Abstract
The purpose of this study was to demonstrate the robustness of our prior computerized texture analysis method for breast cancer risk assessment, which was developed initially on a limited dataset of screen-film mammograms. This current study investigated the robustness by (1) evaluating on a large clinical dataset, (2) using full-field digital mammograms (FFDM) as opposed to screen-film mammography, and (3) incorporating analyses over two types of high-risk patient sets, as well as patients at low risk for breast cancer. The evaluation included the analyses on the parenchymal patterns of women at high risk of developing of breast cancer, including both BRCA1/2 gene mutation carriers and unilateral cancer patients, and of women at low risk of developing breast cancer. A total of 456 cases, including 53 women with BRCA1/2 gene mutations, 75 women with unilateral cancer, and 328 low-risk women, were retrospectively collected under an institutional review board approved protocol. Regions-of-interest (ROIs), were manually selected from the central breast region immediately behind the nipple. These ROIs were subsequently used in computerized feature extraction to characterize the mammographic parenchymal patterns in the images. Receiver operating characteristic analysis was used to assess the performance of the computerized texture features in the task of distinguishing between high-risk and low-risk subjects. In a round robin evaluation on the FFDM dataset with Bayesian artificial neural network analysis, AUC values of 0.82 (95% confidence interval [0.75, 0.88]) and 0.73 (95% confidence interval [0.67, 0.78]) were obtained between BRCA1/2 gene mutation carriers and low-risk women, and between unilateral cancer and low-risk women, respectively. These results from computerized texture analysis on digital mammograms demonstrated that high-risk and low-risk women have different mammographic parenchymal patterns. On this large clinical dataset, we validated our methods for quantitative analyses of mammographic patterns on FFDM, statistically demonstrating again that women at high risk tend to have dense breasts with coarse and low-contrast texture patterns.
Keywords: Computerized texture analysis, Breast cancer risk assessment, Mammographic parenchymal patterns, Full-field digital mammograms, Quantitative imaging analysis
Introduction
Breast cancer is the most commonly diagnosed cancer among women besides skin cancer, and about one in eight women will develop invasive breast cancer over the course of her lifetime in the USA. An estimated 39,520 breast cancer deaths were expected in 2011 [1]. Currently, mammography is still the best available imaging modality for breast cancer detection, and it can detect breast cancer at an early stage. The combination of early detection through screening, improvements in treatment, and increased awareness resulted in decreased breast mortality among women since 1990 [1].
Radiographic density or mammographic breast density (percentage density) refers to the amount of fibroglandular area relative to the whole breast area on a mammographic image. This reflects the breast composition and relative amount of fibroglandular tissue, connective tissue, and fat. BI-RADS classifies a mammogram into one of four categories based on its percentage breast density [2].
Mammographic parenchymal patterns have been studied extensively to demonstrate the relationship between breast density and the risk of developing of breast cancer [3–8]. Numerous studies have shown that increased mammographic breast density may yield as high as a four- to sixfold increase in risk of developing breast cancer [9–11]. Extensive mammographic density is strongly associated with the risk of breast cancer detected by screening or between screening tests, and a substantial fraction of breast cancers can be attributed to this risk factor [12].
Computerized texture analysis has been applied on mammographic images to characterize mammographic parenchyma and estimate breast density in order to assess the risk of developing breast cancer. Byng et al. calculated skewnesss extracted from the image histogram and a fractal texture measure in the classification of mammographic parenchyma, and the relationship to breast cancer risk [13]. Manduca et al. performed texture analysis on a case–control study on screen-film mammograms. The texture features showed a similar predicting power for breast cancer risk assessment (area under curve(AUC) = 0.58–0.60) as that obtained with just breast percent density (AUC = 0.58) [14]. Wei et al. also performed a case–control study with computer-extracted texture features on screen-film mammograms; the AUC of 0.74 was obtained by using combined texture features as the decision variable for differentiating case subjects from control subjects in their study [15].
We also have developed computerized image analysis methods with which to characterize breast parenchyma for breast cancer risk assessment using digitized screen-film mammograms (SFM) [16–20]. Our results showed that women at high risk of developing breast cancer tended to have dense breasts and their mammographic parenchymal patterns were coarse and low in contrast. An AUC value of 0.91 was obtained in the task of differentiating between BRCA1/2 gene mutation carriers and low-risk women using a dataset of 172 screen-film mammograms, in which 30 subjects were gene mutation carriers and 142 subjects were women at low risk [17].
The purpose of our current study was to demonstrate the robustness of our prior computerized texture analysis method for breast cancer risk assessment, which was developed initially on a limited dataset of screen-film mammograms. This current study investigated the robustness by (1) evaluating on a large clinical dataset, (2) using full-field digital mammograms (FFDM) as opposed to screen-film mammography, and (3) incorporating analyses over two types of high-risk patient sets, as well as patients at low risk for breast cancer.
In this study, quantitative image analysis was performed on a region of interest (ROI) on FFDM. A series of features (mathematical descriptors) was extracted from each ROI to characterize the mammographic parenchymal patterns of the image. The evaluation comprised women at high risk of developing breast cancer, including both BRCA1 and BRCA2 gene mutation carriers and unilateral cancer patients, and women at low risk of developing breast cancer. Receiver operating characteristic (ROC) analysis [21, 22] was used to assess the performance of computer-extracted features in the task of differentiating the high-risk women from the low-risk women.
Materials and Methods
Database
Two high-risk datasets were included in this study: FFDM images from BRCA1 or BRCA2 gene mutation carriers and FFDM images from women with unilateral breast cancer.
FFDM images were obtained from BRCA1/2 gene mutation carriers who had been recruited from the cancer risk clinic at the University of Chicago Medical Center, yielding a total of 53 gene mutation carriers. These gene mutation carriers were tested at a Clinical Laboratory Improvement Amendments-approved laboratory under an Institution Review Board (IRB) approved protocol. Their ages ranged from 21 to 72 years, with an average age of 40.2 years, a median of 40, and a standard deviation of 11.8 years. The FFDM images were reviewed by an expert mammographer and used only if no detectable malignant or benign abnormalities were observed.
The other high-risk dataset included FFDM images from women with unilateral breast cancer. A total of 75 unilateral cancer cases were included in this study. Ages ranged from 25 to 89 years, with a mean age of 55.8 years, a median of 55 years, and a standard deviation of 15.0 years. Only the mammograms at the cancer diagnosis date were used in this study, and the contralateral mammograms from these unilateral cancer patients were used for the computerized image analysis. These mammograms were also reviewed by an expert mammographer and were included in the study only if no detectable abnormalities were observed in the contralateral image.
The low-risk dataset included FFDM images from women who had undergone screening mammography in the Department of Radiology at the University of Chicago Medical Center between 2006 and 2008. These low-risk women had no family history of breast or ovarian cancer, had no prior history of breast cancer or breast benign disease, never had a benign breast biopsy, and had a lifetime risk of developing breast cancer of less that 10% based on the Gail breast cancer risk assessment model [23]. Under these conditions, FFDM images from 328 women were included in this study as the low-risk group. Ages ranged from 32 to 89 years, with a mean age of 58.4 years, a median age of 58 years, and a standard deviation of 11.9 years.
Since age is a very important breast cancer risk factor, an age-matched subset analysis was also performed in order to minimize the possible bias due to the difference in age distribution. Age matching was performed using a 1:4 ratio to include as many patients in the study as possible, i.e., one high-risk subject was age-matched with four low-risk subjects at 5-year intervals for the study. Using this age-matching criterion, 136 low-risk cases were randomly selected and age-matched with 34 BRCA1/BRCA2 gene mutation carriers. In addition, 268 low-risk cases were randomly selected and age-matched with 67 unilateral cancer cases. These two age-matched data subsets were used for the age-matching study.
The full-field digital mammograms used in this study were retrospectively collected at the University of Chicago Medical Center under an IRB approved protocol. All images were obtained with a GE (Waukesha, WI, USA) Senographe 2000D FFDM system. The FFDM images were acquired at 12-bit quantization with a pixel size of 100 × 100 μm. ROIs of 256 × 256 pixels were manually selected from the central breast region behind the nipple in the craniocaudal projection of mammographic images. The details of the ROI extraction have been described elsewhere [24].
Computerized Feature Extraction
Computerized feature extraction was performed on each ROI to assess the mammographic parenchymal patterns. These computer-extracted texture features were based on (a) local composition (density-related measures), (b) gray-level histogram analysis, (c) spatial relationship among gray-levels, (d) fractal analysis, including DBC (fractal dimension based on box-counting method) and DM (fractal dimension based on Minkowski method), (e) edge frequency analysis, and (f) Fourier analysis, including RMS variation, first moment of the power spectrum, and power law β from power spectral analysis. Detailed descriptions of these characteristic features can be found elsewhere [16–20, 24, 25].
Linear stepwise feature selection was performed using the Wilks Lambda criterion [26, 27] to select a subset of features for the classification task. A Bayesian artificial neural network [28] (BANN) was then used to merge these selected features in an iterated leave-one-case-out analysis.
Performance Evaluation
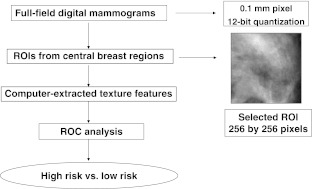
ROC analysis [21, 22, 29] was used to determine the performance of each individual texture feature and the merged classifier output in the task of distinguishing between mammographic parenchymal patterns of high-risk patients and parenchymal patterns of subjects in the low-risk group. Here, the area under the fitted ROC curve (AUC value) was used as a figure of merit to assess the potential usefulness of these computer-extracted texture features in characterizing parenchymal patterns and breast cancer risk. Statistical Z tests were performed to assess the statistical significance of the difference between the estimated AUC value of each individual feature and an AUC of 0.5 (equivalent to random guessing). The level of statistical significance for the difference of two AUC values was calculated using the ROCKIT computer program [29]. The Holm t test was applied for multiple comparison corrections [30]. The overall computerized analysis scheme is shown in Fig. 1.
Fig. 1.
Computerized texture analysis method for breast cancer risk assessment on FFDM
Results
BRCA1/BRCA2 Gene Mutation Carriers Versus Low-Risk Group
Using the database of 53 BRCA1/2 gene mutation carriers and 328 low-risk women, a total of 45 features were extracted from each individual ROI. The AUC performance of the individual features in the task of distinguishing mammographic parenchymal patterns of BRCA1/BRCA2 gene mutation carriers from those of the low-risk women in terms of AUC values was found to be between 0.52 and 0.76.
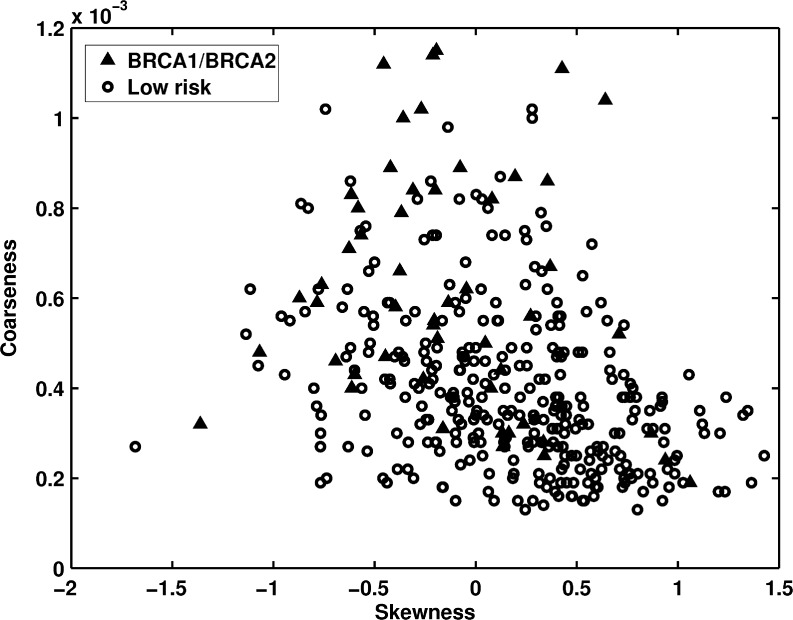
Figure 2 shows the distribution between skewness and coarseness for BRCA1/BRCA2 gene mutation carriers and low-risk women. The skewness measure is related to the mammographic density in the breast, and a dense ROI would have a small value of the skewness measure. The coarseness feature was calculated based on a Neighborhood Gray-Tone Difference Matrix (NGTDM) and represents the coarseness measure of the mammographic image. It considers the difference between the gray levels of all pixels in the region and a particular gray level in the region, and also the mean gray level of the surrounding neighbors. In a coarse texture, the gray level values of neighboring pixels tend to be similar, so the difference between the gray level of the pixels and the mean gray level of the neighborhood is small. Larger coarseness values correspond to coarser mammographic parenchymal patterns within the image. AUC values of 0.66 and 0.75 were obtained in the task of distinguishing between gene mutation carriers and the low-risk group with skewness and coarseness features as decision variables, respectively.
Fig. 2.
The distribution of skewness versus coarseness between BRCA1/BRCA2 gene mutation carriers and the low-risk women
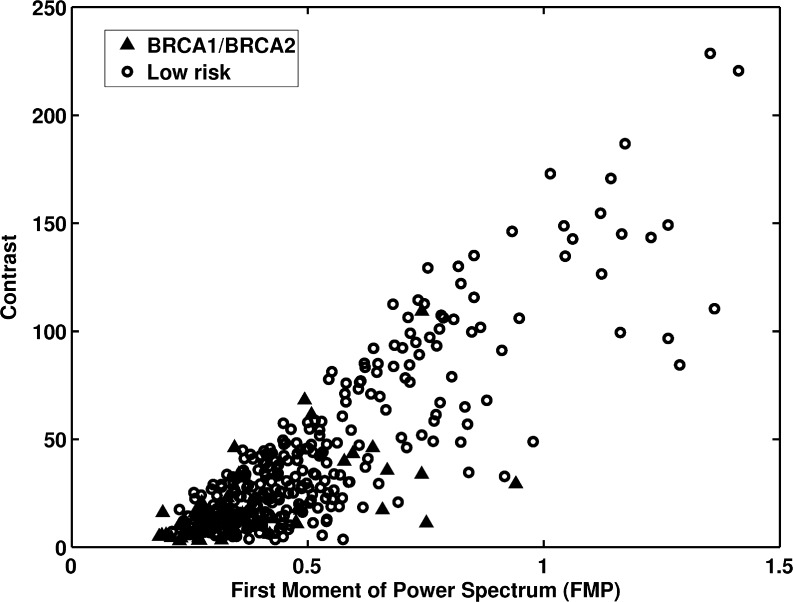
Figure 3 shows the distribution of first moment of power (FMP) spectrum and contrast for the two groups. The FMP and contrast yielded AUC values of 0.70 and 0.75 in the task of distinguishing between the two groups. The FMP was calculated from the two-dimensional Fourier transform of the ROI and was used to characterize the mammographic parenchymal pattern’s spatial frequency content. Like coarseness, FMP represents the coarseness measure of the mammographic image. A small FMP value indicates a dominance of lower spatial frequency content and coarser texture.
Fig. 3.
The distribution of first moment of power spectrum versus contrast between BRCA1/BRCA2 gene mutation carriers and the low-risk women
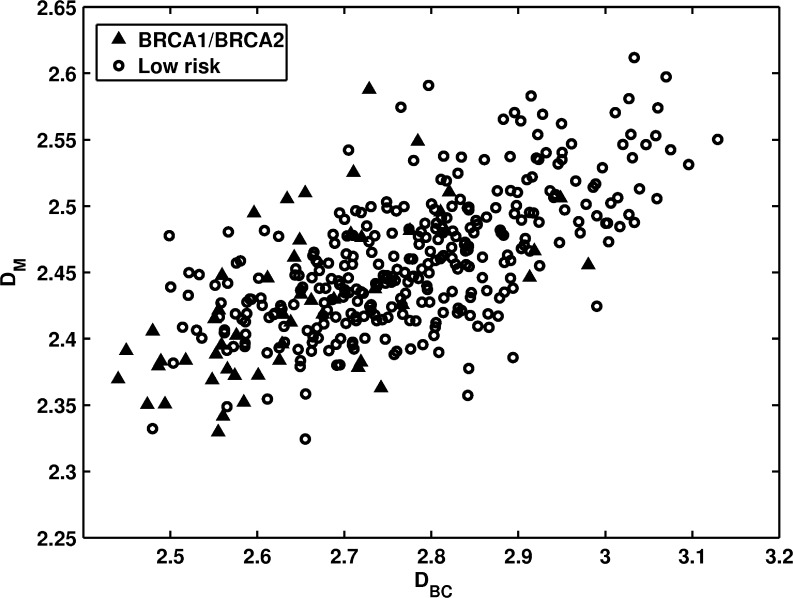
The distribution of the gene mutation carriers and the low-risk women in terms of two fractal features is shown in Fig. 4. These two fractal dimensions were computed using different algorithms. DBC is based on a box-counting method by calculating surface areas using different ruler sizes (image resolutions). DM is based on a Minkowski algorithm by calculating volumes at different scales through performing morphological operations. Gene mutation carriers (high-risk group) exhibited lower fractal dimension values on both fractal features than individuals at low-risk. AUC values of 0.76 and 0.67 were obtained in the task of distinguishing between the two groups using the box-counting and Minkowski fractal features as decision variables, respectively.
Fig. 4.
The distribution of DBC versus DM between BRCA1/BRCA2 gene mutation carriers and the low-risk women
By applying leave-one-case-out (round-robin) stepwise feature selection [31], six texture features were selected as a subset of features for the classification task. The selected features characterized the coarseness, homogeneity, randomness, and nonlinearity of the mammographic texture patterns of the images. A BANN was used as a classifier to merge these six features using a leave-one-case-out analysis. An AUC value of 0.82 (SE = 0.03; 95% CI [0.75, 0.88]) was obtained using the output of the BANN as the decision variable in the task of distinguishing between gene mutation carriers and the low-risk group.
As mentioned earlier, age matching was performed with a 1:4 ratio and a 5-year interval, yielding 34 gene mutation carriers age-matched with 136 low-risk women. AUC values between 0.51 and 0.71 were obtained when using individual features as decision variables in the task of distinguishing between the two groups using the age-matched dataset. An AUC value of 0.81 (SE = 0.04; 95% CI [0.71, 0.89]) was obtained when using BANN output as the decision variable for the merged feature from ROC analysis.
Unilateral Cancer Women Versus Low-Risk Group
For the unilateral cancer patients, only the normal, i.e., lesion-free, mammograms from the contralateral breast were included in the analysis. The individual texture feature performances in the task of distinguishing between unilateral cancer patients and the low-risk group were evaluated using ROC analysis. AUC values of 0.51 to 0.68 were obtained by using individual texture features as decision variables in the task of distinguishing between the two groups.
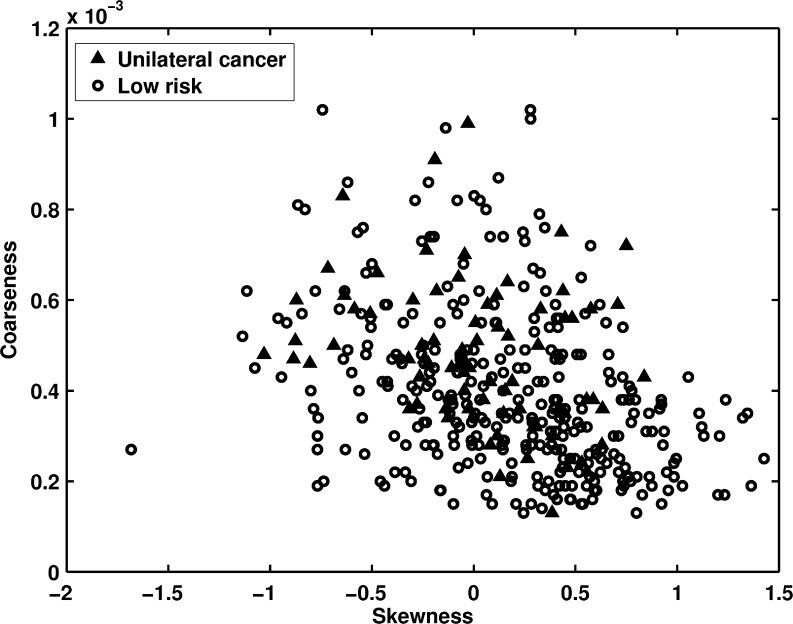
Figure 5 shows the distribution of skewness versus coarseness texture features between the contralateral breasts of unilateral cancer patients and the low-risk group. Again, the smaller skewness and higher coarseness values were observed for the unilateral cancer patients (high-risk group). AUC values of 0.60 and 0.66 were obtained when using skewness and coarseness as decision variables in the task of distinguishing the two groups, respectively.
Fig. 5.
The distribution of skewness versus coarseness between unilateral cancer patients and the low-risk women
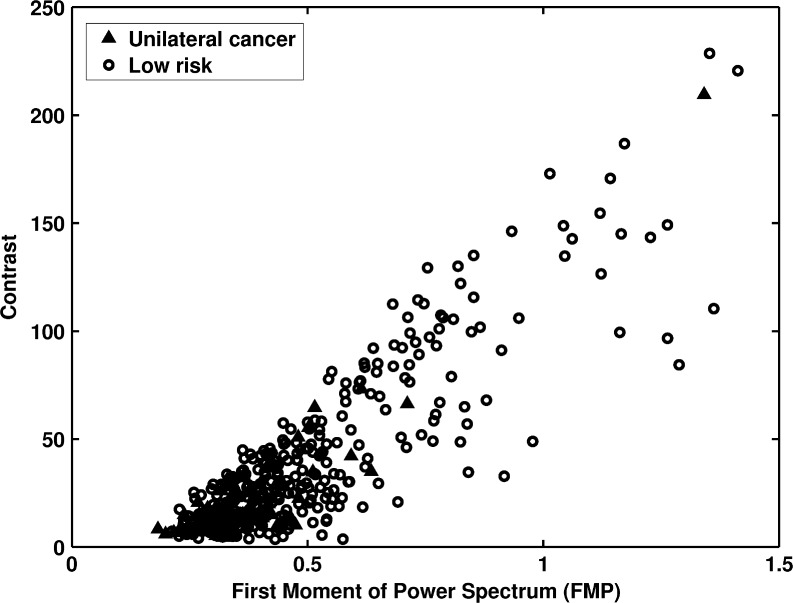
Figure 6 shows the distribution of the FMP versus contrast measures between the two groups. Lower contrast and smaller FMP values were observed for the unilateral cancer patients of the high-risk group. The mammographic images of the unilateral cancer patients had coarser textures than those of the low-risk group, and their mammographic images tended to be lower in contrast. The FMP and contrast features yielded AUC values of 0.68 and 0.67 in the task of distinguishing between the two groups, respectively.
Fig. 6.
The distribution of first moment of power spectrum versus contrast between unilateral cancer patients and the low-risk women
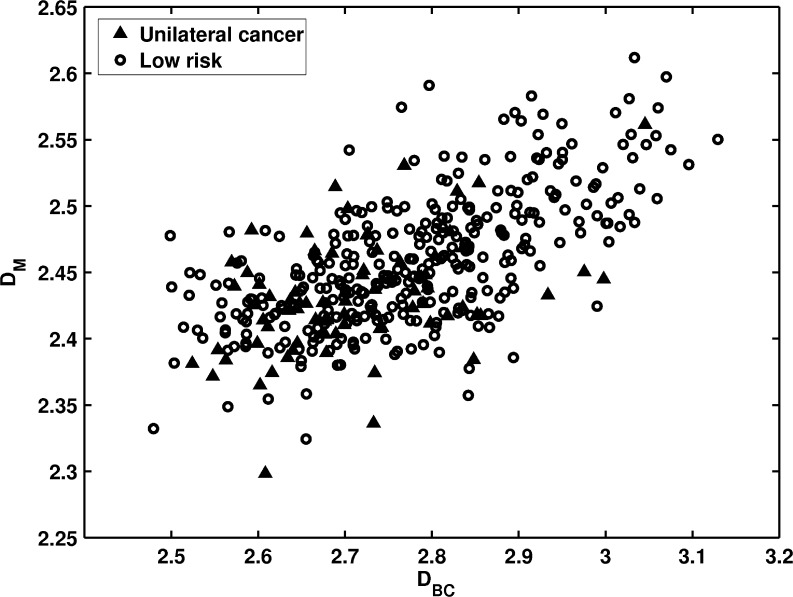
Figure 7 shows the distribution of two fractal features between the unilateral cancer patients and the low-risk group. For both fractal features, smaller fractal dimension measures were observed in the unilateral cancer high-risk group. This indicates that the mammographic patterns are coarser for high-risk women. For the task of distinguishing between the two groups, the box-counting dimension DBC and the Minkowski dimension DM yielded AUC values of 0.65 and 0.67, respectively.
Fig. 7.
The distribution of DBC versus DM between unilateral cancer patients and the low-risk women
The BANN classifier was applied to merge the texture features selected from the linear stepwise feature selection method with a leave-one-case-out analysis. The output generated from the BANN was used as a decision variable for ROC analysis. An AUC value of 0.73 (SE = 0.03; 95% CI [0.67, 0.78]) was obtained in the task of distinguishing the two groups using merged features.
An age-matched analysis was also performed on the task of differentiating the unilateral cancer patients from the low-risk group. Sixty-seven unilateral cancer patients were randomly age-matched with 268 low-risk women to form the age-matched dataset. AUC values of 0.51 to 0.69 were obtained in terms of individual feature performance in the task of differentiating between the two groups in the age-matched dataset. An AUC value of 0.70 (SE = 0.04; 95% CI [0.63, 0.77]) was obtained when using merged features from the BANN classifier in a leave-one-case-out analysis.
Combined High-Risk Women Versus Low-Risk Group
ROC analysis was also performed after combining both BRCA1/2 gene mutation carriers and unilateral cancer patients into one high-risk group. AUC values of 0.53 to 0.71 were obtained by using individual texture features as decision variables in the task of distinguishing between the high-risk group and the low-risk group. Using the BANN to merge the features using a leave-one-case-out analysis, an AUC value of 0.75 (SE = 0.03; 95% CI [0.69, 0.79]) was obtained.
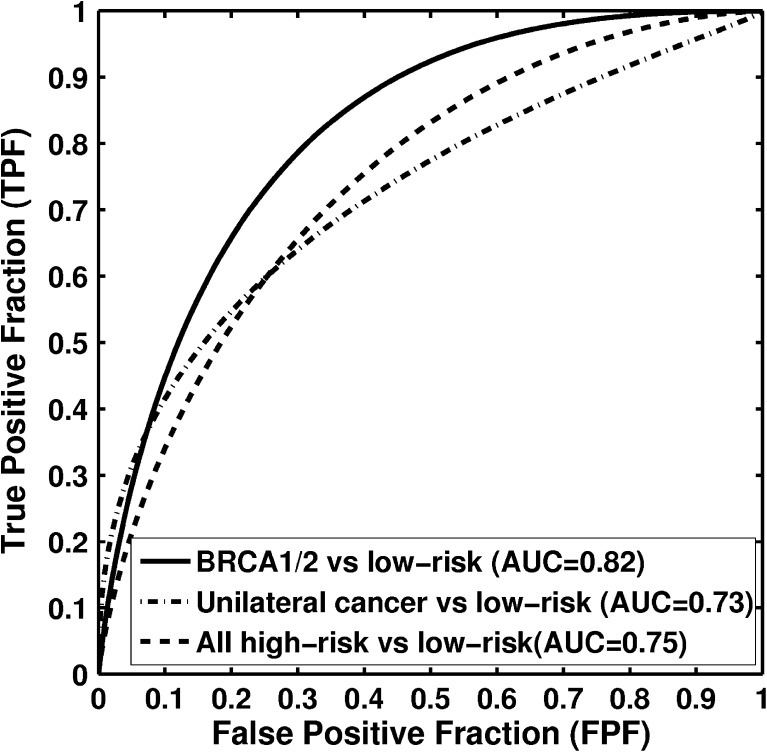
The ROC analysis results are summarized in Table 1. Figure 8 shows the ROC curves for the tasks of distinguishing between BRCA1/2 gene mutation carriers and low-risk women, unilateral cancer patients and low-risk women, and the combined high-risk women dataset and low-risk women.
Table 1.
Performances for the computerized texture analysis on the breast cancer risk assessment on FFDM
| Datasets | Total number of cases | Number of high-risk cases | Number of low-risk cases | AUC ± SE | 95% CI |
|---|---|---|---|---|---|
| BRCA1/2 vs. low risk | 381 | 53 | 328 | 0.82 ± 0.03 | [0.75, 0.88] |
| BRCA1/2 vs. low risk (age-matched) | 170 | 34 | 136 | 0.81 ± 0.04 | [0.71, 0.89] |
| Unilateral cancer vs. low risk | 403 | 75 | 328 | 0.73 ± 0.03 | [0.67, 0.78] |
| Unilateral cancer vs. low risk (age-matched) | 335 | 67 | 268 | 0.70 ± 0.04 | [0.63, 0.77] |
| High risk vs. low risk | 456 | 128 | 328 | 0.75 ± 0.03 | [0.69, 0.79] |
Leave-one-case-out (round robin) analysis was performed on FFDM
AUC area under ROC curve, SE standard error, CI confidence interval
Fig. 8.
Receiver-operating characteristic (ROC) curves indicating the performance of computerized parenchymal analysis performed on full-field digital mammograms (FFDM) in the task of distinguishing between high-risk and low-risk women
Discussion and Conclusion
In this study, we used computerized texture analysis on FFDM to obtain numerical descriptors of breast parenchymal patterns. Our study on FFDM showed that high-risk women, i.e., either BRCA1/2 gene mutation carriers or unilateral cancer patients, and low-risk women have different mammographic parenchymal patterns. Women at high risk for breast cancer tend to have dense breasts, and their mammographic parenchymal patterns are coarse and low in contrast, which agrees with findings from our previous parenchymal analysis studies on screen-film mammography.
It should be noted that the feature sets selected during iterations of the stepwise feature selection process were generally stable, yielding six or seven features for the different classification tasks. While the selected features (i.e., mathematical descriptors) differed for different classification tasks, similar parenchymal characteristics were chosen.
For the features investigated in this paper, for instance, although FMP and coarseness both measured the “coarseness” of the images, they were calculated from different methods: one was based on a Fourier transformation, and the other one was based on the NGTDM method. They measured a similar (but not exactly the same) physical characteristic of the images. Of the two fractal features, one was calculated based on a box-counting method, and the other one was based on morphological operations. Both features agreed that high-risk women have lower fractal dimensions, and thus coarse texture. This, again, reflects the robustness of our computerized texture analysis methods.
We also noticed that the classification performance in terms of AUC value in the task of distinguishing between BRCA1/2 gene mutation carriers and low-risk women (AUC = 0.82) was statistically higher (p value = 0.0374) than the AUC (AUC = 0.73) for distinguishing between unilateral cancer patients and low-risk women (95% CI for the AUC difference [0.0054, 0.1784]). Since we did not have BRCA1/2 mutation testing information in the unilateral cancer patients, the reasons for the classification performance difference were not fully understood. Additional investigations are required to confirm and/or explain this result.
The classification performance in terms of AUC value in the task of differentiating between BRCA1/2 mutation carriers and the low-risk women was lower in this study on FFDM (AUC = 0.82; 95% CI [0.75, 0.88] than in our previous studies with SFM (AUC = 0.91; 95% CI [0.85, 0.97]. We believe that several factors may contribute to this performance difference, including (1) different patient population, ideally we would like to have same subjects with both SFM and FFDM; (2) different dataset, 172 SFM cases versus 381 FFDM cases; and (3) different mammographic acquisition technology, SFM versus FFDM. The further study is needed to fully understand this difference.
In this study, we evaluated the robustness of our image-based risk assessment methods by (1) evaluating with a large clinical dataset, (2) using FFDM as opposed to screen-film mammography, and (3) incorporating analyses over two types of high-risk patient sets (BRCA1 and BRCA2 gene mutation carriers and patients with unilateral cancer). Our results demonstrate that our method, initially developed on digitized screen-film mammography, shows similar findings on the parenchymal patterns of women at high and low risk as presented on FFDM. We believe that the computer-extracted image markers may potentially be used alone or together with clinical measures, as well as biomarkers, for use in identifying women at high risk for breast cancer, and those women would benefit from more aggressive screening.
Acknowledgments
This research was supported in part by the University of Chicago Breast SPORE P50-CA125183, DOE grant DE-FG02-08ER6478, NIH S10 RR021039, and P30 CA14599. M. L. Giger is a stockholder in R2 Technology/Hologic and shareholder in Quantitative Insights, and receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer Statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Breast Imaging Reporting and Data System (BI-RADS) 4. Reston, Va: American College of Radiology; 2003. [Google Scholar]
- 3.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. Am J Roentgenol. 1976;126:1130–1139. doi: 10.2214/ajr.126.6.1130. [DOI] [PubMed] [Google Scholar]
- 4.Boyd NF, Martin LJ, Stone J, Greenberg, et al. Mammographic densities as a marker of human breast cancer risk and their use in chemoprevention. Curr Oncol Rep. 2001;3:314–321. doi: 10.1007/s11912-001-0083-7. [DOI] [PubMed] [Google Scholar]
- 5.Brisson J, Diorio C, Mâsse B. Wolfe’s parenchymal pattern and percentage of the breast with mammographic densities: redundant or complementary classifications? Cancer Epidemiol Biomarkers Prev. 2003;12:728–732. [PubMed] [Google Scholar]
- 6.Boyd NF, Lockwood GA, Martin LJ, et al. Mammographic densities and breast cancer risk. Breast Dis. 1998;10:113–126. doi: 10.3233/bd-1998-103-412. [DOI] [PubMed] [Google Scholar]
- 7.Boyd NF, Martin LJ, Stone J, et al. Mammographic densities as a marker of human breast cancer risk and their use in chemoprevention. Cancer Prev. 2001;3:314–321. doi: 10.1007/s11912-001-0083-7. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson C, Warren R, Bingham SA, et al. Mammographic patterns as a predictive biomarker of breast cancer risk: effect of tamoxifen. Cancer Epidemiol Biomarkers Prev. 1999;8:863–866. [PubMed] [Google Scholar]
- 9.Saftlas AF, Hoover RN, Brinton LA, et al. Mammographic densities and risk of breast cancer. Cancer. 1991;67:2833–2838. doi: 10.1002/1097-0142(19910601)67:11<2833::AID-CNCR2820671121>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 10.Byrne C, Schairer C, Wolfe JN, et al. Mammographic features and breast cancer risk: Effects with time, age, and menopause status. J Natl Cancer Inst. 1995;87:1622–1629. doi: 10.1093/jnci/87.21.1622. [DOI] [PubMed] [Google Scholar]
- 11.Boyd NF, Byng J, Jong R. Quantitative classification of mammographic densities and breast cancer risk: Results from the Canadian National Breast Screening Study. J Natl Cancer Inst. 1995;87:670–675. doi: 10.1093/jnci/87.9.670. [DOI] [PubMed] [Google Scholar]
- 12.Boyd NF, Guo H, Martin LJ, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356:227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 13.Byng JW, Yaffe MJ, Lockwood GA, et al. Automated analysis of mammographic densities and breast carcinoma risk. Cancer. 1997;80:66–74. doi: 10.1002/(SICI)1097-0142(19970701)80:1<66::AID-CNCR9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 14.Manduca A, Carston MJ, Heine JJ, et al. Texture features from mammographic images and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:837–845. doi: 10.1158/1055-9965.EPI-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei J, Chan HP, Wu YT, et al. Association of computerized mammographic parenchymal pattern measure with breast cancer risk: a pilot case–control study. Radiology. 2011;260:42–49. doi: 10.1148/radiol.11101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huo Z, Giger ML, Wolverton DE, et al. Computerized analysis of mammographic parenchymal patterns for breast cancer risk assessment: feature selection. Med Phys. 2000;27:4–12. doi: 10.1118/1.598851. [DOI] [PubMed] [Google Scholar]
- 17.Huo Z, Giger ML, Olopade OI, et al. Computerized analysis of digitized mammograms of BRCA1 and BRCA2 gene mutation carriers. Radiology. 2002;225:519–526. doi: 10.1148/radiol.2252010845. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Giger ML, Olopade OI, et al. Computerized texture analysis of mammographic parenchymal patterns of digitized mammograms. Acad Radiol. 2005;12:863–873. doi: 10.1016/j.acra.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Giger ML, Olopade OI, et al. Fractal analysis of mammographic parenchymal patterns in breast cancer risk assessment. Acad Radiol. 2007;14:513–521. doi: 10.1016/j.acra.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Giger ML, Olopade OI, et al. Power spectral analysis of mammographic parenchymal patterns for breast cancer risk assessment. J Digit Imaging. 2008;21:145–152. doi: 10.1007/s10278-007-9093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metz CE. ROC methodology in radiographic imaging. Invest Radiol. 1986;21:720–733. doi: 10.1097/00004424-198609000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Metz CE. Some practical issues of experimental design and data analysis in radiological ROC studies. Invest Radiol. 1989;24:234–245. doi: 10.1097/00004424-198903000-00012. [DOI] [PubMed] [Google Scholar]
- 23.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Giger ML, Huo Z, et al. Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: effect of ROI size and location. Med Phys. 2004;31:549–555. doi: 10.1118/1.1644514. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med. 2007;58:562–571. doi: 10.1002/mrm.21347. [DOI] [PubMed] [Google Scholar]
- 26.Huberty CJ: Applied Discriminant Analysis, John Wiley and Sons, Inc, 1994
- 27.Lachenbruch PL. Discriminant Analysis. London, England: Hafer; 1975. [Google Scholar]
- 28.Kupinski MA, Edwards DC, Giger ML, et al. Ideal observer approximation using Bayesian classification neural networks. IEEE Trans Med Imaging. 2001;20:886–899. doi: 10.1109/42.952727. [DOI] [PubMed] [Google Scholar]
- 29.ROC software, http://www-radiology.uchicago.edu/krl/roc_soft6.htm
- 30.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 31.Johnson RA, Wichern DW. Applied multivariate statistical analysis. 3. Englewood Cliffs, NJ: Prentice-Hall; 1992. [Google Scholar]