Abstract
Nephrolithiasis is a multi-factorial disease which, in the majority of cases, involves the renal deposition of calcium oxalate. Oxalate is a metabolic end product excreted primarily by the kidney. Previous studies have shown that elevated levels of oxalate are detrimental to the renal epithelial cells; however, oxalate renal epithelial cell interactions are not completely understood. In this study, we utilized an unbiased approach of gene expression profiling using Affymetrix HG_U133_plus2 gene chips to understand the global gene expression changes in human renal epithelial cells [HK-2] after exposure to oxalate. We analyzed the expression of 47,000 transcripts and variants, including 38,500 well characterized human genes, in the HK2 cells after 4 hours and 24 hours of oxalate exposure. Gene expression was compared among replicates as per the Affymetrix statistical program. Gene expression among various groups was compared using various analytical tools, and differentially expressed genes were classified according to the Gene Ontology Functional Category. The results from this study show that oxalate exposure induces significant expression changes in many genes. We show for the first time that oxalate exposure induces as well as shuts off genes differentially. We found 750 up-regulated and 2276 down-regulated genes which have not been reported before. Our results also show that renal cells exposed to oxalate results in the regulation of genes that are associated with specific molecular function, biological processes, and other cellular components. In addition we have identified a set of 20 genes that is differentially regulated by oxalate irrespective of duration of exposure and may be useful in monitoring oxalate nephrotoxicity. Taken together our studies profile global gene expression changes and provide a unique insight into oxalate renal cell interactions and oxalate nephrotoxicity.
Introduction
Oxalate is a metabolic end product that is freely filtered at the glomerulus, undergoes bi-directional transport in the renal tubules, and is excreted primarily by the kidney [1]–[3]. The most common pathological condition involving oxalate is the formation of calcium oxalate stones in the kidney [4]. While very high levels of urinary oxalate are observed only in subjects with primary hyperoxaluria, a majority of idiopathic kidney stone patients only show a mild elevation in urinary oxalate [5]–[9] In addition several other conditions associated with oxalate deposits are: renal cysts in acquired renal cystic disease [7], proliferating cells in the kidney [8], hyperplasic thyroid glands [9], and benign neoplasm of the breast [10], [11]. These considerations suggest that the pathological deposition of calcium oxalate is more complex than a simple physical precipitation of calcium oxalate crystals. In 1994, we were the first group to note that oxalate renal cell interactions involved alterations in gene expression [12]. Over the past two decades, studies [13]–[23] have demonstrated that oxalate interactions with renal epithelial cells result in a program of events, including changes in gene expression and cell dysfunction, consistent with cellular stress. Studies from our laboratory demonstrated that oxalate induced changes in renal cells (viz. re-initiation of DNA synthesis) are inhibited by inhibitors of transcription and translation, indicating that the cellular response to oxalate toxicity is dependent on new gene expression and protein synthesis [24]. Moreover, cells of the renal tubular epithelium are exposed to an environment with variable and elevated concentrations of the oxalate and must be able to adapt to oxalate stress. Indeed we have shown that many signal transduction pathways, including p38 MAPK and JNK, are activated in renal epithelial cells in response to oxalate and COM crystals [14], [24]. However, the genetic response of renal epithelial cells to oxalate exposure remains ambiguous.
HK-2 cells are a line of human proximal tubular epithelial cells immortalized by using the E6/E7 genes of human papilloma virus [HPV 16] [25]. These cells retain the characteristics of proximal renal tubular epithelium and have been used successfully as an in vitro model system to represent the human kidney epithelial cells. Previous studies have identified a number of stress and inflammation related genes whose expression is increased in renal epithelial cell cultures responding to oxalate [12], [18], [22]. However, these studies provide limited information on the gene expression program orchestrated in renal epithelial cells in response to increased levels of oxalate. In general it is believed that the effect of oxalate in renal epithelial cells is mediated, at least in part, through the regulation of gene transcription. Therefore to gain insights into molecular events associated with oxalate nephrotoxicity we used HK2 cells and cDNA microarray technology, which allows for the simultaneous analysis of multiple gene expression patterns, to evaluate changes in the global gene expression in renal epithelial cells in response to oxalate [26]. We used Affymetrix hg_u133_plus 2 gene arrays comprised of 54000 probe sets and 1,300,000 distinct oligonucleotide features representing 47,000 transcripts and variants including 38,500 well-characterized human genes. Preliminary studies [27], presented in this report, shows that exposure to oxalate elicits a specific gene expression response: oxalate exposure regulates expression of a much larger number of genes than previously thought. We also demonstrated that in addition to upregulating gene expression oxalate exposure results in the down-regulation of expression of a large set of genes. Moreover, our analysis of this gene expression data also reveals an array of twenty new genes that are differentially regulated by oxalate and could be useful for monitoring oxalate nephrotoxicity. Our studies provide a large amount of the data that may help improve our understanding of the pathogenesis associated with hyperoxaluria in general, and more specifically, with oxalate renal cell interactions.
Results
Treatment with oxalate causes unprecedented changes in gene expression in renal epithelial cells
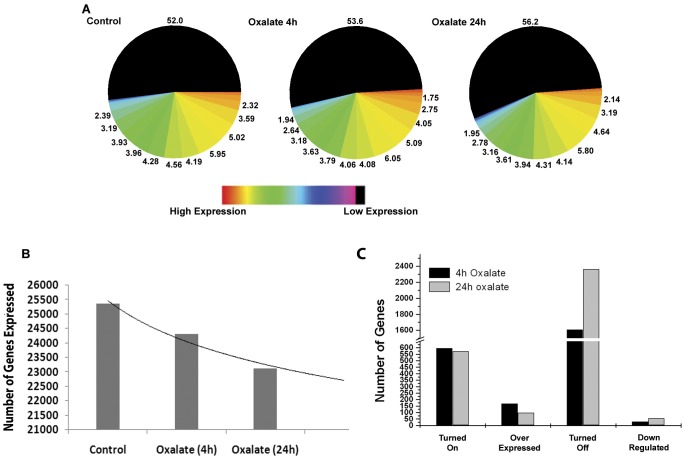
Affymetrix gene chip analysis for changes in gene expression in HK-2 cells upon oxalate exposure identified global changes in the number of genes that are differentially expressed (Figure 1A). We analyzed changes in gene expression upon short term, 4 hours, and long term, 24 hours, exposure of HK-2 cells to oxalate. Of the 47,000 gene expression transcripts and variants analyzed on the gene chip, 26211 transcripts are expressed in the control compared to 25107 transcripts upon short term (4 h) and 23935 transcripts upon long term exposure (24 h) to oxalate (Fig. 1B). These results show that oxalate exposure to the renal cells is associated with the transcriptional silencing of over 8.55 of all transcripts. These results reveal that oxalate exposure, in addition to turning on gene expression, is associated with the significant suppression of gene expression in renal epithelial cells.
Figure 1. Exposure of HK-2 cells to oxalate results in changes in global gene expression.
(A) Levels of gene expression are color coded to depict high to low expression and the numbers represent percentages of genes at each expression level. Representative images from duplicate experiments are shown. (B) Quantitative representation of translational suppression following exposure to Oxalate. (C) Quantitative analysis of differential gene expression in HK2 cells following oxalate exposure. Each data point represents mean of two individual experiments.
Effects of oxalate on the modulation of gene expression in HK2 cells are selective
Even though there is a major reduction in the number of genes that were expressed upon oxalate exposure, the results presented in figure 1C indicate that about 600 genes that were expressed upon oxalate exposure were not expressed in the control cells at all. In addition, these results show over 150 genes are over-expressed in response to oxalate exposure. New genes are turned on in as little as 4 hours exposures to oxalate and are observed even after 24 hours of oxalate exposure. The results presented in figure 1C and table 1 show that oxalate exposure results in the complete silencing of some 1104 transcripts in as little as 4 hours and 2,276 transcripts in 24 hours of exposure. Taken together these results demonstrate the complex nature of oxalate interactions with renal cells and demonstrate that oxalate selectively turns on some sets of genes, while selectively turning off others. This data also suggests that oxalate induced changes seen in renal epithelial cells may be driven by the modulation of gene expression.
Table 1. Changes in gene expression upon exposure of HK-2 cells to oxalate.
| Treatment | Genes Absent | Genes Strongly Expressed | Genes Mildly Expressed |
| Control | 28464 | 25367 | 844 |
| Oxalate (4 h) | 29568 | 24312 | 795 |
| Oxalate (24 h) | 30780 | 23102 | 883 |
Oxalate differentially regulates the expression of specific genes associated with molecular function, biological processes as well as cellular components
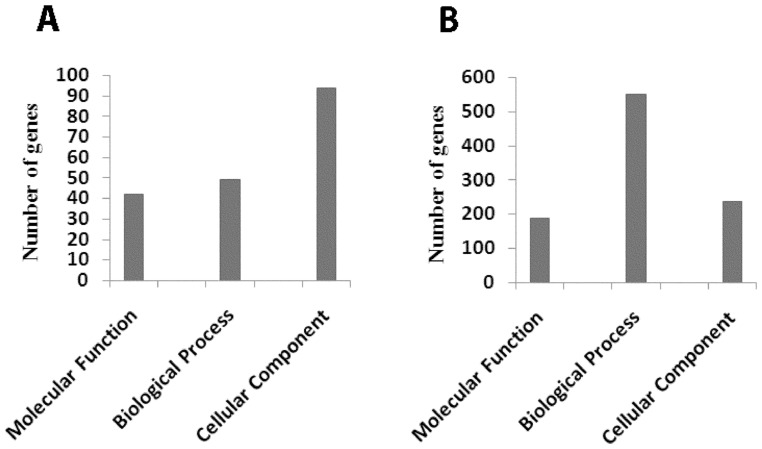
Genes that were either up or down regulated in the oxalate treated cells were categorized based on gene ontology using GenMAPP software. The results presented in Fig. 2A and Table 2 lists genes which are up-regulated greater than fivefold in HK2 cells in response to oxalate exposure. When categorized based on gene ontology (using GenMAPP software), we observed that of the 6,725 genes associated with molecular functions, oxalate exposure resulted in the upregulation of 152 genes. Similarly, of the 16,467 genes associated with biological processes oxalate exposure resulted in the upregulation of 105 genes; and among the 10,435 genes associated with various cellular components oxalate exposure results in the upregulation of 94 of them.
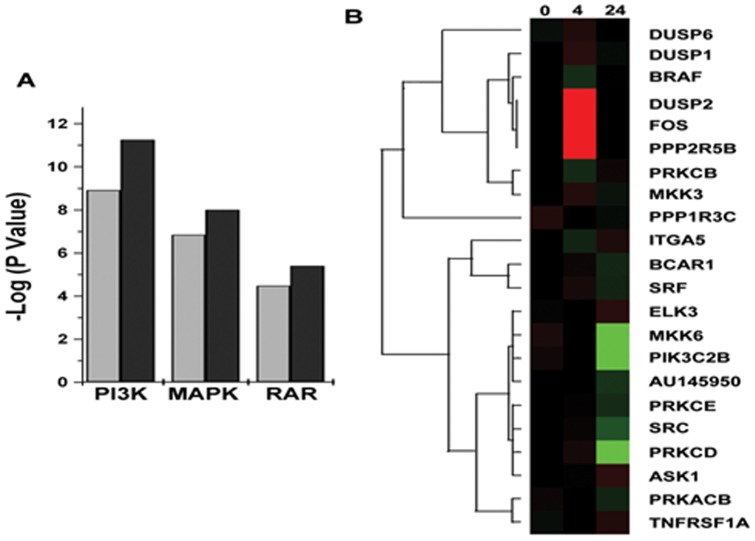
Figure 2. Exposure to oxalate modulates signaling pathways in HK-2 cells.
Changes in the expression of genes involved in various signaling pathways upon exposure of HK-2 cells to oxalate, as determined by Affymetrix gene chip analysis. (A) Changes in gene expression of genes involved in signal transduction pathways after exposure to oxalate. (B) Heat map and cluster diagram represent changes in expression of genes that constitute the Mitogen Activated Protein Kinase (MAPK) pathway. Each data point is representative of the median expression level of two independent gene chip arrays. The Cluster diagram and dendogram was generated using BRB-Array Tools software with a minimum fold change less than 20% of expression data and have at least a 1.5 fold change in either direction from the gene's median expression value.
Table 2. Genes that show an average change more than 5 fold up-regulation compared to control (p value<0.05) upon oxalate exposure grouped based on gene ontology using GenMAPP.
| 2.1 Molecular Function | ||||||||
| Go ID | GO Name | Genes | Genes | Genes | % Changed | % Present | Z Score | p Value |
| Changed | Measured | in GO | ||||||
| 30528 | transcription regulator activity | 15 | 1012 | 1398 | 1.482213 | 72.38913 | 3.664 | 0 |
| 16941 | natriuretic peptide receptor activity | 1 | 1 | 1 | 100 | 100 | 12.651 | 0.001 |
| 19955 | cytokine binding | 3 | 44 | 87 | 6.818182 | 50.57471 | 5.244 | 0.003 |
| 4888 | transmembrane receptor activity | 8 | 429 | 1294 | 1.864802 | 33.15302 | 3.347 | 0.003 |
| 4907 | interleukin receptor activity | 2 | 17 | 32 | 11.76471 | 53.125 | 5.854 | 0.004 |
| 19965 | interleukin binding | 2 | 20 | 38 | 10 | 52.63158 | 5.344 | 0.004 |
| 42610 | CD8 receptor binding | 1 | 1 | 1 | 100 | 100 | 12.651 | 0.006 |
| 3700 | transcription factor activity | 10 | 630 | 936 | 1.587302 | 67.30769 | 3.182 | 0.007 |
| 4872 | receptor activity | 11 | 752 | 1799 | 1.462766 | 41.801 | 3.047 | 0.007 |
| 42609 | CD4 receptor binding | 1 | 2 | 2 | 50 | 100 | 8.89 | 0.008 |
| 4184 | lysine carboxypeptidase activity | 1 | 1 | 2 | 100 | 50 | 12.651 | 0.009 |
| 5142 | interleukin-11 receptor binding | 1 | 1 | 1 | 100 | 100 | 12.651 | 0.009 |
| 51117 | ATPase binding | 1 | 2 | 3 | 50 | 66.66666 | 8.89 | 0.009 |
| 4126 | cytidine deaminase activity | 1 | 2 | 5 | 50 | 40 | 8.89 | 0.009 |
| 4896 | hematopoietin/interferon-class (D200-domain) | 2 | 33 | 54 | 6.060606 | 61.11111 | 3.984 | 0.01 |
| cytokine receptor activity | ||||||||
| 45029 | UDP-activated nucleotide receptor activity | 1 | 2 | 2 | 50 | 100 | 8.89 | 0.011 |
| 16494 | C-X-C chemokine receptor activity | 1 | 3 | 10 | 33.33333 | 30 | 7.214 | 0.011 |
| 5006 | epidermal growth factor receptor activity | 1 | 3 | 7 | 33.33333 | 42.85714 | 7.214 | 0.011 |
| 19958 | C-X-C chemokine binding | 1 | 3 | 11 | 33.33333 | 27.27273 | 7.214 | 0.011 |
| 19957 | C-C chemokine binding | 1 | 4 | 16 | 25 | 25 | 6.208 | 0.012 |
| 16493 | C-C chemokine receptor activity | 1 | 4 | 16 | 25 | 25 | 6.208 | 0.012 |
| 60089 | molecular transducer activity | 13 | 1071 | 2228 | 1.213819 | 48.07002 | 2.603 | 0.012 |
| 4871 | signal transducer activity | 13 | 1071 | 2228 | 1.213819 | 48.07002 | 2.603 | 0.012 |
| 4911 | interleukin-2 receptor activity | 1 | 2 | 3 | 50 | 66.66666 | 8.89 | 0.015 |
| 19976 | interleukin-2 binding | 1 | 2 | 3 | 50 | 66.66666 | 8.89 | 0.015 |
| 43533 | inositol 1\,3\,4\,5 tetrakisphosphate binding | 1 | 2 | 2 | 50 | 100 | 8.89 | 0.016 |
| 43178 | alcohol binding | 1 | 2 | 2 | 50 | 100 | 8.89 | 0.016 |
| 3704 | specific RNA polymerase II transcription | 2 | 31 | 41 | 6.451613 | 75.60976 | 4.139 | 0.016 |
| factor activity | ||||||||
| 46592 | polyamine oxidase activity | 1 | 2 | 2 | 50 | 100 | 8.89 | 0.017 |
| 46983 | protein dimerization activity | 5 | 226 | 294 | 2.212389 | 76.87075 | 3.078 | 0.017 |
| 8528 | peptide receptor activity\, G-protein coupled | 2 | 32 | 115 | 6.25 | 27.82609 | 4.06 | 0.018 |
| 1653 | peptide receptor activity | 2 | 32 | 115 | 6.25 | 27.82609 | 4.06 | 0.018 |
| 43548 | phosphoinositide 3-kinase binding | 1 | 4 | 4 | 25 | 100 | 6.208 | 0.019 |
| 16647 | oxidoreductase activity\, acting on the CH-NH | 1 | 3 | 3 | 33.33333 | 100 | 7.214 | 0.02 |
| group of donors\, oxygen as acceptor | ||||||||
| 15065 | uridine nucleotide receptor activity | 1 | 3 | 3 | 33.33333 | 100 | 7.214 | 0.02 |
| 4383 | guanylate cyclase activity | 1 | 5 | 9 | 20 | 55.55556 | 5.518 | 0.02 |
| 3707 | steroid hormone receptor activity | 2 | 34 | 59 | 5.882353 | 57.62712 | 3.912 | 0.021 |
| 8022 | protein C-terminus binding | 2 | 40 | 45 | 5 | 88.88889 | 3.532 | 0.022 |
| 19911 | structural constituent of myelin sheath | 1 | 3 | 3 | 33.33333 | 100 | 7.214 | 0.023 |
| 4370 | glycerol kinase activity | 1 | 4 | 4 | 25 | 100 | 6.208 | 0.023 |
| 4879 | ligand-dependent nuclear receptor activity | 2 | 36 | 62 | 5.555555 | 58.06452 | 3.775 | 0.024 |
| 5244 | voltage-gated ion channel activity | 3 | 99 | 181 | 3.030303 | 54.69613 | 3.066 | 0.024 |
| 5179 | hormone activity | 2 | 39 | 112 | 5.128205 | 34.82143 | 3.59 | 0.025 |
| 8060 | ARF GTPase activator activity | 1 | 4 | 4 | 25 | 100 | 6.208 | 0.027 |
| 1637 | G-protein chemoattractant receptor activity | 1 | 7 | 27 | 14.28571 | 25.92593 | 4.604 | 0.03 |
| 4950 | chemokine receptor activity | 1 | 7 | 27 | 14.28571 | 25.92593 | 4.604 | 0.03 |
| 1948 | glycoprotein binding | 1 | 4 | 6 | 25 | 66.66666 | 6.208 | 0.032 |
| 5521 | lamin binding | 1 | 5 | 5 | 20 | 100 | 5.518 | 0.032 |
| 16564 | transcriptional repressor activity | 4 | 194 | 236 | 2.061856 | 82.20339 | 2.578 | 0.032 |
| 32395 | MHC class II receptor activity | 1 | 5 | 22 | 20 | 22.72727 | 5.518 | 0.033 |
| 19956 | chemokine binding | 1 | 8 | 29 | 12.5 | 27.58621 | 4.279 | 0.036 |
| 19899 | enzyme binding | 4 | 209 | 250 | 1.913876 | 83.6 | 2.403 | 0.038 |
| 5261 | cation channel activity | 3 | 129 | 246 | 2.325581 | 52.43903 | 2.48 | 0.044 |
| 42826 | histone deacetylase binding | 1 | 10 | 11 | 10 | 90.90909 | 3.777 | 0.048 |
| 5328 | neurotransmitter\:sodium symporter activity | 1 | 8 | 20 | 12.5 | 40 | 4.279 | 0.049 |
| 5326 | neurotransmitter transporter activity | 1 | 8 | 22 | 12.5 | 36.36364 | 4.279 | 0.049 |
| 5102 | receptor binding | 6 | 418 | 742 | 1.435407 | 56.33423 | 2.162 | 0.049 |
The results presented in Fig. 2B and Table 3 reveal a list of genes which were down-regulated greater than fivefold in HK2 cells in response to oxalate exposure. When categorized based on gene ontology (using GenMAPP software), we observed that of the 5,230 genes associated with molecular functions oxalate exposure resulted in the down-regulation of 189 genes. Similarly, of the 16,467 genes associated with biological processes oxalate exposure resulted in the down-regulation of 552 genes; and among the 11,905 genes associated with various cellular components oxalate exposure resulted in the down-regulation of 239 genes.
Table 3. Genes that show an average change more than 5 fold down-regulation compared to control (p value<0.05) upon oxalate exposure grouped based on gene ontology using GenMAPP.
| 3.1 Molecular Function | ||||||||
| Go ID | GO Name | Genes | Genes | Genes | % Changed | % Present | Z Score | p Value |
| Changed | Measured | In GO | ||||||
| 4095 | carnitine O-palmitoyl- | 2 | 3 | 4 | 66.66666 | 75 | 7.244 | 0.002 |
| transferase activity | ||||||||
| 16416 | O-palmitoyltransferase activity | 2 | 3 | 4 | 66.66666 | 75 | 7.244 | 0.002 |
| 16406 | carnitine O-acyltransferase activity | 2 | 5 | 6 | 40 | 83.33334 | 5.471 | 0.003 |
| 42805 | actinin binding | 2 | 3 | 4 | 66.66666 | 75 | 7.244 | 0.004 |
| 5522 | profilin binding | 2 | 5 | 5 | 40 | 100 | 5.471 | 0.007 |
| 4527 | exonuclease activity | 5 | 46 | 60 | 10.86957 | 76.66666 | 3.739 | 0.007 |
| 17076 | purine nucleotide binding | 18 | 1369 | 1735 | 1.314828 | 78.9049 | −2.845 | 0.007 |
| 5083 | small GTPase regulator activity | 11 | 182 | 173 | 6.043956 | 105.2023 | 3.211 | 0.009 |
| 8171 | O-methyltransferase activity | 2 | 8 | 9 | 25 | 88.88889 | 4.159 | 0.01 |
| 8108 | UDP-glucose\:hexose-1-phosphate | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.012 |
| uridylyltransferase activity | ||||||||
| 17124 | SH3 domain binding | 3 | 17 | 18 | 17.64706 | 94.44444 | 4.09 | 0.013 |
| 3712 | transcription cofactor activity | 13 | 274 | 314 | 4.744525 | 87.26115 | 2.538 | 0.014 |
| 8374 | O-acyltransferase activity | 3 | 23 | 33 | 13.04348 | 69.69697 | 3.32 | 0.017 |
| 8408 | 3′-5′ exonuclease activity | 3 | 24 | 33 | 12.5 | 72.72727 | 3.218 | 0.017 |
| 48156 | tau protein binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.018 |
| 50749 | apolipoprotein E receptor binding | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.018 |
| 5355 | glucose transporter activity | 2 | 9 | 14 | 22.22222 | 64.28571 | 3.869 | 0.018 |
| 15085 | calcium ion transporter activity | 2 | 9 | 13 | 22.22222 | 69.23077 | 3.869 | 0.019 |
| 47066 | phospholipid-hydroperoxide | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.021 |
| glutathione peroxidase activity | ||||||||
| 3714 | transcription corepressor activity | 7 | 109 | 125 | 6.422019 | 87.2 | 2.734 | 0.021 |
| 50501 | hyaluronan synthase activity | 1 | 1 | 3 | 100 | 33.33333 | 6.352 | 0.022 |
| 16564 | transcriptional repressor activity | 10 | 194 | 236 | 5.154639 | 82.20339 | 2.503 | 0.022 |
| 4671 | protein-S-isoprenylcysteine O- | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.023 |
| methyltransferase activity | ||||||||
| 15149 | hexose transporter activity | 2 | 10 | 15 | 20 | 66.66666 | 3.62 | 0.023 |
| 15145 | monosaccharide transporter | 2 | 10 | 15 | 20 | 66.66666 | 3.62 | 0.023 |
| activity | ||||||||
| 19968 | interleukin-1\ II\, blocking binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.024 |
| 4910 | interleukin-1\, II\, blockingactivity | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.024 |
| 5118 | sevenless binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.025 |
| 26 | alpha-1\,2-mannosyltransferase | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.025 |
| 4244 | mitochondrial inner membrane | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.025 |
| peptidase activity | ||||||||
| 45518 | interleukin-22 receptor binding | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.026 |
| 15227 | acyl carnitine transporter activity | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.026 |
| 5536 | glucose binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.027 |
| 16209 | antioxidant activity | 3 | 27 | 46 | 11.11111 | 58.69565 | 2.944 | 0.027 |
| 15646 | permease activity | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.028 |
| 15196 | L-tryptophan transporter activity | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.028 |
| 15216 | purine nucleotide transporter activity | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.028 |
| 51370 | ZASP binding | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.029 |
| 51374 | FATZ 1 binding | 1 | 1 | 1 | 100 | 100 | 6.352 | 0.029 |
| 51373 | FATZ binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.029 |
| 5088 | Ras guanyl-nucleotide exchange | 5 | 74 | 88 | 6.756757 | 84.09091 | 2.437 | 0.03 |
| factor activity | ||||||||
| 30346 | protein phosphatase 2B binding | 1 | 1 | 2 | 100 | 50 | 6.352 | 0.034 |
| 42903 | tubulin deacetylase activity | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.035 |
| 16409 | palmitoyltransferase activity | 2 | 12 | 14 | 16.66667 | 85.71429 | 3.214 | 0.036 |
| 46873 | metal ion transporter activity | 4 | 45 | 63 | 8.888889 | 71.42857 | 2.831 | 0.036 |
| 166 | nucleotide binding | 26 | 1577 | 1994 | 1.6487 | 79.08727 | −2.154 | 0.036 |
| 30197 | extracellular matrix constituent\, | 1 | 2 | 3 | 50 | 66.66666 | 4.38 | 0.038 |
| lubricant activity | ||||||||
| 900 | translation repressor activity | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.039 |
| 42835 | BRE binding | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.039 |
| 8451 | X-Pro aminopeptidase activity | 1 | 2 | 3 | 50 | 66.66666 | 4.38 | 0.04 |
| 30554 | adenyl nucleotide binding | 16 | 1109 | 1393 | 1.442741 | 79.61235 | −2.233 | 0.04 |
| 4307 | ethanolaminephospho- | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.042 |
| transferase activity | ||||||||
| 15266 | protein channel activity | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.045 |
| 8321 | Ral guanyl-nucleotide exchange | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.046 |
| factor activity | ||||||||
| 30275 | LRR domain binding | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.047 |
| 15215 | nucleotide transporter activity | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.048 |
| 15016 | [heparan sulfate]-glucosamine N- | 1 | 2 | 4 | 50 | 50 | 4.38 | 0.048 |
| sulfotransferase activity | ||||||||
| 15082 | di-\, tri-valent inorganic cation | 3 | 30 | 43 | 10 | 69.76744 | 2.707 | 0.048 |
| transporter activity | ||||||||
| 51393 | alpha-actinin binding | 1 | 2 | 3 | 50 | 66.66666 | 4.38 | 0.049 |
| 15173 | aromatic amino acid transporter | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.049 |
| 3880 | C-terminal protein carboxyl | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.049 |
| methyltransferase activity | ||||||||
| 5128 | erythropoietin receptor binding | 1 | 2 | 3 | 50 | 66.66666 | 4.38 | 0.049 |
| 4908 | interleukin-1 receptor activity | 1 | 2 | 7 | 50 | 28.57143 | 4.38 | 0.049 |
Taken together these results reveal that oxalate exposure differentially modulates genes required for molecular functions, biological pathways, and cellular components. Moreover, the number of genes up-regulated in each functional group is far less than the number of genes down regulated. Surprisingly, oxalate exposure significantly suppresses the expression of 552 genes, while only upregulating 105 genes associated with biological processes. These results show the molecular network of gene expression that is associated with cellular dysfunction modulated by oxalate.
Oxalate exposure modulates gene expression by regulating transcription factors
To further gain insights into how oxalate controls differential gene expression, we analyzed the gene expression of transcription regulatory genes in response to oxalate exposure in HK2 cells. The results presented in Table 4 show the effects of oxalate exposure on the expression of genes involved in the regulation of transcription. These results show that oxalate exposure upregulates genes associated with the suppression of gene expression and at the same time down-regulates genes associated with transcription. Further assignment of these transcriptional regulatory genes by gene ontology groups (Table 4) reveals how oxalate influences gene expression of various transcription regulatory genes. These results reveal the selective nature of oxalate interactions with transcription regulatory machinery and provide a mechanistic link as to how oxalate treatment regulates changes in global gene expression in HK-2 cells.
Table 4. Genes involved in regulation of transcription in HK-2 cells after exposure to oxalate.
| GO ID | GO Name | GO | Genes | Genes | Genes | % Changed | % Present | Z Score | p Value |
| Type | Changed | Measured | in GO | ||||||
| Up-Regulated | |||||||||
| 30528 | transcription regulator activity | F | 15 | 1012 | 1398 | 1.482213 | 72.38913 | 3.664 | 0 |
| 3700 | transcription factor activity | F | 10 | 630 | 936 | 1.587302 | 67.30769 | 3.182 | 0.006 |
| 3704 | specific RNA polymerase II | F | 2 | 31 | 41 | 6.451613 | 75.60976 | 4.139 | 0.016 |
| transcription factor activity | |||||||||
| 16564 | transcriptional repressor activity | F | 4 | 194 | 236 | 2.061856 | 82.20339 | 2.578 | 0.032 |
| 6357 | regulation of transcription from RNA | P | 6 | 371 | 474 | 1.617251 | 78.27004 | 2.486 | 0.024 |
| polymerase II promoter | |||||||||
| 45892 | negative regulation of transcription\, | P | 4 | 181 | 218 | 2.209945 | 83.02752 | 2.745 | 0.027 |
| DNA-dependent | |||||||||
| 42994 | cytoplasmic sequestering of | P | 1 | 7 | 10 | 14.28571 | 70 | 4.604 | 0.041 |
| transcription factor | |||||||||
| 42992 | negative regulation of transcription | P | 1 | 8 | 12 | 12.5 | 66.66666 | 4.279 | 0.048 |
| factor import into nucleus | |||||||||
| Down-Regulated | |||||||||
| 3712 | transcription cofactor activity | F | 13 | 274 | 314 | 4.744525 | 87.26115 | 2.538 | 0.014 |
| 3714 | transcription co-repressor activity | F | 7 | 109 | 125 | 6.422019 | 87.2 | 2.734 | 0.021 |
| 16564 | transcriptional repressor activity | F | 10 | 194 | 236 | 5.154639 | 82.20339 | 2.503 | 0.022 |
| 40029 | regulation of gene expression | P | 4 | 41 | 47 | 9.756098 | 87.23404 | 3.064 | 0.012 |
| 122 | negative regulation of transcription | P | 7 | 121 | 153 | 5.785124 | 79.08497 | 2.424 | 0.02 |
| from RNApolymerase II promoter | |||||||||
| 45892 | negative regulation of transcription\, | P | 9 | 181 | 218 | 4.972376 | 83.02752 | 2.255 | 0.031 |
| DNA-dependent | |||||||||
| 5672 | transcription factor TFIIA complex | C | 1 | 2 | 2 | 50 | 100 | 4.38 | 0.04 |
Oxalate exposure modulates expression of gene associated with Signal transduction pathways
The results presented in Figure 3A show that several genes associated with signal transduction pathways, PI3 Kinase, MAP kinase and Retinoic acid Receptor pathways, show significantly altered expressions. In general, more genes associated with these three pathways are up-regulated in renal epithelial cells as early as 4 hours into oxalate exposure and some of these genes remain elevated for up to 24 hours. The results presented in Figure 3B show the heat MAP of this data specifically listing the genes associated with these pathways that are differentially regulated in HK2 cells upon exposure to oxalate.
Figure 3. Regulation of Gene expression associated with Molecular functions, Biological Processes or Cellular component.
Gene expression was analyzed with respect to gene ontology Groups and the number of genes that showed more than fivefold changes within each ontology group were identified. A)Genes that show an average change more than 5 fold up-regulation compared to control (p value<0.05) upon oxalate exposure grouped based on gene ontology using GenMAPP ; B) Genes that show an average change more than 5 fold down-regulation compared to control (p value<0.05) upon oxalate exposure grouped based on gene ontology using GenMAPP.
Genes that are always differentially expressed in renal cells after exposure to oxalate may serve as gene expression signature of oxalate nephrotoxicity
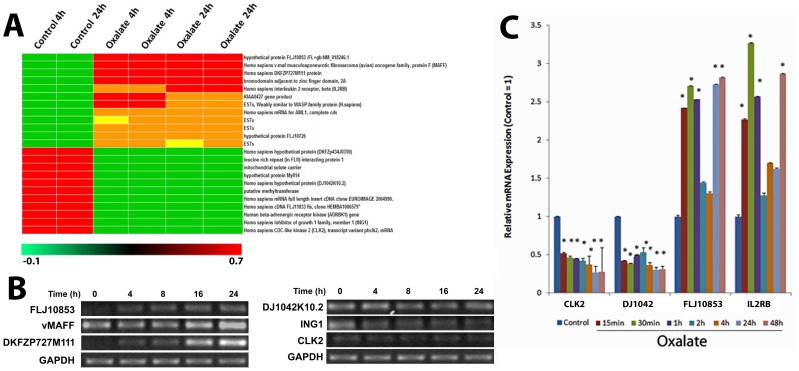
The results presented in figure 4 A shows a heat-map of set of twenty genes that are differentially modulated in HK-2 cells upon exposure to oxalate regardless of the duration of exposure. These include genes that are expressed either only in the oxalate treated HK-2 cells (absent in control) or only in control cells (absent in oxalate treated HK-2 cells). The expression of the differentially expressed genes was confirmed in parallel experiments using relative quantitative RT-PCR (Fig. 4B). We also evaluated the duration of expression of these genes in response to oxalate exposure. For these experiments we evaluated the quantitative changes by using real-time PCR. The results of these studies show a time dependent change in the mRNA levels of the genes tested [Fig. 4C]. These results also shows that these genes are turned on as early as 15 minutes into oxalate exposure and remain differentially expressed for a long period of time [over 48 hours]. Taken together these results suggest that transcriptional profiling using this set of genes may be useful to monitor oxalate nephrotoxicity.
Figure 4. Gene Expression Signature of Oxalate Nephrotoxicity.
(A) Gene expression changes highly significant in HK-2 cells exposed to oxalate compared to control cells and their fold changes. (B) HK-2 cells were treated with Oxalate for various time points and RT-PCR was used to identify differences in mRNA levels with GAPDH as a loading control. Left panel shows genes that are highly up-regulated and the right panel shows genes that are down-regulated upon oxalate exposure. C) Real Time Quantitative PCR analysis of time dependent changes in expression of two upregulated and two down regulated genes.
Discussion
Our studies describe genome wide changes in the transcript expression of renal epithelial cells in response to oxalate. Specifically, these studies show the upregulation of 750 genes and their transcripts and the down regulation of 2,276 genes and their transcripts that have not been reported before. More importantly, these studies identified the differential expression of genes that regulates transcription machinenary and thus may serve as drivers in oxalate induced transcriptional changes associated with oxalate nephropathy, including nephrolithiasis. Nephrolithiasis is a multi-factorial disorder which, in a majority of patients, results in renal deposition of calcium oxalate. While chronic hyperoxaluria is a common finding in patients with inherited metabolic conditions of primary-hyperoxaluria, a majority of patients generally show only a mild elevation in urinary oxalate. High urinary levels of oxalate, as seen in patients with hyperoxaluria, are generally associated with interstitial nephritis, extracellular fibrosis, and may eventually result in kidney failure if left uncorrected [28]. The development of a kidney stone depends on many extrinsic and intrinsic factors, which are not completely understood. The disease is multi-factorial and the nature of urine milieu is complex. As such, focusing on specific urinary components associated with urolithaisis is critical. Previous studies have shown that oxalate exposure to the renal epithelial cells promotes cellular dysfunction, which has been shown to promote renal crystal retention [5], [29]–[31]. Oxalate renal interactions are complex and poorly understood. While many previous studies have evaluated the over expression of a few genes in renal epithelial cells in response to oxalate, our results presented here profiled oxalate renal cell interactions in an unbiased fashion using microarray gene expression technology. Our studies revealed that in addition to promoting the expression of specific genes, oxalate exposure to renal epithelial cells is also associated with silencing the expression of many genes. We also show that oxalate differentially regulates the expression of genes in each ontology group: molecular functions, biological processes, and cellular components. Our studies further demonstrate that oxalate exposure differentially effects the expression of transcription regulatory genes, thus suggesting that the modulation of transcription regulatory genes might be a potential regulatory mechanism of modulating gene expression by oxalate. In addition, we identified a set of twenty genes differentially regulated by oxalate which might serve as a useful marker for monitoring oxalate nephrotoxicity.
Gene expression at any given time point represents a snapshot of a cells molecular machinery. Our results [Fig. 1A, B] show that oxalate exposure is associated with decrease in mRNA levels of several genes in all systems. These results show that oxalate exposure to the renal cells for 24 hours is associated with the net inhibition of the expression of 2,276 of genes. This unprecedented suppression of a large number of genes in renal epithelial cells suggests a program of the suppression of transcriptional activity. However, previous studies by have shown that the exposure of renal epithelial cells to oxalate results in the up-regulation of several genes [12], [15], [18], [20], [20], [22,23]. Thus, the notion of a general transcriptional repression is too simplistic of an explanation. Further analysis of our gene expression data (Fig. 1 C) reveals that although, over all, less genes are expressed in oxalate exposed cells for either 4 hour or 24 hour durations (26, 211 (control)vs. 25,107 (oxalate-4 h) and 23,935 (oxalate-24 hours), a number of genes (∼750 genes) are up-regulated. These findings demonstrate that oxalate exposure differentially and selectively regulates gene expression. Moreover, expression profiling at 4 hour and 24 hour intervals following oxalate exposure suggest that gene expression changes in response to oxalate are in part governed by the duration of exposure.
The studies presented here have identified changes in the expression of genes that are responsible for increased transcription, lending credibility to the earlier observations. HK-2 cells exposed to oxalate show extensive changes in gene expression over a wide spectrum of functions. In addition to genes that have been already implicated, many unique genes were found to be either expressed or inhibited upon oxalate exposure. We observed that some genes are differentially affected depending on the duration of oxalate exposure. HK-2 cells exposed to oxalate for 24 hours show differential gene expression changes compared to HK-2 cells exposed for only 4 hours. This suggests that in addition to the amount of oxalate present in the cell's vicinity, the duration of exposure to oxalate also plays an important role in modulating gene expression in renal cells following oxalate exposure.
Differential gene expression as a result of oxalate exposure can stimulate different responses in the cells directly exposed to oxalate that may ultimately lead to either survival or cell death depending on the concentration and the duration of exposure to oxalate. Results first reported by us [12], that have since been confirmed by several others, have shown that oxalate is toxic to renal epithelial cells. Analysis of the gene expression data using gene ontology software reveals that oxalate exposure inhibits the expression of over hundreds of genes that are required for cellular functions, which would suggest that oxalate might broadly impact cellular functions. Thus, renal cell dysfunction in response to oxalate may be in part driven by altered gene expression; however, the present study design does not permit the separation of causal and bystander genes. It is also possible that some of the changes in gene expression are causal, while other changes are bystander effects. In other words, whether the changes in gene expression play a causal role in oxalate nephrotoxicity or whether the gene expression changes are a result of dysfunction need to be addressed in additional studies.
Our analysis of gene expression data in renal epithelial ells in response to oxalate exposure revealed that 43 transcription regulatory genes are overexpressed while 51 transcription regulatory genes are down regulated by oxalate (Table 4). Given the enormous implications in regulation of cellular function by the modulation of gene expression, specific mechanisms are in place in eukaryotic cells that regulate gene expression. Transcription regulatory machinenary in eukaryotes involves specific transcription factors and transcription inhibitors; proteins that are required to turn on and turn off the expression of particular genes. These considerations point to a possible mechanism of how oxalate differentially regulates the gene expression of such a large number of genes.
Given that oxalate is a metabolic end product in humans that cannot be further metabolized, such large scale changes in gene expression in renal epithelial cells in response to high oxalate levels points to an indirect mechanism of action, which may involve the interaction of oxalate with the cell membrane or in intracellular components. The primary site of oxalate action in cell remains unknown. Irrespective of primary site of action, one of the most common means by which cells sense changes is by activating the signal transduction pathways, especially the stress signal pathways. The stress associated signals are transduced through a series of proteins that are activated by phosphorylation/dephosphorylation steps and are finally turned into transcription factors, causing changes in gene expression. Though the present study design does not allow for the identification of activity changes due to phosphorylation, we identified changes in the gene expressions of upstream activators of several signaling pathways. Proteins like Ras, Fas and MKK are highly up-regulated as a result of oxalate exposure. These proteins are known to play important roles in JNK/SAPK signaling and p38 MAPK signaling. These results are in agreement with previous studies, by us [14], [15], [ and 24] and others [32], that identified an active role for Stress Activated Protein Kinases in oxalate renal cell interactions. We also identified changes in the expression of genes associated with retinoic Acid Receptor Signaling Pathway. Clearly additional studies are required to evaluate the functional consequence of these gene expression changes.
In summary, our study is the first attempt at profiling the Genome-wide expression changes in human renal epithelial cells as a result of exposure to oxalate. Results from our study point to complex and intricate mechanisms, including differential gene expression, in renal epithelial cells in response to oxalate exposure. Clearly further studies are required to completely understand the implications of the plethora of changes in gene expression occurring as a result of oxalate exposure in renal epithelial cells. We must separate and characterize the genes that are derived from the by-stander effect and identify the genes whose altered expression is responsible for oxalate nephrotoxicity.
Concise Methods
Cell culture
Human Kidney Epithelial Cells, HK-2, were procured from ATCC and maintained in a DMEM medium supplemented with 10% Fetal Bovine Serum and antibiotics. Before Oxalate treatments, cells were serum starved for 16 to 20 hours. Media components were procured form Invitrogen Corporation and all other chemicals were procured from Sigma-Aldrich.
Microarray analysis using Affymetrix Gene Chip
HK2 cells were incubated in the presence of 1 mM Sodium Oxalate for different amounts of time and all cellular RNA was isolated using a RNEasy Kit (Qiagen). RNA was quantified with a NanoDrop ND-1000 spectrophotometer and tested for quality using an Agilent Bioanalyzer 2100 (Agilent) before being used for microarray analysis. cRNA was prepared using 50 ng of RNA and then hybridized to an Affymetrix Human genome U133 Plus 2.0 gene array comprising of 11 independent replicate sets for each message Hybridization, staining, and post-hybridization washes were completed according to the manufacturer's recommendations (Affymetrix). Following hybridization, gene arrays were processed with a GeneChip fluidics station 450 and double staining was captured using a gene array scanner 3000. All experiments were designed to comply with the Minimum Information About a Microarray Experiment [MIAME; http://www.mged.org/index.html] guidelines and were interpreted by independent verification [33].
Data analysis and bioinformatics
Hybridization intensities were quantified from the data image files using Gene Chip Operating Software algorithms (GCOS1.2, Affymetrix) with global scaling. Data analysis was performed using a Data Mining Tool [DMT 3.1, Affymetrix] and a GeneSpring 7.2 (Silicon Genetics). Cell Intensity files were processed into expression values for all the 55,000 probe sets (transcripts) on each array and following the respective normalization step. Differentially expressed genes were selected if they passed Welch's t test and parametric test (variance not assumed equal, P<0.05) and showed at least 2-fold changes between control and oxalate treated sets. Global gene expression was visualized by STAGE, a Smart Tool for Gene Expression analysis developed in house (Bhat S and Koul H). Differentially expressed genes were classified according to the Gene Ontology functional category (GenMAPP v2). Cluster and Heatmap images were generated using BRB-Array tools [34], freely available from http://linus.nci.nih.gov/BRB-ArrayTools.html.
Reverse transcriptase PCR
1 µg RNA was used to synthesize cDNA by using iScript, a cDNA synthesis Kit [Bio-Rad Laboratories], and reverse transcriptase. PCR was performed with gene specific primers using Platinum Taq Polymerase (Invitrogen) and separating the products on a 1% agarose gel. Primers were procured from Integrated DNA Technologies and primer sequences used are described in Table 4.
Real-time qPCR
Melt curve analysis was included to assure that only one PCR product was formed. Primers were designed to generate a PCR amplification product of 100–550 bp. Only primer pairs yielding unique amplification products without primer dimer formation were subsequently used for real-time PCR assays. Expression was related to the control gene [GAPDH], which did not change under any of the experimental conditions studied.
The real-time PCR reaction mixture was prepared in a Light Cycler 480 (Roche Diagnostics), a Multiwell 96-well plate containing 10 µM of each primer, 10 µl of 2× master mix, and 1 µl of cDNA template in a final reaction volume of 20 µl. The real-time PCR amplification was performed using the specific primers as shown in Table 1, using the following cycle parameters: enzyme activation at 95°C for 10 min; 45 cycles of 95°C for 10 s, 63°C for 10 s and 72°C for 10 s. Following the amplification phase, a cooling step was performed at 4°C for 10 s (ramp rate of 1.5°C/s). Acquisition of the fluorescence signal was performed using the Mono Hydrolysis Probe setting [483–523 nm] following the 72°C extension phase of each cycle. GAPDH primers were included to normalize variation from sample to sample. All experiments were repeated three times using three independent preparations of cDNA.
Acknowledgments
We are grateful to Genomics and Microarray Core Laboratory and the University of Colorado Anschutz Medical Campus for their support with Affymetrix Microarray work. We gratefully acknowledge Neil Koul for editing the manuscript.
Funding Statement
This work was supported in part by a research grant to Hari K. Koul from the National Institutes of Health (NIH) (NIH-DK-RO1-54084) and Department of Surgery Academic Enrichment Funds (HKK). HKK is also supported in part by VA Merit Award-01BX001258 and NIH/National Cancer Institute R01CA161880 grants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Knight TF, Senekjian HO, Taylor K, Steplock DA, Weinman EJ (1979) Renal transport of oxalate: effects of diuretics, uric acid, and calcium. Kidney Int 16: 572–576. [DOI] [PubMed] [Google Scholar]
- 2. Koul H, Ebisuno S, Renzulli L, Yanagawa M, Menon M, et al. (1994) Polarized distribution of oxalate transport systems in LLC-PK1 cells, a line of renal epithelial cells. Am J Physiol 266: F266–F274. [DOI] [PubMed] [Google Scholar]
- 3. Kuo SM, Aronson PS (1996) Pathways for oxalate transport in rabbit renal microvillus membrane vesicles. J Biol Chem 271: 15491–15497. [DOI] [PubMed] [Google Scholar]
- 4.Robertson WG, Peacock M, Heyburn PJ, Marshall RW, Rutherford A, et al. (1979) The significance of mild hyperoxaluria in calcium stone-formation. In: Rose GA, Robertson WG, and Watts REW, eds. Oxalate in Human Biochemistry and Clinical Pathology. London: The Welcome Foundation Ltd. p. 173.
- 5.Koul H, Koul S (2008) Molecular basis of Urolithiasis: Role of crystal retention. In: Evan AP, Lingeman JE, McAteer JA, Williams JC Jr, editors. Renal stone disease 2, 2nd International Urolithiasis Research Symposium. Melville, NY: American Institute of Physics. pp 120–125.
- 6. Hoppe B, Beck BB, Milliner DS (2009) The primary hyperoxalurias. Kidney Int 75 12: 1264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ono K, Yasukohchi A, Kikawa K (1987) Pathogenesis of acquired renal cysts in hemodialysis patients. The role of oxalate crystal deposition in renal tubules. ASAIO Trans 33: 245–249. [PubMed] [Google Scholar]
- 8. Lieske JC, Spargo BH, Toback FG (1992) Endocytosis of calcium oxalate crystals and proliferation of renal tubular epithelial cells in a patient with type 1 primary hyperoxaluria. J Urol 148 5: 1517–9. [DOI] [PubMed] [Google Scholar]
- 9. Hackett RL, Khan SR (1988) Presence of calcium oxalate crystals in the mammalian thyroid gland. Scanning Microsc 2: 241–246. [PubMed] [Google Scholar]
- 10. Radi MJ (1989) Calcium oxalate crystals in breast biopsies. An overlooked form of microcalcification associated with benign breast disease. Arch Pathol Lab Med 113: 1367–1369. [PubMed] [Google Scholar]
- 11. Truong LD, Cartwright J Jr, Alpert L (1992) Calcium oxalate in breast lesions biopsied for calcification detected in screening mammography: incidence and clinical significance. Mod Pathol 5: 146–152. [PubMed] [Google Scholar]
- 12. Koul H, Kennington L, Nair G, Honeyman T, Menon M, et al. (1994) Oxalate-induced initiation of DNA synthesis in LLC-PK1 cells, a line of renal epithelial cells. Biochem Biophys Res Commun 205: 1632–1637. [DOI] [PubMed] [Google Scholar]
- 13. Koul H, Menon M, Scheid C (1996) Oxalate and renal tubular cells: A complex interaction. Ital J Electrol Metab 10: 67–74. [Google Scholar]
- 14. Koul H, Menon M, Chaturvedi L, Koul S, Sekhon A, et al. (2002) Activation of the p38-MAP kinase Signal transduction Pathways by COM-crystals. J Biol Chem 277 39: 36845–36852. [DOI] [PubMed] [Google Scholar]
- 15. Koul S, Chaturved LS, Sekhon A, Bhandari A, Menon M, et al. (2002) Effect of oxalate on the re-initiation of DNA synthesis in LLC-PK1 cells do not involve p42/44 MAP kinase activation. Kidney Int 61: 525–533. [DOI] [PubMed] [Google Scholar]
- 16. Bhandari A, Koul S, Sekhon A, Pramanik SK, Chaturvedi LS, et al. (2002) Effects of oxalate on HK-2 cells, a line of proximal tubular epithelial cells from normal human kidney. J Urol 168 1: 253–259. [PubMed] [Google Scholar]
- 17. Lieske JC, Hammes MS, Hoyer JR, Toback FG (1997) Renal cell osteopontin production is stimulated by calcium oxalate monohydrate crystals. Kidney Int 51: 679–686. [DOI] [PubMed] [Google Scholar]
- 18. Koul S, Huang M, Bhat S, Maroni P, Meacham RB, et al. (2008) Oxalate exposure provokes HSP 70 response in LLC-PK1 cells, a line of renal epithelial cells: protective role of HSP 70 against oxalate toxicity. Urol Res 36: 1–10. [DOI] [PubMed] [Google Scholar]
- 19. Koul S, Fu S, Koul H (2003) Oxalate exposure promotes reinitiation of the DNA synthesis and apoptosis of HK-2 cells, a line of human renal epithelial cells. Ann N Y Acad Sci 1010: 292–295. [DOI] [PubMed] [Google Scholar]
- 20. Habibzadegah-Tari P, Byer K, Khan SR (2005) Oxalate induced expression of monocyte chemoattractant protein-1 (MCP-1) in HK-2 cells involves reactive oxygen species. Urol Res 33: 440–447. [DOI] [PubMed] [Google Scholar]
- 21. Jeong BC, Kwak C, Cho KS, Kim BS, Hong SK, et al. (2005) Apoptosis induced by oxalate in human renal tubular epithelial HK-2 cells. Urol Res 33: 87–92. [DOI] [PubMed] [Google Scholar]
- 22. Huang MY, Chaturvedi LS, Koul S, Koul HK (2005) Oxalate stimulates IL-6 production in HK-2 cells, a line of human renal proximal tubular epithelial cells. Kidney Int 68 2: 497–503. [DOI] [PubMed] [Google Scholar]
- 23. Iida S, Peck AB, Byer KJ, Khan SR (1999) Expression of bikunin mRNA in renal epithelial cells after oxalate exposure. J Urol 162: 1480–1486. [PubMed] [Google Scholar]
- 24. Chaturvedi L, Koul S, Sekhon A, Bhandari A, Menon M, et al. (2002) Oxalate Selectively Activates p38 Mitogen-activated Protein kinase and c-Jun N-terminal kinase Signal transduction Pathways in Renal Epithelial Cells. J Biol Chem 277 15:13321–30. [DOI] [PubMed] [Google Scholar]
- 25. Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, et al. (1994) HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int 45: 48–57. [DOI] [PubMed] [Google Scholar]
- 26. Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470. [DOI] [PubMed] [Google Scholar]
- 27.Koul S, Bhat S, Chandhoke PS, Meacham RB, Koul HK (2006) Defining the molecular signature of oxalate nephrotoxicity: Global changes in gene expression by oxalate inhuman kidney epithelial cells. Journal of Urology 175 (4), Supplement: 499–499, Meeting Abstract: 1547.
- 28. Wharton R, D'Agati V, Magun AM, Whitlock R, Kunis CL, et al. (1991) Acute deterioration of renal function associated with enteric hyperoxaluria. Clin Nephrol 34: 116–121. [PubMed] [Google Scholar]
- 29. Verkoelen CF, van der Boom BG, Houtsmuller AB, Schröder FH, Romijn JC (1998) Increased calcium oxalate monohydrate crystal binding to injured renal tubular epithelial cells in culture. Am J Physiol 274: F958–F965. [DOI] [PubMed] [Google Scholar]
- 30. Koul HK, Koul S, Fu S, Santosham V, Seikohn A, et al. (1999) Oxalate: From crystal formation to crystal retention. Journal of the American Society of Nephrology 10 14: S417–S421. [PubMed] [Google Scholar]
- 31. Wiessner JH, Hasegawa AT, Hung LY, Mandel GS, Mandel NS (2001) Mechanisms of calcium oxalate crystal attachment to injured renal collecting duct cells. Kidney Int 59: 637–644. [DOI] [PubMed] [Google Scholar]
- 32. Han HJ, Lim MJ, Lee YJ (2004) Oxalate inhibits renal proximal tubule cell proliferation via oxidative stress, p38 MAPK/JNK, and cPLA2 signaling pathways. Am J Physiol Cell Physiol 287: C1058–C1066. [DOI] [PubMed] [Google Scholar]
- 33. Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29: 365–371. [DOI] [PubMed] [Google Scholar]
- 34. McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, et al. (2002) Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics 18: 1462–1469. [DOI] [PubMed] [Google Scholar]