Abstract
Digital Imaging and Communications in Medicine (DICOM) is the dominant standard for medical imaging data. DICOM-compliant devices and the data they produce are generally designed for clinical use and often do not match the needs of users in research or clinical trial settings. DicomBrowser is software designed to ease the transition between clinically oriented DICOM tools and the specialized workflows of research imaging. It supports interactive loading and viewing of DICOM images and metadata across multiple studies and provides a rich and flexible system for modifying DICOM metadata. Users can make ad hoc changes in a graphical user interface, write metadata modification scripts for batch operations, use partly automated methods that guide users to modify specific attributes, or combine any of these approaches. DicomBrowser can save modified objects as local files or send them to a DICOM storage service using the C-STORE network protocol. DicomBrowser is open-source software, available for download at http://nrg.wustl.edu/software/dicom-browser.
Keywords: Digital imaging and communications in medicine (DICOM), Workflow, Image viewer, Imaging informatics
Background
DICOM is the dominant standard format for medical imaging data [1, 2]. The imaging devices that produce DICOM-formatted data, the PACS systems that store these data, the workstations used to view images, and the workflows best supported by all of these systems are strongly oriented towards clinical applications. Research projects using medical imaging data, including clinical trials, have their own specialized needs that diverge from workflows designed for clinical settings. For example, limitations of clinical PACS systems and the need to run post-processing and analysis software often lead users to store their DICOM data on general-purpose file systems, where there is no common convention for data organization.
Images acquired in a clinical setting and used in research studies often must have at least some protected health information (PHI) removed from the image metadata. Some information, such as study date, that could in principle be used to identify the subject, might be needed for data analysis; other attributes, such as patient name, can and should be removed. Operating procedures at some clinical imaging facilities may inject PHI into fields that are not generally considered sensitive. Images acquired in research facilities may contain some PHI and often use standard fields such as patient name in nonstandard ways. The DICOM standard includes deidentification schemes (the attribute confidentiality profiles [2, section PS 3.15]), but the variability in image metadata content and in the specific requirements of each research study creates a need for custom deidentification, pseudonymization, or, generally, metadata conditioning procedures.
We set out to write software that would smooth the transition between clinically oriented DICOM and the specialized workflows of research imaging. Our goals included an easy-to-use interface for locating and identifying DICOM files, summary views of DICOM metadata from multiple studies at varying levels of the patient–study–series–instance hierarchy, and tools for modifying DICOM metadata that mix fully automated modification, guided manual input, and ad hoc, manual manipulation.
Methods
DicomBrowser is written in the Java language and runs on any operating system with an installed Java virtual machine, version 1.5 or later, including Windows, Mac OS X, and Linux. The package includes three programs: DicomBrowser, a graphical user interface (GUI) program for interactively viewing and modifying DICOM metadata; DicomSummarize, a command-line program that generates a spreadsheet of metadata from DICOM files; and DicomRemap, a command-line program that applies metadata changes specified in a script file and/or in a DicomSummarize spreadsheet.
DicomBrowser is released under the BSD open-source license, with the source code available in a Mercurial version control system repository at http://hg.xnat.org/dicombrowser. It uses several open-source libraries, notably including dcm4che [3] for DICOM operations; HSQLDB [4] for a condensed, relational, internal representation of the DICOM metadata; and ImageJ [5] for image display.
DicomBrowser GUI
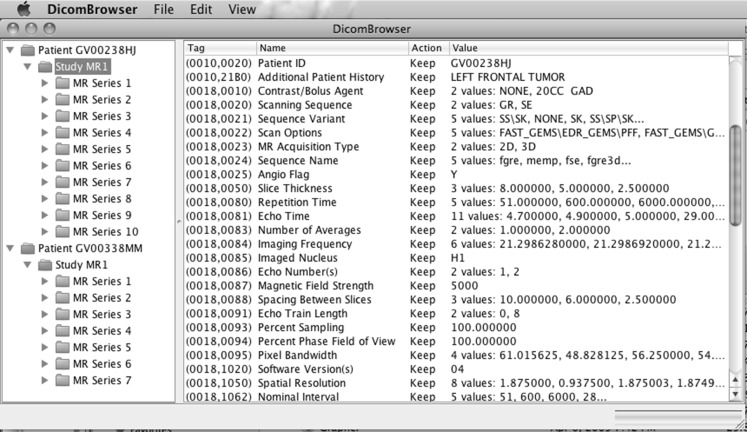
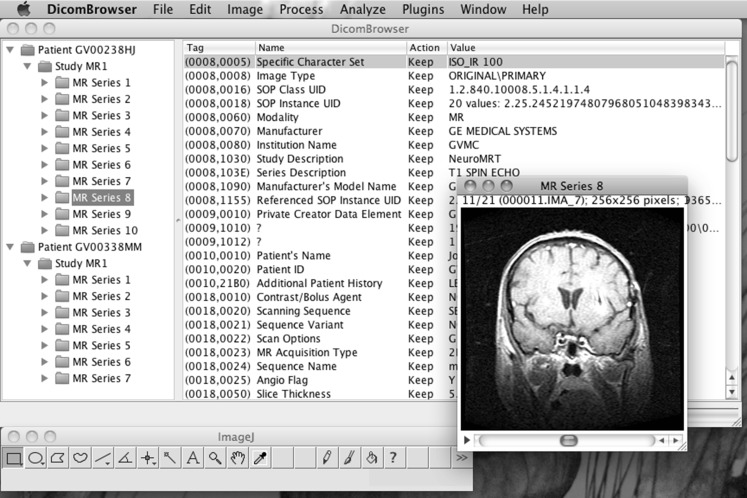
The DicomBrowser GUI presents two views of the DICOM metadata, as shown in Fig. 1. The left side of the main DicomBrowser window is a tree view of the patient–study–series–instance hierarchy of all loaded DICOM objects; items in this view can be expanded or collapsed to show or hide the lower levels of the hierarchy. The right side of the main window is a summary of the metadata for all objects selected in the tree view at left. This summary includes, for each attribute, its numeric tag, the human-readable attribute description, an indicator of whether the value has been changed, and the value or values of that attribute in the selected objects. Attribute values can be changed by clicking on the value field and typing a replacement value. A selected attribute can be deleted via a menu option. Changes made to the DICOM metadata are applied only to those objects selected in the left side tree view; also, although changes are immediately visible in the metadata summary, the actual DICOM objects are modified only when exported. Modified objects can be saved to disk or sent via the DICOM networking protocol to a C-STORE storage service, such as a PACS. The image data for selected objects can be viewed using the embedded ImageJ image viewer by using the View/View Selected Images menu item (Fig. 2).
Fig. 1.
DicomBrowser graphical interface
Fig. 2.
Using ImageJ to view image data
Metadata Modification Scripts
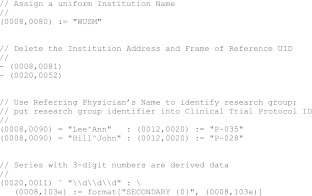
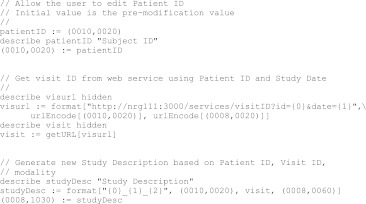
We developed a language, DicomEdit, for specifying changes to DICOM metadata. The language grammar was specified in Extended Backus-Naur Form [6] and translated to Java code by the ANTLR parser generator [7]. DicomEdit scripts are plain text files that can be written and modified using any text editor. An example DicomEdit script is shown divided into parts in Figs. 3, 4, and 5.
Fig. 3.
Metadata modification script, part 1: operations and conditions
Fig. 4.
Metadata modification script, part 2: user-defined variables
Fig. 5.
Metadata modification script, part 3: UID reassignment
Script Elements
DicomEdit scripts are composed from four elemental types:
String literals contain a concrete value.
Tags represent a DICOM metadata attribute and are expressed in the same notation used throughout the DICOM standard. (0008,0080), for instance, is the institution name attribute.
Identifiers are names that can represent user-defined variables or built-in functions or generators. They consist of a sequence of letters, digits, and the underscore character _, except that no identifier may begin with a digit.
Operators establish the syntactical structure of a DicomEdit statement and define its semantics. The simplest operators are symbols, representing operations such as value assignment, attribute deletion, and value comparisons (Table 1). Some operators are words: for example, echo prints a value to the program's standard output stream. The operators describe and hidden control the display of user-defined variables; this is described in more detail below.
Table 1.
DicomEdit symbolic operators
| Operator | Name | Usage | Description |
|---|---|---|---|
| := | Assignment | (gggg,eeee) := A | The attribute (gggg,eeee) is assigned the value obtained by evaluating A. |
| := | Initialization | v := A | The user-defined variable v is initialized with the value obtained by evaluating A. |
| - | Deletion | - (gggg,eeee) | The attribute (gggg,eeee) is removed. |
| : | Constraint application | A : B | Operation B is applied only if constraint A is satisfied. |
| = | Equality constraint | A = B | Evaluates true if A and B evaluate to equivalent string values. |
| ~ | Pattern constraint | A ~ B | Evaluates true if A evaluates to a string that matches the regular expression B. |
Values
A value is a string produced by evaluating any expression. Several expression types can be evaluated: string literals, i.e., string values explicit in the script; tags, which evaluate to the string representation of the corresponding DICOM attribute; user-defined variables, which evaluate to the value that they have been assigned; and built-in function and generator calls, both described in detail below.
Operations
An operation specifies a change to an attribute value. There are two types of operation: assignment, which changes an attribute value or assigns a value to a previously unset attribute, and deletion, which removes an attribute from the object. Assignment is specified by the := infix operator, while deletion is specified by the - prefix operator (Fig. 3, first and second blocks).
Constraints and Conditional Operations
A constraint defines to which DICOM objects an operation will be applied. There are two types of constraints, equality (=) constraints, which evaluate true only if the operands evaluate to equivalent strings, and pattern (~) constraints, which evaluate true if the left operand matches the regular expression in the right operand. A constraint is followed by the constraint application operator (:) and an operation to be constrained. The operation is applied if and only if the constraint evaluates true.
The third block of Fig. 3 shows assignment operations conditional on equality constraints: the Clinical Trial Protocol ID is assigned only if the referring physician's name is equal to one of the specified values. The fourth block of Fig. 3 shows an assignment operation conditional on a pattern matching constraint; the series description is modified only for series with a three-digit series number.
Built-In Functions
The DicomEdit language includes a number of built-in functions to support complex value construction. A built-in function application evaluates to a value and has the general form:
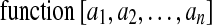 |
The sample script listed in Figs. 3, 4, and 5 includes several examples of using built-in functions to obtain or transform values. The available built-in functions are listed in Table 2.
Table 2.
DicomEdit built-in functions
| Function name | Arguments | Description |
|---|---|---|
| format | Format string, value-1, value-2, … | Formats the given values using the provided format string, using the same formatting syntax as the Java class java.text.MessageFormat. |
| getURL | URL | Retrieves the content of the resource at the provided URL; if a username and password are included in the URL, uses HTTP Basic Authentication. |
| lowercase | String | Converts all uppercase characters in the argument to lowercase. |
| match | Value, regexp, index | Matches the value against the provided regular expression and returns the content of the contained matching group with the given index. This is a more powerful and flexible method for extracting substrings than the index-based substring function. |
| replace | String, target, replacement | Replaces all occurrences of target in the given string with replacement. |
| substring | String, start-index, end-index | Returns a substring of string beginning at character number start-index (where the first character has index 0) and extending to character number end-index-1. |
| uppercase | String | Converts all lowercase characters in the argument to uppercase. |
| urlEncode | String | Encodes the given value so that it can be embedded in a URL. |
User-Defined Variables
Scripts may contain variables, identifiers that represent a value. Variables can be initialized to a particular value using the assignment operator (:=), as shown in Fig. 4.
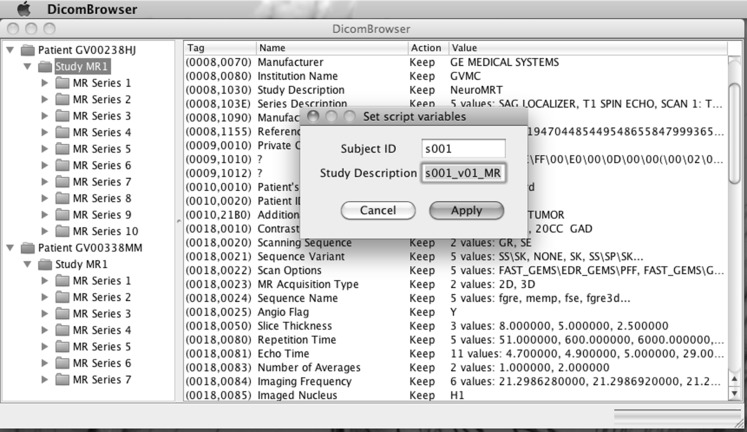
When a script is applied in the DicomBrowser GUI, a dialog box appears to let the user modify the value of each variable used in the script (Fig. 6). The label accompanying the variable in this dialog can be defined by a describe declaration, as shown in Fig. 4 (top block), where the variable patientID is given the label “Subject ID.” If no describe declaration is provided for a variable, then the description is simply the variable name. A variable can also be omitted from the dialog, so that the user cannot see or modify its value, by declaring the variable hidden, as in the middle block of Fig. 4. Hidden variables are useful for holding intermediate results when multiple steps are required to build a value.
Fig. 6.
Dialog allowing the user to set values of user-defined variables from a metadata modification script
Generators
The DicomEdit language includes a feature for replacing unique identifiers (UIDs), the globally unique identifiers ubiquitous in the DICOM standard. Figure 5 shows the use of the new UID generator. DicomEdit's UID generator creates new UIDs from randomly generated Universally Unique Identifiers [8]. UID replacement requires some care, as the hierarchical structure of DICOM data is defined implicitly in the values of the Study Instance UID and Series Instance UID. The DicomEdit UID generator retains the UID-defined structure: for each distinct pre-modification UID u1, u2, … un, the replacement UIDs u1′, u2′, un′ are the same in every modified object. Thus, for example, DICOM objects that were in the same study before script application are still in the same study (with a newly generated UID) afterwards.
Results
We consider two distinct use cases of the DicomBrowser software: using the GUI to identify DICOM files, to modify the metadata, and to send the modified objects to a DICOM archive and using the command-line tools to manage complex metadata changes to many studies.
GUI-Based Manual Metadata Modification
The DicomBrowser GUI is well suited to identifying DICOM data stored in files, to making ad hoc modifications to metadata one study at a time, and to saving the changes to file or to a DICOM archive.
Locating and Reading DICOM Files
A user finds and reads DICOM files using the File/Open… menu item. A standard dialog for opening files appears, and the user selects a directory containing her DICOM data. The program identifies all DICOM files in the selected directory and all its subdirectories, then displays a tree representation of the patient–study–series–instance hierarchy. The user can click the icon next to a patient to “expand” that patient and see a list of contained studies, can similarly expand studies to view the contained series, and can expand each series to view the contained instances. Clicking the icon next to an expanded item “collapses” the item, hiding its contents. More DICOM objects can be added to this view by using the File/Open… menu item again.
Viewing and Editing Metadata
When the user clicks to select an item in the tree view, the metadata are read from all of the contained DICOM files and displayed in the metadata summary table on the right side of the DicomBrowser window. The metadata summary table has one row per attribute and four columns: the attribute's tag (i.e., the 32-bit unsigned number that uniquely identifies the attribute); the name of the attribute, a word describing what action will be applied to that attribute if the data are exported; and an attribute value summary. If all the selected DICOM objects have the same value for an attribute, the value summary is simply that value; otherwise, the summary is a prefix describing how many values are present (e.g., “42 values:”) followed by an enumeration of the values (truncated if there are too many to display).
The simplest way to modify an attribute is to click on its value field. If there is a single existing value for that attribute, the user simply edits the value in place; otherwise, a dialog appears in which the user can either choose one of the existing values from a pull-down menu, enter a new value, enter a blank value (manually or using the “Clear” button), or delete the attribute entirely using the “Delete” button. The “Set” and “Cancel” buttons accept or abort the modification, respectively, and close the edit dialog.
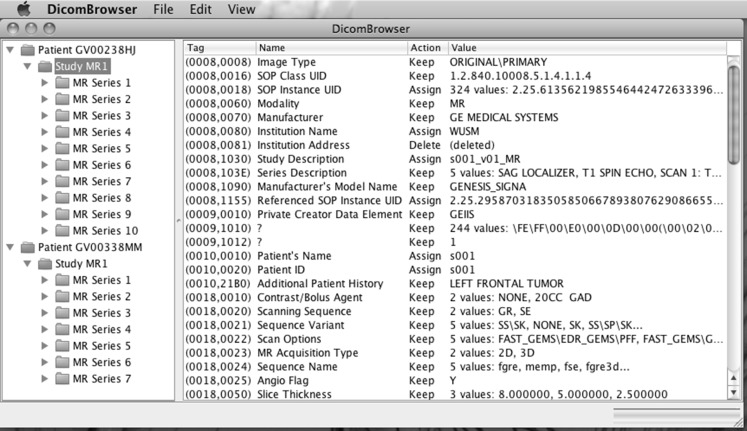
Alternatively, the user can select an attribute by clicking in the tag, name, or action column and then change the attribute by selecting an action from the edit menu: keep retains the original value, clear sets an empty value, and delete removes the attribute. The “Add new attribute…” item in the edit menu allows users to add an attribute not previously present in the object by specifying the attribute tag and value. The “Action” column indicates which rows (attributes) have been modified (Fig. 7). All changes can be unapplied and reapplied using the edit/undo and edit/redo menu items.
Fig. 7.
DicomBrowser metadata summary, with some attributes modified
The metadata summary window shows any metadata changes that have been made, but no changes are made to the original data on disk. Specified changes take effect only when the data are exported, either to the file system (as new files or overwriting the originals) or via DICOM C-STORE.
GUI-Based Scripted Metadata Modification
DicomBrowser can read and apply DicomEdit metadata modification scripts by using the “Apply script…” item in the edit menu. This is a streamlined method for repeatedly making similar metadata changes to many sessions. Any variables defined in the script (except those explicitly declared hidden) are displayed in a dialog box for the user to modify before the script is applied (see Fig. 6).
Command-Line Processing
The DicomBrowser GUI is useful for viewing and modifying metadata in a small number of studies, but manually applying modifications to many studies can be tedious and error prone. Large-scale metadata modification is better done by writing a script describing the changes common to all DICOM objects, then using DicomSummarize to build a spreadsheet of the components that must be modified one patient, study, or series at a time. DicomSummarize reads a configuration file to determine which DICOM attributes should be included, then produces a spreadsheet filled with values taken from the original DICOM objects. Users can specify the metadata changes they want by editing this spreadsheet using a program such as Microsoft Excel or LibreOffice.
In order to build a spreadsheet summary of DICOM metadata, the user must first specify which attributes are to be included in the spreadsheet. This is done in a DicomSummarize configuration XML, which lists for each attribute of interest its tag and its level in the patient/study/series hierarchy. The sample configuration in Fig. 8 specifies a spreadsheet for modifying the patient ID and name, the study ID and study description, and the series description. Each entry in the columns list defines a spreadsheet column containing the original value of that field; those entries with a remap attribute define an additional column where the user can enter a replacement value. Note that this configuration file creates a column for series number (0020,0011), not for modification, but rather so that the user can see the series number when modifying the series description.
Fig. 8.
DicomSummarize configuration file
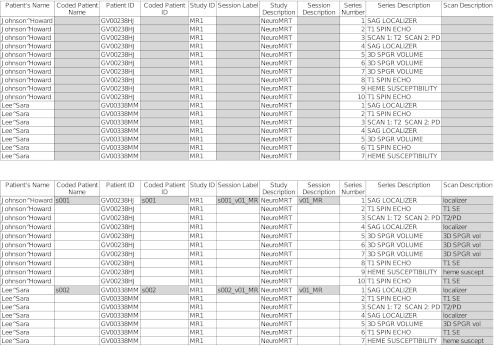
The command-line utility DicomSummarize walks through specified directories, looking for DICOM files, and uses the spreadsheet configuration XML to build a spreadsheet (Fig. 9, top). The user can fill in values in the for-modification columns (shaded in Fig. 9) to specify the changes to be made (Fig. 9, bottom, with user modifications shaded). Finally, the command-line utility DicomRemap reads the modified spreadsheet and applies the specified changes to the DICOM objects, along with any additional changes described in a DicomEdit script. DicomRemap can save the modified DICOM objects to disk or send them to a DICOM storage service using the C-STORE protocol.
Fig. 9.
A DicomSummarize spreadsheet. Top: spreadsheet generated by DicomSummarize; columns to be edited by user are shaded. Bottom: edited spreadsheet, specifying metadata changes to be made; edits are shaded
Discussion
DicomBrowser allows users to view and modify metadata across multiple studies. The process of modifying metadata can be fully interactive and ad hoc, via the DicomBrowser GUI; fully automated, via the application of DicomEdit metadata modification scripts; or partially automated with user input for specific fields, via user-defined variables in a DicomEdit script or remap columns in a DicomSummarize-generated spreadsheet.
The DicomEdit metadata modification language interpreter is a discrete component that can be embedded in other software. For example, the XNAT imaging informatics platform [9] uses DicomEdit to modify the metadata of received DICOM objects. The DicomEdit interpreter is open source and available at http://hg.xnat.org/dicomedit.
DicomEdit Scripts for Deidentification
A key use of DicomEdit scripts is to encapsulate a customizable, reproducible, one-step operation for removing PHI while retaining needed metadata. The DicomEdit language supports not only simple deletion or clearing of potentially sensitive fields, but also sophisticated replacement of values for pseudonymization. Scripts that include user-defined variables generate a dialog asking the user to fill in a value (Fig. 6). Alternatively, complex values such as one-way hashes or other subject or session identifiers can be generated by and retrieved from a web service using the built-in function getURL (Fig. 3). Also, the UID generator retains UID-defined relationships between objects such as study or series membership without leaving any trace of the original UID.
Sample DicomEdit scripts for deidentification are available for download at http://nrg.wustl.edu/software/dicom-browser. Before using these or any scripts for deidentification in a production environment, we recommend testing the script on data from the same facilities that will be producing the data to be deidentified and using multiple viewing methods to verify that the results are satisfactory—e.g., that all PHI has been removed. Our practice is first to examine a few files from each of a few studies in the DicomBrowser GUI, then to examine some different files using dcmdump (from DCMTK [10]), and finally, to examine a few more files using a binary dump utility such as the UNIX program od. We have found that vendor private tags in particular sometimes contain PHI in ways that are obscured by DICOM-aware programs such as DicomBrowser and dcmdump.
Comparison to Other Software
There are many programs, commercial and open source, available for viewing DICOM images and metadata; some allow metadata modification. We describe and compare a few here. This is not a comprehensive list, but we are not aware of any software nearer in functionality to DicomBrowser than the programs enumerated below.
RSNA MIRC
The RSNA MIRC DicomEditor [11] is a GUI-based DICOM image viewer and anonymizer. DICOM images and metadata can be viewed one file at a time. Changes can be made to metadata using the MIRC DICOM Anonymizer language. DicomEditor does not provide a summary view of either the patient–study–series hierarchy or the metadata in multiple files.
The RSNA MIRC Clinical Trial Processor (CTP) [12] is a program that can, among other operations, receive, deidentify, and resend DICOM files via C-STORE and HTTP protocols. The deidentification is customizable using the MIRC DICOM Anonymizer language. The MIRC DICOM Anonymizer includes database support for remembering previously used mappings, allowing preservation of the UID-defined patient–study–series structure. The CTP is useful for fully automated workflows, but it is not designed for interactive viewing or manipulation.
DCMTK/GDCM/dcm4che
DCMTK [10], GDCM [13], and dcm4che [3] are software packages each implementing part or all of the DICOM standard. All include command-line programs for common operations. For example, in DCMTK, the program dcmdump displays metadata for a single file, including sequence contents; dcmodify changes metadata, even in nested attributes; and storescu uses the C-STORE protocol to send data to a DICOM storage server. None of these libraries includes a GUI-based application, nor programs with other DicomBrowser features such as summary views of metadata from multiple files, hierarchy-preserving UID generation, conditional assignments, or scriptable prompting for user-supplied values. Such features could be built on top of any of these libraries; DicomBrowser is based on dcm4che.
PixelMed DicomCleaner
PixelMed [14] is another library implementing the DICOM standard. It includes a GUI utility, DicomCleaner, that has many features comparable to DicomBrowser's. DicomCleaner presents a tree view of the patient–study–series–instance hierarchy and displays some additional metadata at each level, though it does not provide a complete metadata summary. DicomCleaner allows some metadata modification, controlled via predefined sets of changes (e.g., “Remove patient characteristics”), unlike DicomBrowser's support for arbitrary operations on individual attributes. DicomCleaner offers both file-based and DICOM networking-based import and export; it also supports blacking out specific regions of the image data.
OsiriX
OsiriX [15] is a full-featured DICOM image viewing and management program for Mac OS X. In addition to its extensive image viewing capabilities, OsiriX has support for viewing and editing metadata. The metadata viewer shows one file at a time, so the user cannot view the complete set of original values as in DicomBrowser. As each modification is made, it is explicitly assigned to the patient, study, series, or image (instance) level. There is also an anonymize panel, which allows several tags to be modified in one operation. There is no support for scripted modifications.
OsiriX has a persistent database that stores metadata for all files that have been loaded, essentially serving as an internal PACS. The DicomBrowser GUI does use a database for managing metadata, but the contents are deleted when the application window is closed. DicomBrowser's transient database is better suited to workflows where each study is loaded once, viewed, modified, and finally exported. A comparable workflow in OsiriX would require the user to explicitly remove objects from the database.
ImageJ
ImageJ [5] is a general-purpose image viewing, processing, and analysis tool capable of handling DICOM images. It has a plugin architecture allowing it to be extended. Plugins are available to perform operations on DICOM files including sorting viewed files by study and series and performing a simple anonymization. No existing plugins provide patient, study, or series-level views of the metadata, nor support viewing or modifying metadata beyond a few predefined fields. In principle, any functionality implemented as an ImageJ plugin is also available in DicomBrowser, since DicomBrowser includes ImageJ for image viewing.
Features for Future Development
The DicomBrowser tools currently provide only limited support for the DICOM SQ (sequence) value representation. Sequences are invisible in the DicomBrowser GUI. In DicomEdit scripts, top-level attributes of type SQ can be ignored or deleted, but no changes can be made individually to attributes nested within a sequence. Adding sequence support is especially important for multiframe DICOM objects, since metadata for individual frames are stored in sequences.
Neither the DicomEdit interpreter nor the DicomBrowser GUI offers any explicit support for private data elements, though they can be operated on using the numeric tag like any other attribute. In practice, we have found that each vendor private element gets assigned the same numeric tag from one file to the next, even across studies, so the lack of explicit support has not yet been a problem. In general, DicomEdit operates only on top-level attributes using numeric tags, with no support for content-based tag block assignment or for any pattern matching on tags.
The DicomBrowser GUI displays only the DICOM dataset and in particular does not display the file metainformation attributes. Any files written by DicomBrowser do include a newly generated part 10 header, complete with file metainformation.
All of the DicomBrowser tools read data only from files. There is no support for getting data from a storage service using DICOM network services such as C-FIND, C-MOVE, or C-GET. No type checking or other validation is performed on metadata changes, so it is possible to use DicomBrowser to generate DICOM objects with invalid attribute values.
DicomBrowser does not directly support any modification of the image pixel data. Some devices do add text to the image data, and in principle, one could use DicomBrowser's embedded ImageJ image manipulation capabilities to modify the image data, but an operation comparable to DicomCleaner's Blackout would be a useful addition.
Conclusion
The DicomBrowser software package includes GUI and command-line programs for viewing and flexibly modifying DICOM metadata. Metadata changes can be specified manually in the DicomBrowser GUI, in a spreadsheet generated by DicomSummarize, or via scripts written in the DicomEdit language. DicomBrowser is open-source software, available for download at http://nrg.wustl.edu/software/dicom-browser.
References
- 1.National Electrical Manufacturer's Association: Digital Imaging and Communications in Medicine (DICOM). Available at: http://dicom.nema.org. Accessed 8 September 2011
- 2.National Electrical Manufacturer's Association: Digital Imaging and Communications in Medicine (DICOM) standard. Available at: ftp://medical.nema.org/medical/dicom/2011. Accessed 8 September 2011
- 3.dcm4che Open source clinical image and object management. Available at: http://dcm4che.org. Accessed 8 September 2011
- 4.HyperSQL. Available at: http://hsqldb.org. Accessed 8 September 2011
- 5.Rasband WS: U.S. National Institutes of Health. ImageJ. Available at: http://imagej.nih.gov/ij. Accessed 8 September 2011
- 6.ISO/IEC 14977: Information technology—syntactic metalanguage—Extended BNF, Geneva, Switzerland: ISO/IEC, 2006
- 7.ANTLR. Available at: http://www.antlr.org. Accessed 8 September 2011
- 8.ITU-T X.667, Generation and registration of universally unique identifiers (UUIDs) and their use as ASN.1 object identifier components, Geneva, Switzerland: International Telecommunications Union, 2004
- 9.Marcus DS, Olsen TR, Ramaratnam M, Bucker RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 10.OFFIS DCMTK DICOM Toolkit. Available at: http://dicom.offis.de/dcmtk.php.en. Accessed 8 September 2011
- 11.RSNA MIRC DicomEditor. Available at: http://mircwiki.rsna.org/index.php?title=DicomEditor. Accessed 8 September 2011
- 12.RSNA MIRC Clinical Trial Processor (CTP). Available at: http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Accessed 8 September 2011
- 13.Grassroots DICOM (GDCM). Available at: http://sourceforge.net/projects/gdcm. Accessed 8 September 2011
- 14.PixelMed Java DICOM Toolkit. Available at: http://www.pixelmed.com. Accessed 8 September 2011
- 15.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17:205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]