):563–568. doi: 10.1016/j.autrev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Hidalgo-Carcedo C, Hooper S, Chaudhry SI, Williamson P, Harrington K, Leitinger B, et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol. 2011 Jan;13(1):49–58. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper S, Gaggioli C, Sahai E. A chemical biology screen reveals a role for Rab21-mediated control of actomyosin contractility in fibroblast-driven cancer invasion. Br J Cancer. Jan 19;102(2):392–402. doi: 10.1038/sj.bjc.6605469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: A) Schematic outline of E3 ligase and DUB screening. B) F-actin organisation in Detroit 562 and SCC12 transfected with Traf6 and ZA20D1 siRNA. C) i) Quantification of the long axis of Detroit 562 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. ii) Quantification of the percentage of Detroit 562 cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. iii) Quantification of the long axis of SCC12 cells following mock, control, Traf6 or ZA20D1 depletion. Min, Max, quartiles and mean are shown, >50 cells analysed for each condition, * indicates p<0.01 one way ANOVA. iv) Quantification of the percentage of SCC12 cells with microspikes >7.5μm. >50 cells analysed for each condition, * indicates p<0.01 t-test. D) Invasion assays E) List of 29 genes that gave consistent F-actin cell morphology changes when depleted with multiple independent siRNA together with their requirement for SCC invasion (as judged by siRNA smart-pool mediated depletion in SCC12 cells).
Supplementary Figure 2: IKK inhibitors block IκB degradation. A431 cells were treated with TNFα (31ng/ml) for the times indicated in the presence of either IKK inhibitor (a) or (b). Western blot analysis shows the levels of pIκB, IκB and β-tubulin.
Supplementary Figure 3: A) Validation of Traf6 antibody on formalin-fixed paraffin embedded material. Cells pellets of either mock transfected or Traf6 siRNA#2 or #3 transfected A431 cells were formalin fixed, paraffin embedded and sectioned before staining with a Traf6 antibody or no primary antibody (control). Samples were counter-stained with haematoxylin. B) Traf6 staining of three oral SCC biopsies and three normal oral mucosa biopsies. Left-hand panels show staining in the absence of Traf6 primary antibody. Samples were counter-stained with haematoxylin. C) Quantification of Traf6 staining in multiple regions of oral SCC and normal oral mucosa biopsies. Also, included for comparison is quantification of Traf6 levels in A431 SCC cells. D) Traf6 staining in patient matched oral SCC and normal oral mucosa biopsies. β-catenin staining is also shown to emphasise epithelial and carcinoma tissue. E) Validation of Traf6 antibody. Either mock transfected or Traf6 siRNA#smartpool transfected A431 cells were fixed, paraffin embedded and stained with a Traf6 antibody (white). Samples were counter-stained with phalloidin (red). F) Traf6 staining of one vulval SCC biopsy and one normal vulval mucosa biopsy. Left-hand panels show staining in the absence of Traf6 primary antibody. Samples were counter-stained with haematoxylin. G) Quantification of Traf6 staining in multiple regions of vulval SCC and normal oral mucosa biopsies. Also, included for comparison is quantification of Traf6 levels in A431 SCC cells.
Supplementary Figure 4: A431 but not HNCAF respond to IL-1. Either A431 or HNCAF cells were treated with either IL-1α (1000pg/ml) or BSA for the times indicated. Western blot analysis shows the levels of pIκB, IκB and β-tubulin.
Supplementary Figure 5: Engagement of TLR2 or 4 does not affect SCC12 invasion. SCC12 organotypic cultures were set up in the presence of HNCAF and treated with either 100ng/ml LPS or 100μg/ml DI-C. Graph shows invasion relative to untreated control cultures.
Supplementary Figure 6: mRNA analysis in SCC12 cells. A) QRT-PCR showing IL-1β mRNA levels normalised to GAPDH in SCC12 cells treated with or without 1 hour stimulation with IL-1β (1000pg/ml). B) QRT-PCR showing IL-1β mRNA levels normalised to GAPDH in SCC12 cells transfected with Traf6 siRNA smartpool or control. C) QRT-PCR showing TNFα mRNA levels normalised to GAPDH in SCC12 cells transfected with Traf6 siRNA smartpool or control.