Abstract
CONSTANS (CO) is an important flowering-time gene in the photoperiodic flowering pathway of annual Arabidopsis thaliana in which overexpression of CO induces early flowering, whereas mutations in CO cause delayed flowering. The closest homologs of CO in woody perennial poplar (Populus spp.) are CO1 and CO2. A previous report [1] showed that the CO2/FLOWERING LOCUS T1 (FT1) regulon controls the onset of reproduction in poplar, similar to what is seen with the CO/FLOWERING LOCUS T (FT) regulon in Arabidopsis. The CO2/FT1 regulon was also reported to control fall bud set. Our long-term field observations show that overexpression of CO1 and CO2 individually or together did not alter normal reproductive onset, spring bud break, or fall dormancy in poplar, but did result in smaller trees when compared with controls. Transcripts of CO1 and CO2 were normally most abundant in the growing season and rhythmic within a day, peaking at dawn. Our manipulative experiments did not provide evidence for transcriptional regulation being affected by photoperiod, light intensity, temperature, or water stress when transcripts of CO1 and CO2 were consistently measured in the morning. A genetic network analysis using overexpressing trees, microarrays, and computation demonstrated that a majority of functionally known genes downstream of CO1 and CO2 are associated with metabolic processes, which could explain their effect on tree size. In conclusion, the function of CO1 and CO2 in poplar does not appear to overlap with that of CO from Arabidopsis, nor do our data support the involvement of CO1 and CO2 in spring bud break or fall bud set.
Introduction
The CONSTANS (CO) gene encodes a zinc finger transcription factor that plays a major role in the photoperiodic flowering pathway of the annual and facultative long-day plant Arabidopsis thaliana [2]. Mutations in CO cause delayed flowering under long days in Arabidopsis, but do not affect flowering time relative to wild-type plants grown under short days, suggesting that CO promotes flowering under long days. CO transcripts are present in leaves and shoots and promote flowering in a dosage-dependent manner in Arabidopsis. Constitutive overexpression of CO from the cauliflower mosaic virus (CaMV) 35S promoter induces earlier flowering relative to wild-type plants grown under short or long days [3], [4]. CO transcript abundance follows a circadian rhythm, where high CO mRNA levels coincide with long days, but are also seen in darkness under short days [5]–[7]. CO protein is degraded in darkness by a CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1, an E3 ubiquitin ligase)-dependent mechanism when plants are grown under short days [8], but light stabilizes CO under long days through cryptochrome 2 (cry2) and phytochrome A (phyA) [9]. These mechanisms ensure the accumulation of CO protein only under long days, thus enabling flowering.
CO initiates flowering via upregulation of FLOWERING LOCUS T (FT) in the companion cells of leaf phloem in Arabidopsis [6], [10]–[15]. Following induction, the FT protein appears to be translocated through phloem to the shoot apex, where it forms a protein complex with FD (bZIP transcription factor), which upregulates APETALA1 to initiate floral development [16]–[20].
In Arabidopsis, the CO gene family contains CO and 16 CONSTANS-LIKE (COL) genes within the B-box family of zinc finger proteins [21], [22]. Although COL1 and COL2 are under control of a circadian clock, with a peak in transcript levels at dawn, constitutive expression of COL1 or COL2 did not cause early- or late-flowering phenotypes in Arabidopsis [23], indicating that the function of these genes does not overlap with that of CO. The COL3 protein, which physically interacts with the COP1 protein, appears to be a positive regulator of root growth, because col3 mutants show reduced formation of lateral roots [24]. The col3 mutant flowers early under both long and short days, suggesting that COL3 does not promote flowering, but may be a general repressor instead. Moreover, reduced shoot branching was observed on the col3 mutant under short days, indicating that COL3 regulates shoot branching in a day-length-dependent manner. COL5 transcription is under circadian regulation and is detected in vascular tissues, but its role in controlling flowering time is unclear [25]. Constitutive expression of COL9 resulted in plants with delayed flowering, whereas mutants with reduced COL9 transcription flowered early under long days, suggesting that COL9, like COL3, is a floral repressor [26]. These studies show that COL genes are functionally unrelated to CO, with respect to photoperiodic flowering regulation, suggesting that they may have other roles in controlling growth and development.
CO homologs have been isolated from other annual or herbaceous plants such as Japanese morning glory [Pharbitis nil [27], [28]]; rice [Oryza sativa [29]]; potato [Solanum tuberosum ssp. andigena [30]]; wheat [Triticum aestivum [31]]; and ryegrass [Lolium perenne [32]]. These homologous proteins appear to be functionally conserved in photoperiodic flowering, although their coding sequences may be diverging evolutionarily across species. For example, the rice CO homolog (Hd1) inhibits transition to flowering under long days, but promotes it under short days in this short-day plant [29], [33], [34].
In woody perennials, CO homologs have been cloned and characterized for expression [35]–[39]. Two of 18 poplar (Populus spp.) CO-like genes [CO1 (POPTR 0017s14410.1) and CO2 (POPTR 0004s10800.1)] closely cluster with Arabidopsis CO, COL1, and COL2 phylogenetically (see tree in Figure S1). Along with FLOWERING LOCUS T1 (FT1; POPTR_0008s07730.1), CO2 was shown to be part of a mechanism by which poplar controls reproductive onset and fall bud set by sensing critical day lengths [1]. Transcriptional repression of CO2 via RNA interference (RNAi) in poplar appeared to cause sensitivity to short days, initiating early growth cessation and bud set [1]. If CO2 and/or CO1 are functionally conserved in poplar relative to CO in Arabidopsis [3], [4], their constitutive overexpression should induce early reproductive onset and delay fall bud set. To test this hypothesis, we conducted physiological and genetic experiments of CO1 and CO2, including expression analysis, in poplar. Our long-term field experiments showed no evidence for involvement of these genes, singly or in combination, in reproductive onset, spring bud break, or fall bud set, suggesting that CO1 and CO2 in poplar are not functional orthologs of Arabidopsis CO.
Results
CO1 and CO2 Transcripts are Most Abundant during the Growing Season
To conduct transcript analyses reliably via quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), we designed and tested gene-specific primers for CO1 and CO2. The forward and reverse primers for CO1 and CO2 differed by five and seven nucleotides, respectively, out of 28 (Figure S2A). The primer pairs spanned the only intron present in both genes to ensure that genomic DNA, if any remained in the RNA extracts, would not be amplified. When PCR analysis was conducted using plasmid DNA harboring CO1 or CO2 cDNA, no cross-amplification was detected (Figure S2B). Thus, the CO1 and CO2 primer pairs were transcript-specific. Amplicons were cloned, sequenced, and confirmed as CO1 and CO2 (Figure S2B). Given that the sequence of the CO1 and CO2 primer binding sites diverged greatly from the remaining poplar CO genes (Figure S3A–B) and that the overall sequence similarity between the other members of the poplar CO family and CO1 or CO2 is low (45–57%; Figure S3C), we did not expect these primers to amplify the other poplar CO transcripts.
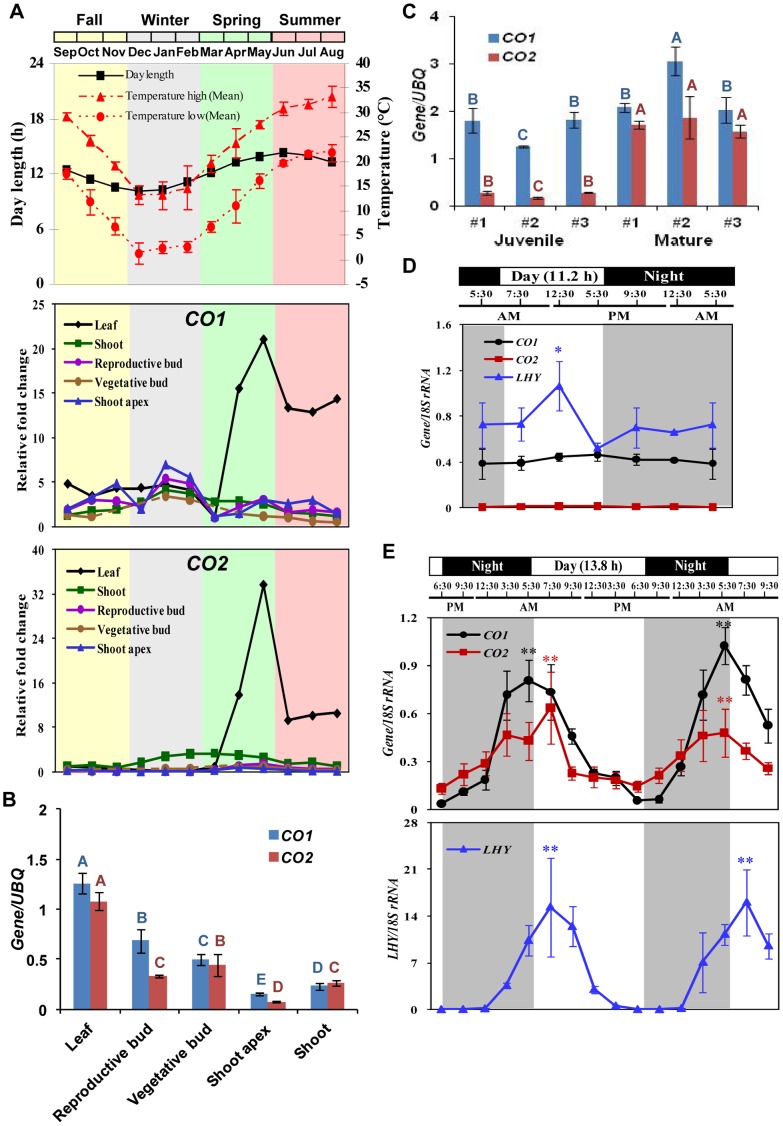
To identify the temporal and spatial expression patterns of CO1 and CO2, we conducted year-round transcript analyses in various tissues of field-grown, wild-type Populus deltoides. Although CO1 was expressed throughout the year at low levels in all five tissues analyzed, its transcripts were most abundant in leaves during the growing season (Figure 1A–B). CO2 was expressed abundantly in leaves in the growing season, but at background levels at other times and in other tissues (Figure 1A–B). Although CO1 transcripts were abundant in leaves of both juvenile and mature trees, CO2 transcripts were significantly (P≤0.001) more abundant in leaves of mature trees than juveniles during the growing season (Figure 1C).
Figure 1. Transcript analysis of CO1 and CO2 via RT-PCR in field-grown P. deltoides.
(A) Average monthly high and low temperatures and daylength in Mississippi (USA) and year-round expression of CO1 and CO2 in mature P. deltoides. CO1 and CO2 graphs show the relative fold change in expression levels normalized against March expression level. Dashed lines indicate missing samples. Error bars show standard deviation about the mean. (B) CO1 and CO2 transcripts were most abundant in leaf tissues of mature P. deltoides sampled in May, but least abundant in the shoot apex. Poplar UBQ was used as an internal control. Letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant (P≤0.001) differences. Error bars indicate SD about the mean. (C) CO1 transcripts were expressed abundantly in juvenile and mature trees (April). However, CO2 transcripts were significantly more abundant in mature and juvenile trees. Letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant (P≤0.001) differences. Error bars indicate SD about the mean. (D) Transcript abundance of CO1 and CO2 did not significantly (P>0.8) fluctuate in leaves sampled in February. LHY transcripts were significantly (P≤0.05) abundant at mid-day in the same tissues. (E) Transcript levels of CO1 and CO2 were significantly (P≤0.001) higher at dawn in leaves sampled in May, whereas LHY was significantly (P≤0.001) more abundant in the morning. *≤0.05 and **≤0.005, statistical significance within time points.
To determine whether expression of CO1 and CO2 fluctuates daily, we analyzed their transcripts in field-grown, wild-type P. deltoides at and after dawn and dusk, and in the middle of the day and the night. When CO1 and CO2 transcripts were analyzed in preformed leaves in February, no significant (P>0.8) differences were found among the six time points analyzed (Figure 1D). In contrast, we detected a significant (P≤0.05) difference in transcript abundance of the positive-control LATE ELONGATED HYPOCOTYL (LHY) gene at midday in the same samples (Figure 1D). In Arabidopsis [40] and poplar [41], LHY shows a circadian expression pattern with a peak in the morning under long days. Conversely, CO1 and CO2 expression showed a rhythm with a periodicity of about 24 h when their transcripts were analyzed in leaves in May. We detected significant differences (P≤0.005) among the 16 time points analyzed over 48 h (Figure 1E). CO1 and CO2 transcripts were significantly (P≤0.001) more abundant at 5∶30 AM (dawn) than at 6∶30 PM (dusk), whereas LHY transcripts were significantly (P≤0.001) more abundant at 7∶30 AM (Figure 1E).
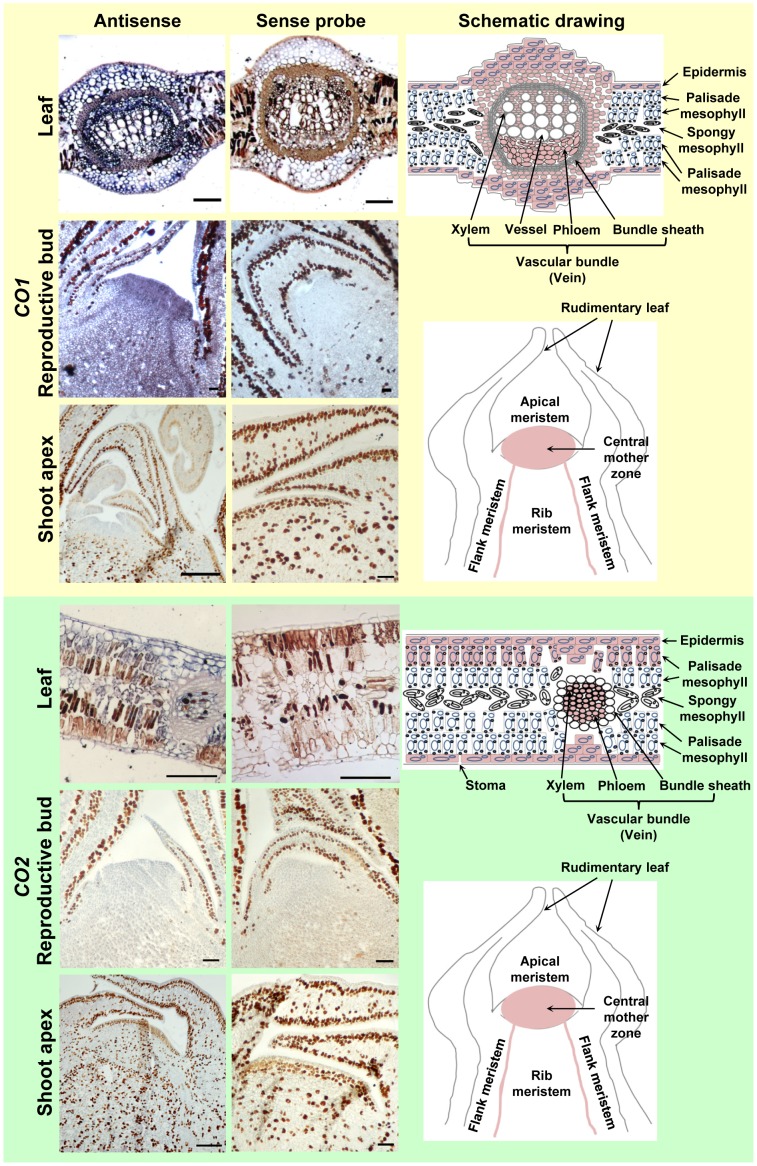
To define where CO1 and CO2 transcripts were expressed, we conducted in situ expression analysis using leaves, reproductive buds, and shoot apices from mature P. deltoides (Figure 2). In leaf tissue, CO1 transcripts were predominantly detected in epidermal, xylem, and phloem cells, as well as in the cells surrounding the vascular bundle. They were not detected in palisade and spongy parenchyma cells. CO1 expression was also largely located in the apical meristem and vasculature of reproductive buds, as well as in the apical meristem and primordial (rudimentary) leaves of the shoot apex. CO2 showed similar expression patterns to CO1, except that CO2 was expressed uniformly throughout the shoot apex. Taken together, these results suggest that transcripts of CO1 and CO2 are most abundant in leaves and show a similar expression rhythm with a peak in transcript levels at dawn during the growing season. Their expression is predominantly confined to epidermal and vascular cells in leaves.
Figure 2. In situ expression analysis of CO1 and CO2 in leaf, reproductive bud, and shoot apex collected during the growing season from mature P. deltoides.
Panels in the first two columns were results from the bright-field image of in situ hybridization and colorimetric detection of CO1 or CO2 transcripts. The antisense probe generated positive signals (dark blue) if present, while the sense probe served as negative control. The third column (schematic drawing) illustrates leaf cross-sections and longitudinal reproductive bud and shoot apex sections where CO1 and CO2 transcripts (pink color) were located, based on visual observations, as well as captured images. Scale bar, 100 µm.
CO1 and CO2 Transcription is not Regulated by Environmental Factors
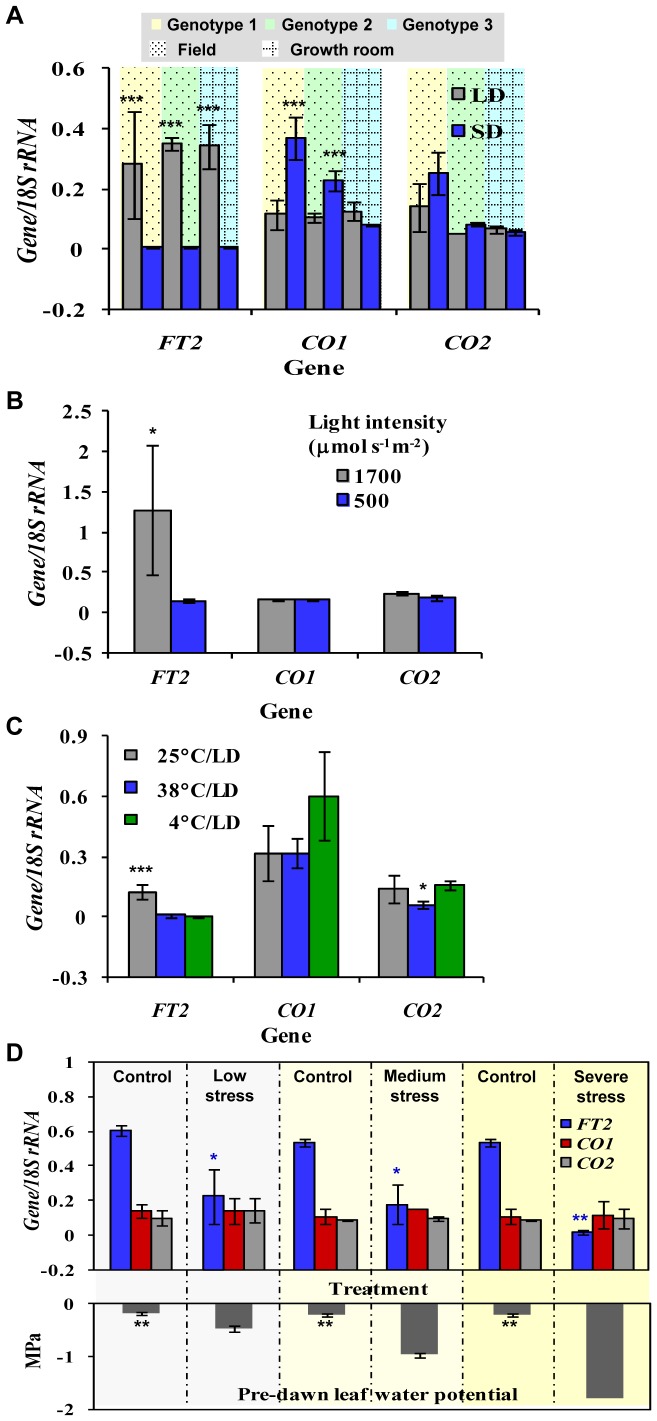
Because CO1 and CO2 are predominantly expressed in leaves during the growing season, and leaves often respond robustly to stress [42], we tested whether day length, light intensity, temperature, and water stress affect CO1 and CO2 transcription in leaves of mature P. deltoides. In the first experiment, shoots with leaves of field-grown trees were maintained under short days (8 h) and ambient long days (12–14 h) from March 25 to May 31, spanning bud break, shoot growth, and expression period of CO1 and CO2. Unlike CO2, CO1 transcripts were significantly (P≤0.0002) higher under short days (Figure 3A). However, when we repeated this experiment in a controlled environment (growth rooms) at 25°C for 42 d in spring, we did not detect a significant (P>0.05) difference between short and long days (Figure 3A). Transcripts of the stress-responsive FLOWERING LOCUS T2 (FT2) gene [43] were significantly (P≤0.0001) less abundant under short days in both field and controlled experiments (Figure 3A). These results suggest that both CO1 and CO2 are not regulated by daylength. However, our data point to an unknown factor that regulates CO1 in the field, but not in a controlled environment.
Figure 3. Environmental regulation of CO1 and CO2 transcription in mature P. deltoides.
(A) Abundance of CO1 transcripts increased (P≤0.0002) under short days (SD) in leaves of field-grown trees (two genotypes), but showed no significant (P>0.05) difference under SD in growth rooms. CO2 expression did not change significantly under SD and long days (LD). FT2 transcripts were significantly (P≤0.0001) less abundant under SD in trees grown both in the field and growth room. Samples were collected 2 h after sunrise or the beginning of the light period. (B) Reduced ambient light intensity did not significantly (P>0.05) affect CO1 and CO2 transcription in field-grown trees. Conversely, transcript abundance of FT2 was significantly reduced (P≤0.008) at the lower light intensity. (C) Temperatures of 38, 25, and 4°C did not significantly (P>0.05) influence the abundance of CO1 and CO2 transcripts under LD, except that CO2 transcripts were significantly (P≤0.05) fewer at 38°C. FT2 transcription was significantly (P≤0.0005) repressed by 38°C and 4°C. (D) While FT2 transcription decreased significantly (P≤0.05) under low, medium, and severe water stress (predawn leaf water potential in MPa), the levels CO1 and CO2 transcripts were not significantly (P>0.05) affected when compared with controls. *≤0.05, **≤0.005, and ***≤0.0005, statistical significance. Error bars represent standard deviation about the mean.
In the second experiment, we tested whether CO1 and CO2 transcription would respond to a change in light intensity. A reduction in the ambient light intensity from 1,700 to 500 µmol s−1 m−2 via continuous shading of field-grown P. deltoides trees for 19 days in May did not significantly (P>0.05) alter transcript abundance of CO1 and CO2, when compared with control trees (Figure 3B). Reduced light intensity, however, significantly (P≤0.008) decreased the abundance of FT2 transcripts (Figure 3B). In the third experiment, we determined whether temperature stress regulates the transcription of CO1 and CO2. Actively growing, mature P. deltoides trees were subject to 4, 25, and 38°C under long days for 14 days in May when CO1 and CO2 transcripts are normally abundant in leaves. The abundance of CO1 transcripts did not significantly (P>0.05) differ among the three temperature regimes, whereas CO2 transcript abundance was significantly (P≤0.05) lower at 38°C (Figure 3C). FT2 transcript abundance was significantly (P≤0.0005) less at both 4 and 38°C than at 25°C (Figure 3C). These results suggest that heat stress represses CO2 transcription, but not that of CO1. In the final experiment, we tested whether water stress influences CO1 and CO2 transcription. Potted rooted cuttings of P. deltoides actively growing under ambient conditions were subject to low (−0.48 MPa), medium (−0.95 MPa), and severe (−1.8 MPa) water stress (predawn leaf water potential) for 19 days in May. Control plants were watered regularly and had a significantly (P≤0.005) higher predawn leaf water potential of about −0.2 MPa (Figure 3D). Transcript abundance of CO1 and CO2 did not differ significantly (P>0.05) among any of the water-stress regimes (Figure 3D). However, FT2 transcription was significantly (P≤0.05) suppressed under the stress regimes (Figure 3D). Collectively, these experiments revealed that while CO1 transcription was increased by an unknown factor under field conditions and CO2 transcription was repressed by heat stress, we did not observe any effects of day length, light intensity, low temperature, and water stress on the regulation of CO1 and CO2 transcription.
Overexpression of CO1 and CO2 Failed to Alter Normal Reproductive Onset and Fall Bud Set
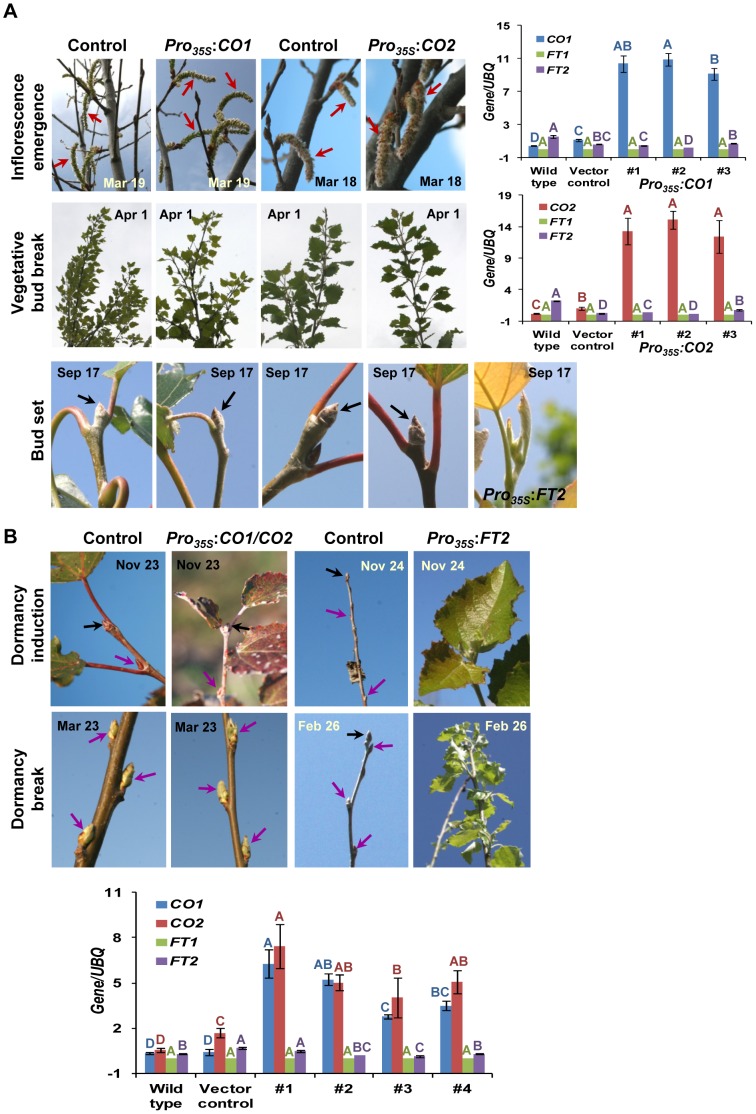
Two binary vectors were used to overexpress the protein-coding regions of CO1 and CO2 under the control of the CaMV 35S promoter, designated Pro35S:CO1 and Pro35S:CO2, respectively. These constructs were independently transformed into poplar clone 717-1B4 (P. alba × P. tremula) via an Agrobacterium-mediated protocol. Transformants and controls were planted in the field and observed for five years, spanning all four seasons annually and in both the juvenile and mature stages of development. The transformants significantly (P≤0.001) overproduced CO1 and CO2 transcripts (Figure 4A). However, unlike controls, no significant (P>0.16) difference was detected in expression of either gene between morning and night in leaves of the same transformants (Figure S3D). Both transformants and controls flowered for the first time at age 5 (Figure 4A), and anthesis occurred at a similar time in March for both. Unlike Pro35S:FT2 trees, which did not set buds in the fall or enter dormancy as long as air temperatures stayed above freezing [43], we did not observe any difference between transformants and controls in spring bud break and fall bud set. Pro35S:CO2 trees formed significantly (P≤0.05) fewer flowers per tree and had significantly (P≤0.05) less height and diameter growth when compared with controls at age 5 (Table 1). When we measured the same traits on a cohort of Pro35S:CO2 trees that were transformed and regenerated separately, we found similar results (Table S1). Although Pro35S:CO1 trees were significantly (P≤0.05) shorter, the number of reproductive buds per tree and diameter growth were not significantly (P>0.05) different between transformants and controls at age 5 (Table 1).
Figure 4. Ectopic expression of CO1 and CO2 individually (Pro35S:CO1 or Pro35S:CO2) or together (Pro35S:CO1/CO2) in poplar (P. tremula × P alba).
(A) When compared with controls at age 5, Pro35S:CO1 or Pro35S:CO2 trees did not differ in reproductive onset, spring reproductive and vegetative bud break, and fall bud set. Pro35S:FT2 trees showed year-round active growth. Red arrows denote the emerging inflorescence in the spring, whereas black arrows point the dormant terminal vegetative bud in the fall. Unlike wild-type and vector controls, Pro35S:CO1 or Pro35S:CO2 trees (1, 2, and 3) significantly overproduced CO1 or CO2 transcripts when analyzed via qRT-PCR in leaves sampled in April. While the expression of FT1 was undetectable, that of FT2 fluctuated with no clear trend in controls and CO1- or CO2-overexpressing trees. Letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant (P≤0.001) differences. Error bars indicate SD about the mean. (B) When Pro35S:CO1 and Pro35S:CO2 were co-expressed in the same trees, no difference between the transformants and controls was observed in spring bud break and fall bud set in two years. However, Pro35S:FT2 trees showed a non-dormant phenotype. Black arrows indicate the terminal vegetative bud, whereas purple arrows point to the axillary vegetative bud. The axillary vegetative buds were opening and preformed leaves were emerging from the control and co-expressing transgenic trees on March 23. Unlike wild-type and vector-control plants, co-expressing transgenic trees (1, 2, 3, and 4) significantly overproduced CO1 and CO2 transcripts in leaves sampled in April. While the expression of FT1 was undetectable, that of FT2 fluctuated with no clear trend in controls and CO1/CO2 overexpressing trees. Letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant (P≤0.001) differences. Error bars indicate SD about the mean.
Table 1. Field-grown Pro35S:CO1, Pro35S:CO2, and control trees were observed for the onset of reproduction for five years, evaluated for the number of reproductive buds or catkins at age 5, and measured for height, diameter, and shoot growth at age 5.
| CO1 | |||
| Control | Pro35S:CO1 | ||
| Anthesis | Age | 5 | 5 |
| n | 15 | 55 | |
| # of reproductive buds | Count | 30.4 A | 43.5 A |
| n | 15 | 55 | |
| Height | m | 9.08 A | 6.95 B |
| n | 15 | 55 | |
| Diameter | cm | 6.05 A | 5.72 A |
| n | 15 | 55 | |
| Shoot length | cm | 39.54 A | 28.74 B |
| n (tree) | 11 | 61 | |
| n (total shoots) | 294 | 1440 | |
| CO2 | |||
| Control | Pro35S : CO2 | ||
| Anthesis | Age | 5 | 5 |
| n | 14 | 14 | |
| # of flowers | Count | 305.0 A | 68.43 B |
| n | 14 | 14 | |
| Height | m | 7.63 A | 5.39 B |
| n | 13 | 14 | |
| Diameter | cm | 9.71 A | 4.64 B |
| n | 13 | 14 | |
| Shoot length | cm | 22.51 A | 24.63 A |
| n (tree) | 14 | 14 | |
| n (total shoots) | 783 | 835 |
Differing letters to the right of the mean (superscript) within a row represent a statistical difference (P≤0.05) between the average control and average transformant. Height was measured in meter (m), whereas diameter and shoot length were measured in centimeter (cm).
Because mature trees showed abundant expression of both CO1 and CO2 in leaves in the growing season (Figure 1C), high co-expression of CO1 and CO2 in the same tree may be required for normal reproductive onset and fall bud set. To test this hypothesis, we co-transformed the Pro35S:CO1 and Pro35S:CO2 constructs into 717-1B4. Although the co-transformants significantly (P≤0.001) overproduced CO1 and CO2 transcripts, they neither had flowers nor showed any difference in spring bud break or fall bud set, when compared with controls by the end of year 2 in the field (Figure 4B). These results indicate that CO1 and CO2 are not involved in reproductive onset, spring bud break, and fall bud set.
Poplar CO1 Rescues the Late-flowering Phenotype of Arabidopsis co-1 Mutant Plants
Because the second zinc finger region in CO of the Arabidopsis co-1 mutant plant is defective [2], these plants have a late-flowering phenotype under long days. We transformed co-1 Arabidopsis with Pro35S:CO1 to determine if it could restore the wild-type phenotype. Out of 15, three randomly selected transgenic lines (2, 8, and 10) with high levels of CO1 expression were used in experiments. The Pro35S:CO1 construct was able to rescue the late-flowering phenotype of co-1 Arabidopsis under long days in all three lines (Figure S4A–B). The transformants flowered significantly (P≤0.001) earlier and had fewer leaves than the co-1 mutant plants, but flowered significantly (P≤0.001) later and had more leaves than wild-type controls. The Pro35S:CO1 transformants and wild-type plants flowered within 23.5 and 22.7 days, respectively, whereas co-1 mutants flowered within 29.9 days. Pro35S:CO1 transformants and wild-type plants formed 16.7 and 15.7 leaves, respectively, at flowering, whereas the co-1 mutants formed 25.6 leaves. We then sought to determine whether CO1 upregulates the Arabidopsis FT gene (AtFT). We analyzed the expression of AtFT in wild-type, co-1 mutant, and Pro35S:CO1 plants at three developmental stages (4- and 6-leaf stages, and bolting). A gradual increase in the amount of AtFT transcript was detected in both wild-type and co-1 mutants from the four-leaf to the bolting (flowering) stages, whereas the expression level of AtFT was generally high at all stages in all three Pro35S:CO1 transgenic lines (Figure S4C). These results suggest that poplar CO1 functions similarly to CO in Arabidopsis under long days.
Genes Downstream of CO1 and CO2 are Predominantly Associated with Metabolism
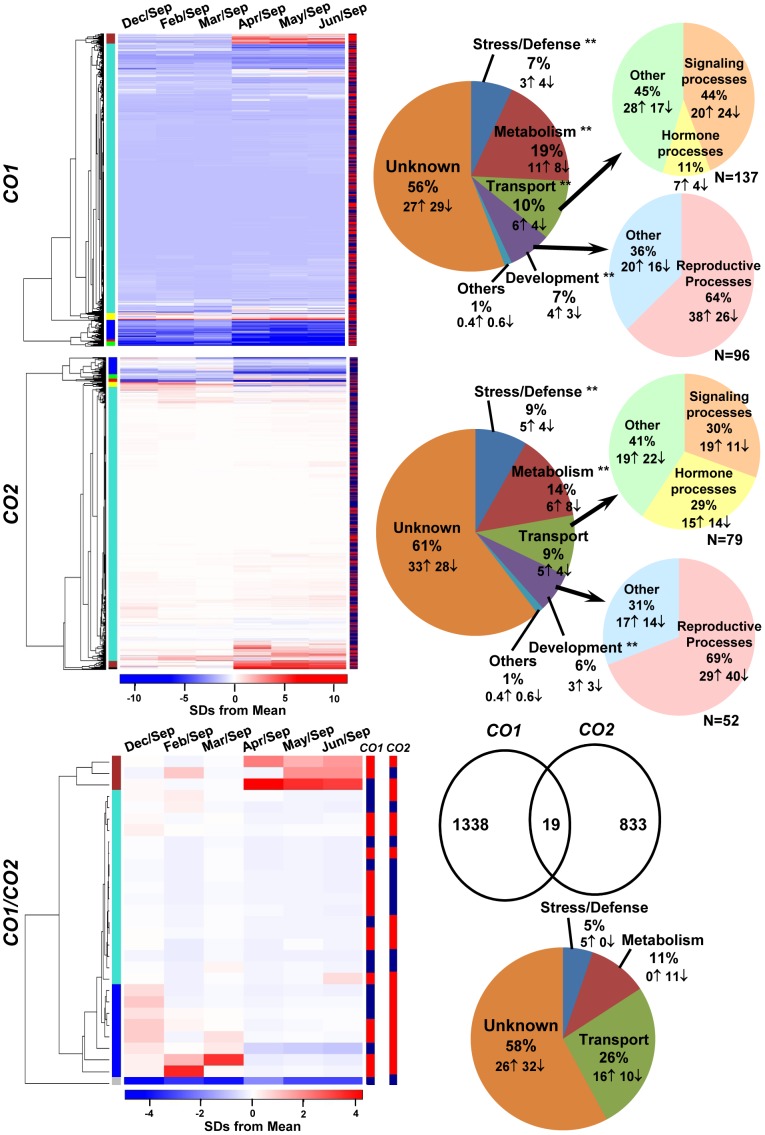
To determine the biological processes CO1 and CO2 are involved in regulating, we conducted microarray experiments comparing leaf transcript profiles of Pro35S:CO1 and Pro35S:CO2 trees with controls. We then evaluated year-round expression of the downstream genes of CO1 and CO2 using another set of previously produced microarray data from leaves of non-transgenic P. deltoides [43]. Cluster analysis and functional classification revealed that a considerable number of genes that act downstream of CO1, CO2, and CO1/CO2 had unknown functions: 56%, 61%, and 58%, respectively (Figure 5; Tables S2, S3, S4). A majority of the downstream genes with assigned putative functions were associated with metabolism: 19% for CO1 and 14% for CO2. A hypergeometric statistical test confirmed that the gene ontology (GO) term “metabolism” associated with genes downstream of CO1 or CO2 was significantly (P≤0.001) over-represented in the microarray data (Figure 5). Other, smaller groups included transport, stress/defense, and development. Although the “development” group made up a small portion of the entire list (7% of CO1, and 6% of CO2), a majority of this group was associated with reproductive processes, including genes similar to Arabidopsis GIGANTEA (GI, POPTR_0005s21870.1); FLAVIN-BINDING, KELCH REPEAT, F BOX protein 1 (FKF1, POPTR_0008s13460.1); circadian responses; and others that are involved in flower development. However, the GO term “reproduction” associated with genes downstream of CO1 or CO2 was not significantly (P>0.05) over-represented according to the hypergeometric test. While a small group of genes downstream of CO1, CO2, and CO1/CO2 was abundantly expressed in the growing season (brown module), others often showed slight fluctuations in expression in leaves.
Figure 5. Transcripts downstream of CO1 and CO2 and their year-round transcript levels were identified in mature P. deltoides via microarray.
Log2 fold-change of each time point relative to the baseline time point (September or Sep) was calculated. Clusters to the left of the heatmaps represent modules and the columns to right of the heatmaps represent the up- (red) and down-regulation (blue) of downstream genes. Months relative to September are above the heatmaps. The pie charts to the right of each heatmap show the functional categorization of GO Biological Process terms. N = number of genes. The Venn diagram shows the number of genes that were common to both CO1 and CO2 (CO1/CO2) datasets, and the pie chart below the diagram shows the GO categorization of CO1/CO2 transcripts. Up (↑) and down (↓) arrows represent partitioning of overall percentage in each pie. “**” denotes the GO term is significantly (P≤0.001, except “development” for genes downstream of CO1 P≤0.006) over-represented in the microarray data when a hypergeometric test was conducted.
Discussion
Based on phylogenetic analyses, CO1 and CO2 in poplar are the closest structural orthologs of CO in Arabidopsis. A previous report [1] showed that the CO2/FT1 regulon controls the onset of reproduction and induction of growth cessation and bud set in poplar. This inference is partly based on observations on trees containing CO2 RNAi constructs showing earlier than normal growth cessation and bud set when they were transferred from long to short days. It is noteworthy that the sequences for the CO1 and CO2 RNAi tag differed by only 9.5%. Therefore, the RNAi construct was expected to knockdown both transcripts. Based on the results, we hypothesized that increased expression of CO1 and CO2 should alter normal reproductive onset and bud set. However, our long-term field trials showed that overexpression of CO1 and CO2 singly or together in poplar does not alter normal reproductive onset, spring bud break, or fall bud set.
Recent findings of Hsu et al. [43] demonstrated that FT1 expression in response to cold (e.g., 4°C) induces the onset of reproduction, as opposed to the findings by Böhlenius et al. [1], who showed that FT1 is expressed and induces reproductive onset during the growing season. If the poplar CO2/FT1 regulon is analogous to the Arabidopsis CO/FT in expression, regulation, and function, as Böhlenius et al. [1] concluded, not only should CO2 overexpression induce the early onset of reproduction, as did CO [3], [4], but also CO2 should be normally and abundantly expressed in leaves in winter along with FT1. However, our data show that not only did overexpression of CO2 not induce early reproduction, but it was not expressed in winter. CO2 was abundantly expressed in leaves only during the growing season. Although CO1 shows a low level of expression in winter, its overexpression does not induce early or late reproductive onset in poplar. Overexpression of CO1 was able to rescue the late-flowering phenotype of the Arabidopsis co-1 mutant plants under long days, albeit at a slower rate than with wild-type plants, indicating that CO1 functions somewhat similarly to CO in Arabidopsis. However, CO1 appears not to be a strong inducer of flowering in Arabidopsis when compared to the overexpression of CO in various mutant backgrounds, such as gi-3, lhy, or fha-1 [6], or to the overexpression of P. nil CO in the co-1 Arabidopsis [28]. Furthermore, our microarray experiments revealed that CO1 and CO2 in poplar are not involved in regulatory networks in the same way as CO in Arabidopsis. For example, CO triggers flowering via upregulation of FT in Arabidopsis leaves [6], [10]–[15], but we did not detect FT-like genes downstream of CO1 or CO2. GI and FKF1 were downstream of CO1 and CO2. In the long-day Arabidopsis flowering pathway, GI forms a protein complex with FKF1, which then binds the CO promoter to regulate its transcription [44]. These observations suggest that the molecular networks controlling reproductive onset may have been modified in poplar and that poplar CO1 and CO2 do not appear to be functional orthologs of Arabidopsis CO.
What, then, are the functions of CO1 and CO2? We observed that CO1- and CO2-overexpressing trees were shorter than controls. In addition, CO2-overexpressing trees grew less in diameter and formed fewer flowers. CO1 and CO2 transcripts were most abundant in leaves in the growing season and showed a diurnal rhythm that peaked at dawn, similar to the COL1 and COL2 circadian expression pattern in Arabidopsis [23]. Our physiological experiments showed that while CO1 transcription was increased by an unknown cue under field conditions and CO2 transcription was repressed by heat stress, other environmental factors, such as day length, light intensity, low temperature, and water stress, did not significantly regulate CO1 or CO2 transcription when their transcripts were consistently measured in the morning. Moreover, our microarray and computation analyses revealed that many known downstream genes of CO1 and CO2 are associated with metabolic processes. Based on this evidence, we hypothesize that CO1 and CO2 are involved in metabolic processes controlling tree size during the growing season. Perhaps, the match between daily CO1 and CO2 rhythms and unidentified environmental factors might contribute to this outcome. Among the functionally characterized CO-like genes in Arabidopsis and other annual plants, COL3 in Arabidopsis is the only gene reported to regulate vegetative production, such as root growth and shoot branching [24]. However, a molecular mechanism has not been identified.
In conclusion, our long-term field observations show that overexpression of two poplar structural orthologs (CO1 and CO2) of Arabidopsis CO does not alter normal reproductive onset, spring bud break, and fall bud set in poplar. CO is critical to regulating reproductive onset under long days in Arabidopsis, but our data indicate that this pathway may have been modified in poplar following the divergence of Arabidopsis and poplar lineages. Given the differences in life-history traits between perennial poplar and annual Arabidopsis, a plausible hypothesis is that poplar either does not use CO function in reproductive onset, or has recruited another gene with a similar function that yet has to be discovered. The fact that CO1- and CO2-overexpressing trees are smaller in size and a majority of known downstream genes of CO1 and CO2 is associated with metabolic processes warrants follow-up experiments on CO1, CO2, and their downstream genes in poplar.
Experimental Procedures
Phylogenetic Analysis
Protein sequences of CO and 16 COLs in Arabidopsis were retrieved from GenBank (http://www.ncbi.nlm.nih.gov). CO homologs were found in the P. trichocarpa genome database (http://www.phytozome.net/poplar.php) using protein-protein BLAST with E values ≤10−5. Multiple alignments were conducted using ClustalX [45]. The resulting alignment was used to generate a phylogenetic neighbor-joining tree via ClustalX. TreeView [46] was used to visualize the tree. Bootstrap analysis was conducted to estimate nodal support based on 1,000 replicates.
Transcript Analysis of CO1 and CO2
The coding sequences for CO1 and CO2 from P. deltoides were aligned to select dissimilar regions for designing gene-specific primers. Primer specificity was tested via PCR using recombinant plasmid DNA containing CO1 or CO2 as described previously [43]. To determine the year-round expression pattern of CO1 and CO2, three independent replications of leaf, shoot, shoot apex, reproductive bud, and vegetative bud tissues from a wild-type, field-grown, sexually mature male P. deltoides tree (30 years old) located near Starkville, MS, USA, were sampled monthly 2 h after sunrise for 12 months. Due to the limited amount of tissue, we pooled the shoot apices into one sample from three replications. Total RNA was isolated as described by Wan and Wilkins [47], which was followed by DNase I digestion and cleanup using the RNeasy Mini Kit (Qiagen; Valencia, CA). Transcript abundance of CO1 and CO2 was analyzed by quantitative real-time (qRT)-PCR using a previously described protocol [43], the Power SYBR Green PCR Master Mix Kit (Applied Biosystems; Foster City, CA), and the 7500 Fast Real-Time PCR system (Applied Biosystems; Foster City, CA). Three technical replicates were performed for each cDNA sample. Poplar UBIQUITIN (UBQ) and 18S rRNA were used as internal controls. Amplicon specificity and primer-dimer formation were monitored by a dissociation curve analysis after each run. Standard curves for CO1, CO2, UBQ, and 18S rRNA were generated by log [cDNA] (represented by the amount of total RNA used in PCR) versus the cycle threshold (CT) using a series of dilutions of first-strand cDNA for each gene. The ratio between CO1 or CO2 and UBQ or 18S rRNA for each sample was calculated using the relative quantitative analysis method [48]. Relative fold change was calculated by normalizing each expression data point for CO1 or CO2 with the expression data point in March. Daily high and low temperature data for 2004 to 2008 were obtained from a nearby weather station (http://ext.msstate.edu/anr/drec/weather.cgi), and a monthly average was calculated over five years. Daylength data were obtained from SunriseSunset (http://www.sunrisesunset.com) for Starkville, MS, USA. The methodology for this section is further described by Hsu et al. [43].
To determine the abundance of CO1 and CO2 transcripts in the shoot apex, vegetative bud (bud #6), reproductive bud (bud #11), shoot, and leaf, a different sexually mature P. deltoides tree was sampled (three independent replicates) 2 h after sunrise in May. To determine the transcript expression of CO1 and CO2 in juvenile and mature P. deltoides, four independent replicates of leaf samples were collected in April from three one-year-old (juvenile) and three mature P. deltoides trees. The juvenile and mature trees were not the same genotype, although they were growing in close proximity to each other. A general linear model was used to test the differences among trees for CO1 or CO2 expression. Means were separated by the Fisher’s protected least significant difference procedure using SAS V9 (SAS Institute; Cary, NC).
To determine whether the abundance of CO1 and CO2 transcripts fluctuated daily, plant material and sampling were as described by Hsu et al. [43], except that we used one mature P. deltoides tree with three independent samples collected at each time point. Poplar LHY was used as a positive control [40], [41], [43]. A general linear model was used to analyze the differences among time points via SAS. Means were separated by the Fisher’s protected least significant difference procedure.
In situ Hybridization
Transcript expression of CO1 and CO2 was determined in the leaf, reproductive bud, and shoot apex. Samples were collected from the same mature tree mentioned above on August 8, 2005 (leaf for CO1), on May 15, 2006 (reproductive bud and shoot apex for CO1 and CO2), and on June 17, 2005 (leaf for CO2). Fixation, dehydration, and clearing of samples were performed according to Jackson [49] with modifications as described in Zhang et al. [50]. Ten-micron sections were sliced with a microtome. Three repeats of the unique sequence from 5′ untranslated region and the beginning of the coding sequence (Table S5) were first PCR-amplified from CO1 or CO2 cDNAs and cloned in tandem into pGEM-T Easy (Promega; Madison, WI). In vitro transcription was conducted with T7 and SP6 RNA polymerase to generate sense or anti-sense mRNA, which was labeled with the DIG RNA Labeling Kit (Roche Diagnostics; Indianapolis, IN) by following the manufacturer’s instructions. Hybridization and detection steps were performed according to Drews et al. [51] and Zhang et al. [50]. For each tissue type and collection time, multiple sections from at least three samples were used for hybridization. Images were taken with an Olympus BX-60 Epi-Fluorescence Microscope equipped with Hamamatsu Orca-100 digital camera.
We did not conduct a cross-reactivity assay for CO1 and CO2 probes in this experiment, because the nucleotide similarity between CO1 and CO2 probes is relatively low (69%). The nucleotide similarity between CO1 or CO2 transcripts and other CO-like transcripts in the poplar genome is also low, ranging from 45% to 57% (Figure S3C). Thus, we expected our probes to be specific for CO1 and CO2 transcripts.
Regulation of CO1 and CO2 Transcription
Daylength, light intensity, temperature, and water stress experiments were performed as described by Hsu et al. [43]. Sample collections were conducted 2 h after sunrise (field) or the beginning of the light period (growth room). Transcript analysis via qRT-PCR was conducted as described above. Poplar FT2 was used as a control in PCR assays because of its involvement in multiple stresses [43]. A general linear model was used to analyze the effect of day length, shade, temperature, or water stress treatment on expression using SAS, and means were separated by the Fisher’s protected least significant difference procedure.
Genetic Manipulation of CO1 and CO2 in Poplar
To determine whether overexpression of CO1 and CO2 alters normal reproductive onset and fall bud set in poplar, the overexpression vectors Pro35S:CO1 and Pro35S:CO2 were constructed. Coding regions of CO1 and CO2 were cloned into the pBI121 binary vector (BD Biosciences; Mountain View, CA) under control of the CaMV 35S promoter. The constructs were individually transformed into 717-1B4 via Agrobacterium tumefaciens (strain C58) [52]. The same poplar clone was also co-transformed with both binary constructs. For co-transformation, the CaMV 35S promoter and coding region of CO1 were cloned into the pCAMBIA1300 binary vector (CAMBIA; Canberra, Australia), which contained hygromycin as a selectable marker. The CO2-containing binary vector was in the pBI121 backbone. The transformants carrying Pro35S:CO1 (19 independent lines), Pro35S:CO2 (9 independent lines), or Pro35S:CO1/CO2 (7 independent lines) were planted along with controls (pBI101 empty-vector or wild-type) at the same developmental stage in the field in a completely randomized design. Each independent line was represented with at least two trees (ramets); and the total number of trees for each construct is provided in Table 1 and Table S1. Transcript abundance of CO1, CO2, FT1, and FT2 was assessed via qRT-PCR in leaf tissues of wild-type, vector-control, Pro35s:CO1, and Pro35s:CO2 trees. Tissues were collected 2 h after sunrise in late April. UBQ was used as an internal control. A general linear model was used to test the differences among trees for CO1, CO2, FT1, and FT2 expression. Means were separated by the Fisher’s protected least significant difference procedure using SAS.
At age 5, Pro35s:CO1, Pro35s:CO2, and control trees were measured for height, diameter, and shoot length. Diameter was measured with a diameter tape at 1 m above ground level. Height was measured with an extendable pole. The first four primary shoots were selected from the apex of the main stem, and the secondary shoots on them were measured. The number of catkins per tree was counted. A t-test was used to detect differences in diameter, height, shoot length, and catkin number between Pro35S:CO1 or Pro35S:CO2 and control trees.
Ectopic Expression of CO1 in Mutant Arabidopsis
Pro35S:CO1 was mobilized into the Arabidopsis co-1 mutant via A. tumefaciens using the floral-dip method [53]. Transformants (15) were selected on ½-strength MS salts containing 50 µg/ml kanamycin. The T3 generation was used for phenotypic assessment. To determine flowering time, Pro35S:CO1 plants along with wild-type (Col-0) and co-1 mutants were grown at 22/19°C (day/night) under long days (16 h) using 27-watt electronic fluorescent flood lights at 115 µmol s−1 m−2. Plants were arranged in a randomized complete block design with four blocks. Each genotype within a block was represented by three plants, for a total of 12 plants per genotype. Flowering time was measured by counting the number of leaves and days from seed sowing to when an inflorescence bud was seen. Analysis of variance was performed in SAS for leaf number and number of days to determine whether significant differences among genotypes could be detected for flowering time. Means were separated by the Fisher’s protected least significant difference procedure.
Leaves from three transgenic lines (#2, #8, and #10), wild-type, and co-1 mutants were collected at the 4-leaf, 6-leaf, and bolting stages. Sample collections were conducted in the morning (8∶00 AM) 2 h from the beginning of the light period. Total RNA was isolated as described above. Expression of CO1 and AtFT was analyzed via traditional RT-PCR as previously described [54]. The 18S rRNA transcript was used as an internal control.
Analysis of CO1 and CO2 Molecular Networks
Three microarray experiments were conducted to identify the genetic networks of CO1 and CO2 using leaves from Pro35S:CO1 and Pro35S:CO2 trees. First, leaf samples were collected in May from four different poplar lines harboring Pro35S:CO1 and four control trees containing the empty vector. Leaf samples within a line were pooled. Eight microarray chips were used for this experiment. Second, leaf samples from four Pro35S:CO2 lines and four vector-control trees were collected as for Pro35S:CO1 lines. Eight microarray chips were used for this experiment. Third, three independent leaf samples from the same mature P. deltoides described above were collected in September 2005, December 2005, February 2005, March 2005, March 2006, April 2006, May 2006, and June 2006, spanning all four seasons, as previously described [43]. Thus, 24 microarray chips were used in all. All Affymetrix GeneChip Poplar Genome Array experiments were conducted as previously described [43], and data were submitted to NCBI GEO (GSE28689 and GSE28693).
To construct clusters of transcripts downstream of CO1 and CO2, probes from CO1 and CO2 microarray data were selected with a log2 expression change of at least 0.5- or 2-fold from control microarray data. The selected probes were mapped over the transcript expression from previous microarray data ([43], Experiment 3, GSE24349). The year-round data were visually represented as the log2 ratio of each time point relative to a baseline time point (September).
Hierarchical clustering was performed on the year-round data that were present in independent CO1 and CO2 datasets. Also, clustering was performed for transcripts common to both CO1 and CO2. In each of the three clustering analyses (i.e., CO1, CO2, and CO1/CO2), co-expression modules were determined using the “Dynamic Hybrid” algorithm [55]. Seven modules were found using the deepSplit = 1 option for both the CO1 and CO2 datasets. Reanalysis of the CO1/CO2 data had only 4 modules using the same deepSplit option. Mapping of Gene Ontology (GO) annotations was performed as detailed in Hsu et al. [43]. This mapping was also conducted on the poplar genome with Arabidopsis ortholog pairings as available at the P. trichocarpa genome database version 2.2, which includes downloadable Arabidopsis annotations. An R script was written to assign these annotations to the poplar genes populating the microarray chips. Following annotation assignments, the number of genes with GO terms pertinent to the four general gene classifications and three sub-classifications in Hsu et al. [43] (e.g., metabolism, stress, reproduction) were counted. The number of genes in each group was then analyzed for over- or under-representation in the entire significant CO1 or CO2 array gene sets. We used methods based on Janz et al. [56] in which the ‘phyper’ function in R calculated a cumulative hypergeometric distribution function and then using the Benjamini-Hochberg correction, and the ‘p.adjust’ R function on the resulting p-values.
Supporting Information
Phylogenetic analysis of CO and CO-like (COL) proteins from Arabidopsis thaliana and poplar ( Populus spp.). The amino acid sequences of zinc finger family proteins, including CO and 16 COL proteins, from A. thaliana (At), 18 COL proteins from P. trichocarpa (POPTR), and CO1 and CO2 proteins from P. deltoides were analyzed using ClustalX and TreeView software. The analysis showed that poplar CO1 and CO2 (or POPTR 0017s14410.1 and POPTR 0004s10800.1, respectively) are the closest homologs of Arabidopsis CO, COL1, and COL2 (gray-boxed). Bootstrap numbers are placed at nodes in the phylogram.
(TIF)
Development and testing of gene-specific primers for analysis of CO1 and CO2 transcripts in poplar. (A) Primer pairs for each transcript were designed based on alignment of nucleotide sequences of CO1 and CO2 cDNAs isolated from P. deltoides. Arrows indicate the locations of forward and reverse primers. (B) PCR amplification was conducted using the designed primer pairs and recombinant plasmids harboring CO1 and CO2 cDNAs. The CO1-specific primer pair only amplified the corresponding region of CO1 cDNA, whereas the CO2-specific primer pair only amplified the corresponding region of CO2 cDNA (the left panel with two gel images). The amplicons were cloned, sequenced, and confirmed as CO1 and CO2 cDNAs (the right panel with nucleotide sequences).
(TIF)
Sequence similarity among 18 poplar CO-like transcripts and constitutive expression of transgenes ( CO1 or CO2 ) in Pro35S : CO1 or Pro35S : CO2 trees. (A and B) Alignment of poplar CO-like transcripts in the region where CO1 and CO2 primers are located. (C) Percent sequence similarity between CO1 or CO2 and other poplar CO family members. (D) Transcript abundance of CO1 in leaves of Pro35S:CO1 trees or CO2 in leaves of Pro35S:CO2 trees was not significantly (P>0.16) different between 7∶30 AM and 9∶30 PM. However, transcripts of CO1 and CO2 in leaves of controls were significantly (P≤0.05) more abundant at 7∶30 AM than at 9∶30 PM. Different letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant differences based on a t test. Error bars indicate SD about the mean.
(TIF)
Ectopic expression of CO1 in A. thaliana and analysis of flowering time under long days. (A) Wild-type (Col-0) and three independent Pro35S:CO1 lines (2, 8, and 10) in the co-1 mutant background were flowered earlier than the co-1 mutant plants. (B) Number of days to flowering and number of leaves at flowering significantly differed (P≤0.001) between Pro35S:CO1 lines in the co-1 mutant background and controls (Col-0 and co-1 mutant plants). Different letters across the bars with the same color indicate that the genotypes significantly differ for flowering time. (C) Abundance of CO1 and AtFT transcripts was analyzed via RT-PCR in wild-type (WT, Col-0), co-1 mutant, and Pro35S:CO1 (2, 8, and 10) plants at three developmental stages of Arabidopsis: 4-leaf, 6-leaf, and bolting. Numbers on the left side represent the size of amplicons in base pair (bp). The 18S rRNA was used as an internal control to verify that similar amounts of cDNA were used in the RT-PCR reaction.
(TIF)
An additional cohort of field-grown Pro35S : CO2 and control trees was observed for the onset of reproduction for five years, evaluated for the number of flowers at age 5, and measured for height, diameter, and shoot growth at age 5. Differing letters to the right of the mean (superscript) within a row represent a statistical difference (P≤0.05) between the average control and average transformant. Height was measured in meter (m), whereas diameter and shoot length were measured in centimeter (cm).
(DOC)
List of CO1, CO2, and CO1/CO2 downstream genes and their associated GO annotations. The list is prepared to match Figure 5.
(XLS)
Normalized expression for all probe sets in the microarray analysis that compared Pro35S:CO1 to controls.
(XLS)
Normalized expression for all probe sets in the microarray analysis that compared Pro35S : CO2 to controls.
(XLS)
List of primers or probes that were used for (q)RT-PCR analyses, vector construction, or in situ hybridization.
(DOC)
Acknowledgments
We thank K.-H. Han, J.-H. Ko, V. Cusimano, L. Vandervelde, T.J. Stoudenmire, J.D. Ellis, H. O’Neal, B.M. Rice, J. Drnevich, D.S. DiLoreto, and K. Deitrick for their assistance.
Funding Statement
This work was funded by National Science Foundation grants DBI-0501890 (CY, GPP, and JEC) and IOS-0845834 (CY). JEC was also partially supported by the World Class University Project R31-2009-000-20025-0 funded by the Ministry of Education, Science, and Technology of South Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, et al. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043. [DOI] [PubMed] [Google Scholar]
- 2. Putterill J, Tobson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- 3. Simon R, Igeno MI, Coupland G (1996) Activation of floral meristem identify genes in Arabidopsis . Nature 384: 59–62. [DOI] [PubMed] [Google Scholar]
- 4. Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roden LC, Song HR, Jackson S, Morris K, Carre IA (2002) Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis . Proc Natl Acad Sci USA 99: 13313–13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, et al. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis . Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- 7. Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis . Nature 419: 308–312. [DOI] [PubMed] [Google Scholar]
- 8. Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, et al. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, et al. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- 10. Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, et al. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis . Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- 11. Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, et al. (1999) Activation tagging of the floral inducer FT . Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 12. Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- 13. Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis . Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- 15. Ayre BG, Turgeon R (2004) Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol 135: 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- 17. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis . Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 18. Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, et al. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis . Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- 19. Jaeger KJ, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis . Curr Biol 17: 1050–1054. [DOI] [PubMed] [Google Scholar]
- 20. Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis . Curr Biol 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- 21. Lagercrantz U, Axelsson T (2000) Rapid evolution of the CONSTANS-LIKE genes in plants. Mol Biol Evol 17: 1499–1507. [DOI] [PubMed] [Google Scholar]
- 22. Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, et al. (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631. [DOI] [PubMed] [Google Scholar]
- 23. Ledger S, Strayer C, Ashton F, Kay SA, Putterill J (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26: 15–22. [DOI] [PubMed] [Google Scholar]
- 24. Datta S, Hettiarachchi GHCM, Deng X-W, Holma M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hassidim M, Harir Y, Yakir E, Kron I, Green RM (2009) Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis . Planta 230: 481–491. [DOI] [PubMed] [Google Scholar]
- 26. Cheng X-F, Wang Z-Y (2005) Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana . Plant J 43: 758–768. [DOI] [PubMed] [Google Scholar]
- 27. Kim S-J, Moon J, Lee I, Maeng J, Kim S-R (2003) Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil . J Exp Bot 54: 1879–1887. [DOI] [PubMed] [Google Scholar]
- 28. Liu J, Yu J, McIntosh L, Kende H, Zeevaart JAD (2001) Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol 125: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, et al. (2000) Hd1, a major photoperiod sensitive quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS . Plant Cell 12: 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez-Garcia JF, Virgos-Soler A, Prat S (2002) Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS . Proc Natl Acad Sci USA 99: 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in rice. Plant J 36: 82–93. [DOI] [PubMed] [Google Scholar]
- 32. Martin M, Storgaard M, Andersen CH, Nielsen KK (2004) Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol Biol 56: 159–169. [DOI] [PubMed] [Google Scholar]
- 33. Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- 34. Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, et al. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Almada R, Cabrera N, Casaretto JA, Ruiz-Lara S, Villanueva EG (2009) VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep 28: 1193–1203. [DOI] [PubMed] [Google Scholar]
- 36. Hattasch C, Flachowsky H, Kapturska D, Hanke M-V (2008) Isolation of flowering genes and seasonal changes in their transcript levels related to flower induction and initiation in apple (Malus domestica). Tree Physiol 28: 1459–1466. [DOI] [PubMed] [Google Scholar]
- 37. Holefors A, Opseth L, Rosnes AKR, Ripel L, Snipen L, et al. (2009) Identification of PaCOL1 and PaCOL2, two CONSTANS-like genes showing decreased transcript levels preceding short day induced growth cessation in Norway spruce. Plant Physiol Biochem 47: 105–115. [DOI] [PubMed] [Google Scholar]
- 38. Jeong D-H, Sung S-K, An G (1999) Molecular cloning and characterization of CONSTANS-LIKE cDNA clones of the Fuji apple. J Plant Biol 42: 23–31. [Google Scholar]
- 39. Yuceer C, Harkess RL, Land Jr SB, Luthe DS (2002) Structure and developmental regulation of CONSTANS-LIKE genes isolated from Populus deltoides . Plant Sci 163: 615–625. [Google Scholar]
- 40. Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, et al. (1998) The late elongated hypocotyl Mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. [DOI] [PubMed] [Google Scholar]
- 41. Takata N, Saito S, Saito CT, Nanjo T, Shinohara K, et al. (2009) Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2 . New Phytol 181: 808–819. [DOI] [PubMed] [Google Scholar]
- 42.Dickson RE, Isebrands JG (1991) Leaves as regulators of stress response. In: Mooney HA, Winner WE, Pell EJ, Chu E, editors. Response of Plants to Multiple Stresses. San Diego, California: Academic Press, Inc. 3–34.
- 43. Hsu C-Y, Adams JP, Kim H, No K, Ma C, et al. (2011) FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA 108: 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawa M, Nusinow DA, Kay SA, maizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis . Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Page RDM (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358. [DOI] [PubMed] [Google Scholar]
- 47. Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12. [DOI] [PubMed] [Google Scholar]
- 48. Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6: 62–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson DP (1991) In situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPhereson M, editors. Molecular Plant Pathology: A Practical Approach. Oxford: Oxford University Press. 163–174.
- 50. Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drews GN, Bowman JL, Meyerowitz EM (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002. [DOI] [PubMed] [Google Scholar]
- 52. Han K, Meilan R, Ma C, Strauss S (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320. [DOI] [PubMed] [Google Scholar]
- 53. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 54. Hsu C-Y, Liu Y, Luthe DS, Yuceer C (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18: 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Langfelder P, Zhang B, Horvath S (2008) Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut library for R. Bioinformatics. 24: 719–720. [DOI] [PubMed] [Google Scholar]
- 56. Janz D, Behnke K, Schnitzler J-P, Kanawati B, Schmitt-Kopplin P, et al. (2010) Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biology 10: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of CO and CO-like (COL) proteins from Arabidopsis thaliana and poplar ( Populus spp.). The amino acid sequences of zinc finger family proteins, including CO and 16 COL proteins, from A. thaliana (At), 18 COL proteins from P. trichocarpa (POPTR), and CO1 and CO2 proteins from P. deltoides were analyzed using ClustalX and TreeView software. The analysis showed that poplar CO1 and CO2 (or POPTR 0017s14410.1 and POPTR 0004s10800.1, respectively) are the closest homologs of Arabidopsis CO, COL1, and COL2 (gray-boxed). Bootstrap numbers are placed at nodes in the phylogram.
(TIF)
Development and testing of gene-specific primers for analysis of CO1 and CO2 transcripts in poplar. (A) Primer pairs for each transcript were designed based on alignment of nucleotide sequences of CO1 and CO2 cDNAs isolated from P. deltoides. Arrows indicate the locations of forward and reverse primers. (B) PCR amplification was conducted using the designed primer pairs and recombinant plasmids harboring CO1 and CO2 cDNAs. The CO1-specific primer pair only amplified the corresponding region of CO1 cDNA, whereas the CO2-specific primer pair only amplified the corresponding region of CO2 cDNA (the left panel with two gel images). The amplicons were cloned, sequenced, and confirmed as CO1 and CO2 cDNAs (the right panel with nucleotide sequences).
(TIF)
Sequence similarity among 18 poplar CO-like transcripts and constitutive expression of transgenes ( CO1 or CO2 ) in Pro35S : CO1 or Pro35S : CO2 trees. (A and B) Alignment of poplar CO-like transcripts in the region where CO1 and CO2 primers are located. (C) Percent sequence similarity between CO1 or CO2 and other poplar CO family members. (D) Transcript abundance of CO1 in leaves of Pro35S:CO1 trees or CO2 in leaves of Pro35S:CO2 trees was not significantly (P>0.16) different between 7∶30 AM and 9∶30 PM. However, transcripts of CO1 and CO2 in leaves of controls were significantly (P≤0.05) more abundant at 7∶30 AM than at 9∶30 PM. Different letters above the bars showing the abundance of CO1 or CO2 transcripts indicate statistically significant differences based on a t test. Error bars indicate SD about the mean.
(TIF)
Ectopic expression of CO1 in A. thaliana and analysis of flowering time under long days. (A) Wild-type (Col-0) and three independent Pro35S:CO1 lines (2, 8, and 10) in the co-1 mutant background were flowered earlier than the co-1 mutant plants. (B) Number of days to flowering and number of leaves at flowering significantly differed (P≤0.001) between Pro35S:CO1 lines in the co-1 mutant background and controls (Col-0 and co-1 mutant plants). Different letters across the bars with the same color indicate that the genotypes significantly differ for flowering time. (C) Abundance of CO1 and AtFT transcripts was analyzed via RT-PCR in wild-type (WT, Col-0), co-1 mutant, and Pro35S:CO1 (2, 8, and 10) plants at three developmental stages of Arabidopsis: 4-leaf, 6-leaf, and bolting. Numbers on the left side represent the size of amplicons in base pair (bp). The 18S rRNA was used as an internal control to verify that similar amounts of cDNA were used in the RT-PCR reaction.
(TIF)
An additional cohort of field-grown Pro35S : CO2 and control trees was observed for the onset of reproduction for five years, evaluated for the number of flowers at age 5, and measured for height, diameter, and shoot growth at age 5. Differing letters to the right of the mean (superscript) within a row represent a statistical difference (P≤0.05) between the average control and average transformant. Height was measured in meter (m), whereas diameter and shoot length were measured in centimeter (cm).
(DOC)
List of CO1, CO2, and CO1/CO2 downstream genes and their associated GO annotations. The list is prepared to match Figure 5.
(XLS)
Normalized expression for all probe sets in the microarray analysis that compared Pro35S:CO1 to controls.
(XLS)
Normalized expression for all probe sets in the microarray analysis that compared Pro35S : CO2 to controls.
(XLS)
List of primers or probes that were used for (q)RT-PCR analyses, vector construction, or in situ hybridization.
(DOC)