Abstract
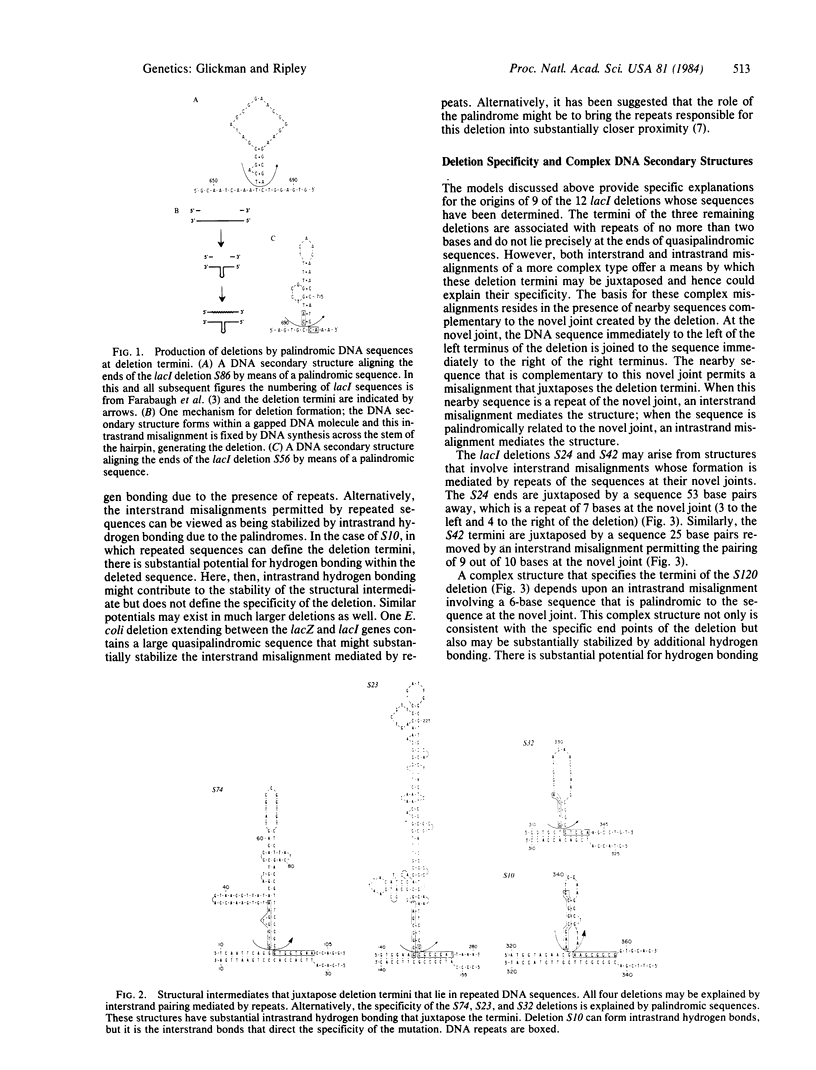
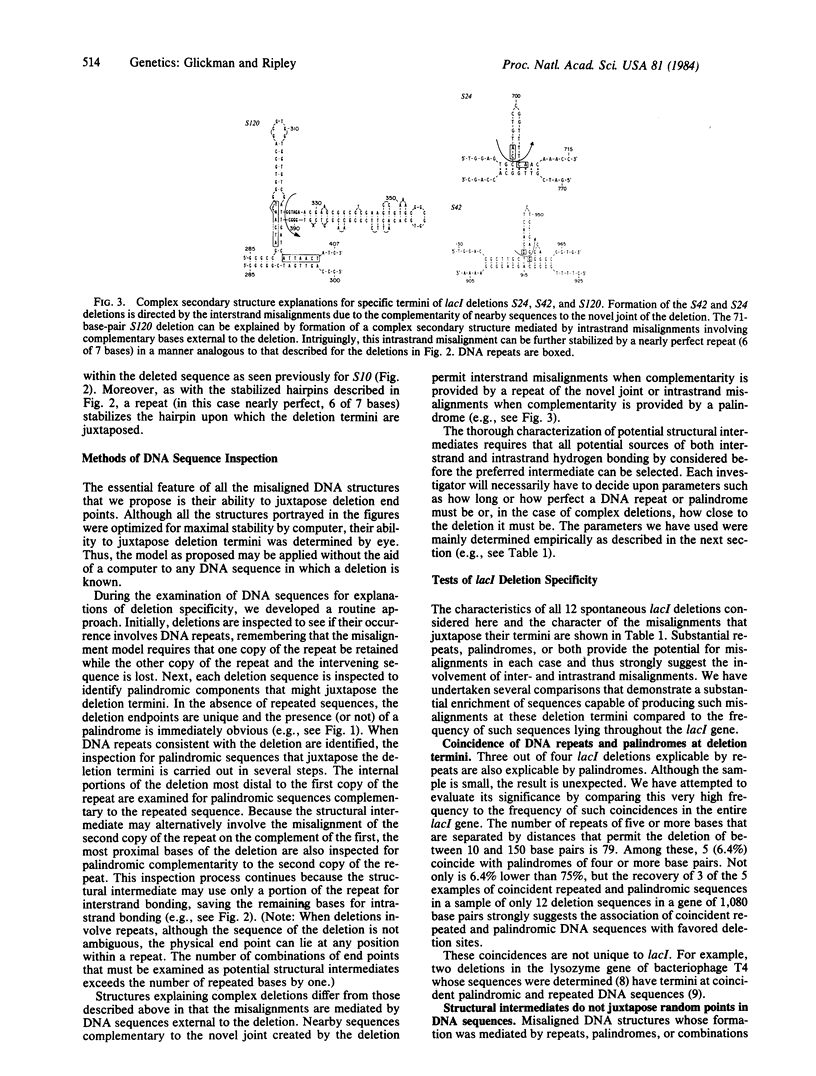
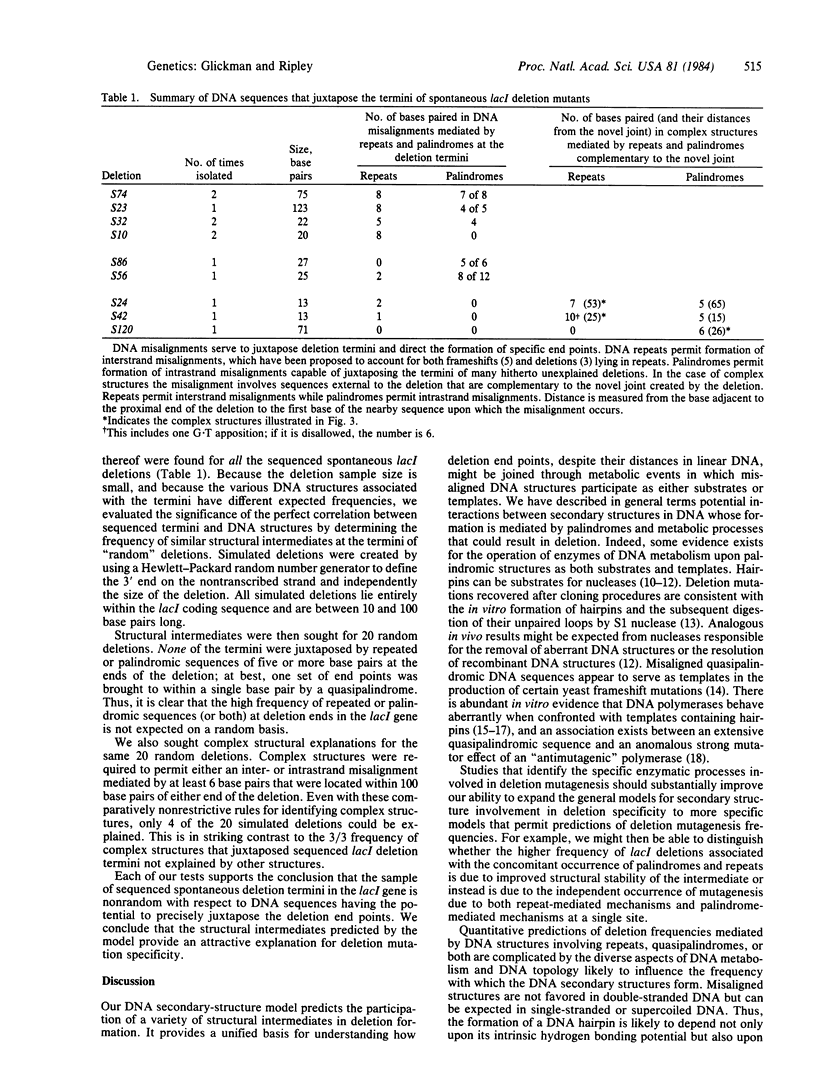
A model is presented for deletion mutations whose formation is mediated by palindromic and quasipalindromic DNA sequences. It proposes that the self-complementarity of palindromes allows the formation of DNA secondary structures that serve as deletion intermediates. The structures juxtapose the end points of the deletion and thus direct deletion specificity. While misaligned DNA intermediates that explain deletion termini occurring in repeated DNA sequences have been described, no explanations have been offered for deletion termini occurring in other sequences. The DNA secondary structures whose formation is mediated by palindromic sequences appear to explain many of these. In this paper, secondary-structure intermediates are described for a series of spontaneous deletions of known sequence in the lacl gene of Escherichia coli. The model is supported by its failure to predict structures that can juxtapose simulated deletion termini in the lacl gene. We have found a strong association between palindromic sequences and repeated sequences at lacl deletion termini that suggests the joint participation of repeated and palindromic DNA sequences in the formation of some deletions. Sequences of deletions in other organisms also suggest the participation of palindromic DNA sequences in the formation of deletions.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961 Mar;47(3):403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Görtz G., Charlier D. Deletions and DNA rearrangements within the transposable DNA element IS2. A model for the creation of palindromic DNA by DNA repair synthesis. Nucleic Acids Res. 1980 Dec 11;8(23):5825–5833. doi: 10.1093/nar/8.23.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. Instability of palindromic DNA in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):409–416. doi: 10.1101/sqb.1981.045.01.055. [DOI] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Saedler H. DNA sequence of the mini-insertion IS2--6 and its relation to the sequence of IS2. Nature. 1978 Oct 19;275(5681):611–617. doi: 10.1038/275611a0. [DOI] [PubMed] [Google Scholar]
- Ikoku A. S., Hearst J. E. Identification of a structural hairpin in the filamentous chimeric phage M13Gori1. J Mol Biol. 1981 Sep 15;151(2):245–259. doi: 10.1016/0022-2836(81)90514-3. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Clayton D. A. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A. 1982 Feb;79(4):983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Owen J. E., Schultz D. W., Taylor A., Smith G. R. Nucleotide sequence of the lysozyme gene of bacteriophage T4. Analysis of mutations involving repeated sequences. J Mol Biol. 1983 Apr 5;165(2):229–248. doi: 10.1016/s0022-2836(83)80255-1. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Porter A. G., Barber C., Carey N. H., Hallewell R. A., Threlfall G., Emtage J. S. Complete nucleotide sequence of an influenza virus haemagglutinin gene from cloned DNA. Nature. 1979 Nov 29;282(5738):471–477. doi: 10.1038/282471a0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W., Shoemaker N. B. Mutator versus antimutator activity of a T4 DNA polymerase mutant distinguishes two different frameshifting mechanisms. Mol Gen Genet. 1983;189(1):113–117. doi: 10.1007/BF00326062. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Ripley L. S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson I. B., Ingram V. M. cDNA sequence of a new chicken embryonic rho-globin. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4782–4785. doi: 10.1073/pnas.78.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R. J., Berns K. I., Tan M., Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A. 1982 Mar;79(6):2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L. A., Gefter M. L. Studies on the mechanism of enzymatic DNA elongation by Escherichia coli DNA polymerase II. J Mol Biol. 1976 May 5;103(1):61–76. doi: 10.1016/0022-2836(76)90052-8. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Todd P. A., Glickman B. W. Mutational specificity of UV light in Escherichia coli: indications for a role of DNA secondary structure. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4123–4127. doi: 10.1073/pnas.79.13.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert G., Tavernier J., Derynck R., Devos R., Fiers W. Molecular mechanisms of nucleotide-sequence rearrangements in cDNA clones of human fibroblast interferon mRNA. Gene. 1981 Nov;15(2-3):215–223. doi: 10.1016/0378-1119(81)90131-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]



