Abstract
Hematopoietic stem cell transplantation (HSCT) is used for treatment of lymphoma. In an attempt to design an efficacious and safe pre-HSCT conditioning regimen, we investigated the cytotoxicity of the combination of busulfan (B), melphalan (M) and gemcitabine (G) in lymphoma cell lines in the absence or presence of drugs that induce epigenetic changes. Cells were exposed to drugs individually or in combination and analyzed by the MTT proliferation assay, flow cytometry, and Western blotting. We used ~IC10 drug concentrations (57 μM B, 1 μM M and 0.02 μM G) which individually did not have major effects on cell proliferation. Their combination resulted in 50% inhibition of proliferation. Reduction to almost half concentration (20 μM B, 0.7 μM M and 0.01 μM G) did not have significant effects, but addition of the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA; 0.6 μM) to this combination resulted in a marked (~65%) growth inhibition. The cytotoxicity of these combinations correlates with the activation of the ATM-CHK2 pathway, phosphorylation of KAP1, epigenetic changes such as methylation and acetylation of histone 3, and activation of apoptosis. The relevance of epigenetic changes is further shown by the induction of DNA methyltransferases in tumor cells with low constitutive levels of DNMT3A and DNMT3B. The addition of 5-aza-2′-deoxycytidine (DAC) to [BMG+SAHA] further enhances cell killing. Overall, BMG combinations are synergistically cytotoxic to lymphoma cells. Epigenetic changes induced by SAHA and DAC further enhance the cytotoxicity. This study provides a rationale for an ongoing clinical trial in our institution using [BMG+SAHA] as pre-HSCT conditioning for lymphoma.
Keywords: DNA alkylator, nucleoside analog, SAHA, lymphoma, drug cytotoxicity Category, Hematological Malignancies or Stem Cell Transplantation
INTRODUCTION
Lymphomas are heterogeneous hematologic neoplasms of the lymphoid system [1]. The intricacy of their pathogenesis and molecular defects require complex treatment modalities, but in spite of recent improvements, a significant proportion of patients still suffer from progressive disease. In this situation, hematopoietic stem cell transplantation (HSCT) is becoming an accepted treatment option [2].
The success of HSCT requires an effective preparative regimen. DNA alkylating agents and nucleoside analogs are commonly used in these regimens [3–6]. These drugs are used in various combinations, and sometimes radiation and/or antibodies are/is added as well. However, an optimized lymphoma-specific conditioning regimen remains to be developed.
Combinations of DNA alkylators and nucleoside analogs have been shown to be efficacious for hematologic disorders [6–8]. DNA alkylators such as busulfan and melphalan have been clinically used since the early 1950’s and 1960’s, respectively [9–10]. It is commonly assumed that their cytotoxic effects are related to the induction of DNA interstrand crosslinks (ICLs), which block replication forks and cause DNA strand breaks when cells attempt to repair the DNA lesions [11]. Nucleoside analogs, on the other hand, inhibit DNA synthesis when incorporated into an elongating nascent DNA strand [12]. Their ability to inhibit ribonucleotide reductase depletes the cellular deoxynucleotide pools and results in a preferential incorporation of the nucleoside analogs into DNA strands, leading to auto-potentiation of the drugs [13]. The differences in their mechanisms of cytotoxicity underscore the potential relevance of combining DNA alkylators and nucleoside analogs. In addition, repair of DNA alkylator-mediated damage is expected to be compromised due to decreased deoxynucleotide pools caused by nucleoside analogs, as shown by Yamauchi et al [14].
The efficacy of DNA alkylator + nucleoside analog combinations is further demonstrated by recent preclinical and clinical studies on the synergistic effects of clofarabine (Clo), fludarabine (Flu) and busulfan (B) combinations in myeloid leukemia [15, 16]. Based on these data we proposed a model whereby Clo and Flu cause chromatin relaxation and facilitate access of busulfan to genomic DNA for alkylation [18]. We recently extended this approach to lymphoma cell lines, also including gemcitabine (G) in the combinations, and observed similar efficacies of [Clo+Flu+B], [Clo+G+B] and [Flu+G+B] in some (but not all) cell types [18].
We used combinations of two nucleoside analogs and a DNA alkylator in the above-mentioned studies. Interestingly, J45.01 cells did not respond well to these combinations. Based on the observed resistance of J45.01 cells to two nucleoside analogs and a DNA alkylator we postulated that an enhanced cytotoxicity would be obtained if we combined one nucleoside analog and two DNA alkylators. In this study we therefore studied the effects of combinations of busulfan (B), melphalan (M) and gemcitabine (G) (i.e., B±M±G) on the J45.01 T-cell lymphoma cell line. Because of the possibility that combining two alkylators, especially at high drug concentrations, might cause excessive normal tissue cytotoxicity it was important that anticancer effects be achievable using reduced doses of the individual drugs. We report in the present study that low drug concentrations of BMG in combination with a chemosensitizer, namely the histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA), do indeed provide very effective killing of lymphoma cells. These findings suggest a role for epigenetic changes in sensitizing lymphoma cells to BMG combinations. Similar responses were observed in myeloid leukemia cell lines, suggesting a generality of the observed effect and providing a basis for a clinical trial on using this four-drug combination as part of pre-transplant therapy for lymphoma patients undergoing HSCT.
MATERIALS AND METHODS
Cell Culture
Lymphoma cell lines J45.01 and Daudi were obtained from the American Type Culture Collection (ATCC, Manassas, VA). KBM7/B5/Bu2506 is a busulfan-resistant CML cell line developed in our laboratory [19]. All cells were cultured in RPMI 1640 (Mediatech, Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Inc., Lawrenceville, GA) and 100 U/ml penicillin and 100 μg/ml streptomycin (Mediatech) at 37°C in a humidified atmosphere of 5% CO2.
Reagents
Busulfan, melphalan and 5-aza-2′-deoxycytidine (DAC) (Sigma-Aldrich, St. Louis, MO) were dissolved in dimethyl sulfoxide (DMSO); gemcitabine (Eli Lilly, Indianapolis, IN) and SAHA (Cayman Chemical Co., Ann Arbor, MI) were dissolved in phosphate-buffered saline (PBS) and ethanol, respectively. The final concentrations of DMSO and ethanol in all experiments did not exceed 0.08% by volume.
Cytotoxicity assay
Cell suspensions were aliquoted (100 μl of 2 × 105 cells/ml) into 96-well plates in the presence of drug(s) or solvent alone and continuously incubated at 37°C for 96 hr. The cells were analyzed for cytotoxicity by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay [20]. The advantage of using a 96-hr treatment in this assay is that the cells proliferate for several cell cycle times without being disturbed prior to staining with MTT. With the relatively short half-lives of the drugs in cell culture medium at 37°C (e.g., busulfan ~16 hr [21]; melphalan ~ 60 min [22]), we assumed there would be negligible residual drug concentrations after a 48-hr exposure. Graphical analyses including calculations of IC20 values (the concentration of drug required for 20% growth inhibition) were done using Prism 5 (GraphPad Software, San Diego, CA).
Flow-cytometric analysis of sub-G1 DNA content
Cells in logarithmic growth phase (5 × 105 cells/ml) were continuously incubated with the indicated concentrations of the drug(s) at 37°C for 48 hr. The cells were centrifuged, resuspended in 70% ethanol in PBS, and fixed at −20°C overnight. Fixed cells were pelleted at 3,000 × g at room temperature, washed with PBS, and treated with 0.25 ml of 500 U/ml RNAse A in PBS containing 1.12% sodium citrate at 37°C for 30 min. After addition of 0.25 ml propidium iodide (PI, 50 μg/ml) solution, the cells were kept in subdued light for at least 1 hr. The DNA content of at least 10,000 cells was analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) and the proportion of cells in the different phases of the cell cycle was determined using CellQuest™ software (Becton Dickinson, Franklin Lakes, NJ). Histograms were analyzed for the proportion of cells with sub-G1 DNA content using ModFit LT (Verity Software House, Topsham, ME).
Apoptosis assay
Cell death by apoptosis following a 48-hr drug exposure was determined by flow cytometric measurements of phosphatidylserine externalization [23] with the Annexin-V-FLUOS staining kit (Roche Diagnostics, Indianapolis, IN) using the FACSCalibur instrument. The extent of cleavage of poly(ADP-ribose) polymerase (PARP) -1 and caspase 3, determined by Western blot, was also used as an indicator of apoptosis.
Determination of caspase activity
J45.01 cells were exposed to the indicated concentrations of drugs for 48 hr, washed with PBS and analyzed for the enzymatic activities of caspases 3 and 9 using colorimetric assay kits from Millipore (Billerica, MA).
Western blot analysis
Cells were incubated with the drug(s) at 37°C for 48 hr, centrifuged, washed with PBS and lysed with cell lysis buffer (Cell Signaling Technology, Danvers, MA). Protein concentrations were determined using a BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL). Western blot analysis was done by separating protein extracts on polyacrylamide-SDS gels and blotting onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Immunoblot analyses by chemiluminescence were done using the Immobilon Western Chemiluminescent HRP Substrate (Millipore, Bedford, MA). All Western blots were performed at least twice, and representative data are shown. All antibodies, their sources and other relevant information are listed in Table 1.
Table 1.
List of primary antibodies, their sources and dilutions
| Antigen | Source/Cat. # | Clone type* | Dilution** |
|---|---|---|---|
|
| |||
| AcH3K9 | Active Motif/39138 | pAb | 3500 |
| AIB1/SR3 | Active Motif/39798 | pAb | 2500 |
| ATM | Santa Cruz Biotech/23921 | mAb | 750 |
| ATR | Cell Signaling/2790 | pAb | 2500 |
| Caspase 3 | Cell Signaling/9661 | pAb | 1500 |
| CHK1 | Cell Signaling/2345 | pAb | 2000 |
| CHK2 | Cell Signlaing/2662 | pAb | 2500 |
| Cyclin E | Cell Signaling/4129 | mAb | 3000 |
| DNMT1 | Santa Cruz Biotech/10222 | pAb | 700 |
| DNMT3A | Cell Signaling/3598 | mAb | 3000 |
| DNMT3B | Santa Cruz Biotech/10236 | pAb | 700 |
| ELP3 | Active Motif/39950 | pAb | 3000 |
| HDAC4 | Cell Signaling/5392 | mAb | 3000 |
| HDAC5 | Cell Signaling/2083 | pAb | 2500 |
| KAP1 | Bethyl Laboratories A300–774A | pAb | 3000 |
| p53 | Santa Cruz Biotech/126 | mAb | 1000 |
| Poly(ADP-ribose) | Alexis Biochemicals | mAb | 3000 |
| PARP1 | Santa Cruz Biotech/8007 | mAb | 1000 |
| P-ATM (Ser1981) | Rockland/200-301-4000 | mAb | 2000 |
| P-ATR (Ser428) | Cell Signaling/2853 | pAb | 2500 |
| P-CHK1 (Ser317) | Cell Signaling/2344 | pAb | 2500 |
| P-CHK2 (Ser19) | Cell Signaling/2666 | pAb | 2500 |
| P-KAP1 (Ser824) | Bethyl Laboratories A300–767A | pAb | 1500 |
| P-p53 (Ser15) | Cell Signaling/9284 | pAb | 2000 |
| P-SMC1 (Ser957) | Novus Biolog/NB100–205 | pAb | 2000 |
| P-SMC1 (Ser966) | Abcam/1276 | pAb | 3000 |
| SMC1 | Cell Signaling/4802 | pAb | 2500 |
| SRC1 | Cell Signaling/2191 | mAb | 2500 |
| XIAP | Cell Signaling/2045 | mAb | 2000 |
| β-ACTIN | Sigma/A5316 | mAb | 10000 |
| γ-H2AX | Upstate Biotech/05–636 | mAb | 3000 |
| 3MeH3K27 | Active Motif/39155 | pAb | 3500 |
pAb: polyclonal antibody; used anti-rabbit IgG (or anti-goat as indicated) for secondary antibody from Bio-Rad Lab
mAb: monoclonal antibody; used anti-mouse IgG for secondary antibody from Bio-Rad Lab
Fold dilution in PBS with 0.05% Tween 20
Global DNA methylation analysis
Immunofluorescence staining with anti-5-methylcytidine antibody and flow cytometry were used to determine the level of DNA methylation according to a previously described protocol [24]. Briefly, J45.01 cells exposed to drugs for 48 hr were washed with PBST-BSA (PBS + 0.1% Tween 20 + 1% bovine serum albumin) then fixed with 0.25% paraformaldehyde (Polysciences, Inc., Warrington, PA) in PBS at 37°C for 10 min. The cell membrane was permeabilized by adding 9 volumes of 88% methanol in PBS and maintaining at −20°C for 0.5 hr – 1.0 hr. Cells were washed twice with PBST-BSA and treated with 2 N HCl at 37°C for 30 min followed by 0.1 M sodium borate (pH 8.5) for 5 min. Samples were treated with 10 μg/ml RNase A in PBST-BSA at 37°C for 30 min and washed twice with PBST-BSA. Cells were resuspended in a blocking solution (10% fetal bovine serum in PBST-BSA) and incubated at 37°C for 20 min followed by incubation with 1 μg/ml anti-5-methylcytidine antibody (AbD Serotec, Raleigh, NC) at 37°C for 45 min. Samples were successively washed three times with PBST-BSA, probed with 1:4,000 diluted anti-mouse IgG conjugated to Alexa Fluor 488 (Invitrogen Corp., Carlsbad, CA), and washed two times with PBST-BSA. A third wash was done by tumbling the samples at 4°C overnight. Cells were stained with 50 μg/ml PI in PBST-BSA for 30 min before flow cytometric analysis using the FACSCalibur instrument.
Statistical analysis
Results are presented as the average of 3–4 independent experiments and statistical analysis was done using a Student’s paired t-Test with a two-tailed distribution.
Results
Busulfan, melphalan and gemcitabine have synergistic inhibitory effects on the proliferation of J45.01 cells and SAHA further enhances these effects
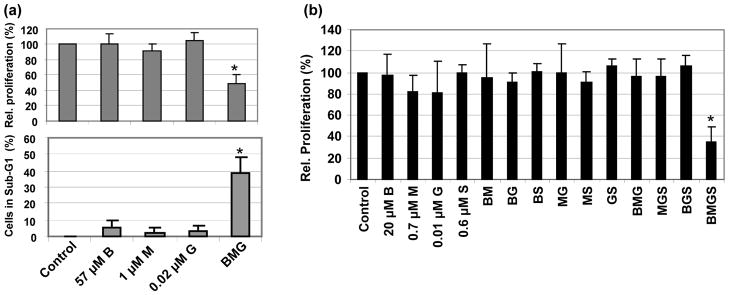
We recently reported greater resistance of J45.01, a T-cell lymphoma cell line, to the combinations of busulfan and two nucleoside analogs when compared with Daudi and U937, which are B-cell lymphoblastic and histiocytic lymphoma cell lines, respectively [18]. In the present study, J45.01 showed greater sensitivity to the combinations of two DNA alkylating agents and a nucleoside analog than it did to the combinations of busulfan and two nucleoside analogs [18]. Exposure of J45.01 cells individually to 57 μM busulfan (B), 1 μM melphalan (M) or 0.02 μM gemcitabine (G) for 96 hr did not show significant effects on cell proliferation or on the proportion of cells in sub-G1 phase (Figure 1a). However, exposure of this cell line to the [B+M+G] combination resulted in 52% inhibition of proliferation and 39% of cells with sub-G1 DNA content, suggesting strong synergism (Figure 1a). These effects were negated when the drug concentrations were reduced by approximately half; thus, exposure of J45.01 cells to the combination of 20 μM B, 0.7 μM M and 0.01 μM G had negligible effects on cell proliferation (BMG in Figure 1b). Addition of 0.6 μM SAHA (S) to this triple-drug combination dramatically inhibited cell proliferation by 65% (BMGS in Figure 1b), even though 0.6 μM SAHA alone had no effect on the proliferation of J45.01 cells. These results suggest that addition of SAHA sensitizes J45.01 cells to the cytotoxic effects of low concentrations of BMG.
Figure 1.
Cytotoxicity of busulfan-melphalan-gemcitabine (BMG) combinations in the absence or presence of SAHA (S). J45.01 cells were continuously exposed to drugs alone, or in combination, for 48 hr (analyzed for cell cycle or sub-G1 DNA content, lower panel in a) or 96 hr (analyzed by the MTT assay for cell proliferation, a (upper panel) and b). Statistically significant difference (P < 0.02) relative to the control is indicated by an asterisk (*). B: busulfan; M: melphalan; G: gemcitabine; S: suberoylanilide hydroxamic acid.
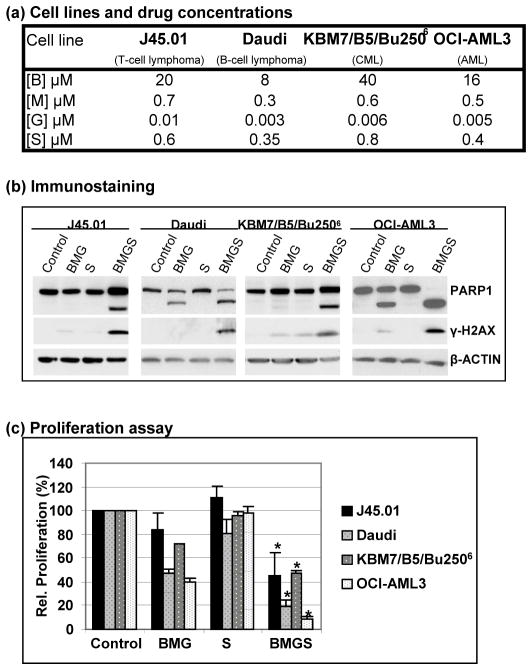
To determine whether the proliferation of other cell lines is similarly inhibited by the BMG±S combinations, we exposed Daudi (B-cell lymphoma), KBM7/B5/Bu2506 (busulfan-resistant CML) and OCI-AML3 (AML) cells to the same drugs at concentrations close to their IC20 values (Figure 2a). The cytotoxicity of BMG±S in these cell lines is shown by the cleavage of PARP1 and phosphorylation of histone 2AX (Figure 2b), indicative of activation of apoptosis and the DNA damage response, respectively; this is further discussed below. SAHA alone did not affect the proliferation of these cells and BMG decreased their relative proliferation to 40%–72% of control (Figure 2c). When SAHA was combined with BMG in these cell lines, cell proliferation further decreased to 8%–32% of control, again suggesting that SAHA sensitizes these cells to the 3-drug combination (Figure 2c). Overall, these results suggest that the HDAC inhibitor SAHA sensitizes both lymphoid and myeloid cancer cell lines to low concentrations of the BMG combination.
Figure 2.
Cytotoxicity of busulfan-melphalan-gemcitabine in the absence (BMG) or presence (BMGS) of SAHA (S) towards other cell lines. Various cell lines were exposed to the indicated drug concentrations, which are close to the IC20 values (a), for 48 hr and analyzed by Western blot for cleavage of PARP1 and phosphorylation of histone 2AX (b) or for 96 hr and analyzed by the MTT assay for cell proliferation (c). Statistically significant difference (P < 0.05) between BMG and BMGS in each cell line is indicated by an asterisk (*). B: busulfan; M: melphalan; G: gemcitabine; S: suberoylanilide hydroxamic acid.
The combination of [B+M+G±SAHA] activates the DNA damage response (DDR) signaling pathway primarily through ATM
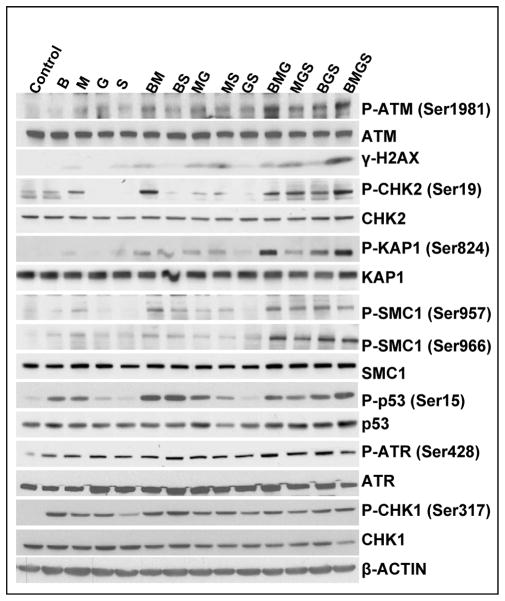
DNA alkylators and nucleoside analogs have been shown to cause DNA strand breaks and other types of damage which activate the DDR when cells attempt to process such DNA lesions [25, 26]. Among the early steps associated with the cellular DDR are activation of the ATM (Ataxia Telangiectasia Mutated) and ATR (ATM and Rad3-related) kinases. As shown in Figure 3, exposure of J45.01 cells to 20 μM B, 0.7 μM M, 0.01 μM G or 0.6 μM SAHA, either alone or in 2-drug combinations, slightly increased the phosphorylation of ATM, a serine-threonine protein kinase which phosphorylates itself (at Ser1981) as well as other substrates. Triple drug combinations such as MGS and BGS further increased the level of P-ATM; however, the highest levels of P-ATM were observed in cells exposed to BMG and BMGS (Figure 3). The efficiency of BMGS in activating the DDR is further indicated by the phosphorylations of histone 2AX (H2AX) and CHK2, both of which are known substrates for the activated ATM kinase [26, 27]. P-CHK2 mediates the cell cycle checkpoint functions of the ATM pathway [28] whereas phosphorylated H2AX (γ-H2AX) is believed to hold broken chromosomal DNA ends in close proximity and recruit DNA damage response proteins [29]. We also looked at KAP1 (KRAB-associated protein-1), which is phosphorylated by ATM following certain types of DNA damage and alters chromatin structure to provide access for DNA repair machinery [30]. The highest level of KAP1 phosphorylation was seen in cells exposed to BMG and BMGS (Figure 3), which is broadly consistent with the pattern of activation of ATM and of other ATM substrates. Other known substrates for the ATM kinase include SMC1 and p53, which are both phosphorylated in response to the various drug combinations used in the present study (Figure 3).
Figure 3.
Activation of the ATM-CHK2 and ATR-CHK1 pathways in cells exposed to various combinations of busulfan-melphalan-gemcitabine-SAHA (B±M±G±S). J45.01 cells were continuously exposed to drugs for 48 hr and total protein extracts were analyzed for phosphorylation of ATM and ATR and their known substrates. B: 20 μM busulfan; M: 0.7 μM melphalan; G: 0.01 μM gemcitabine; S: 0.6 μM suberoylanilide hydroxamic acid.
We also examined the effects of each drug combination on activation of the ATR-CHK1 pathway in J45.01 cells. Figure 3 shows that phosphorylation of ATR at Ser428 occurred in the presence of each individual drug, although this did not significantly increase for any drug combination. Unlike the autophosphorylation of ATM at Ser1981 (which is a widely used marker for ATM activation), none of the known phosphorylations of ATR have been established as reliable markers of ATR activation [31]. We therefore monitored downstream events such as the phosphorylation of CHK1 at Ser317, which appears to be a reasonable indicator of ATR activity [31, 32]. As shown in Figure 3, phosphorylation of CHK1 at Ser317 parallels the phosphorylation of ATR; thus, the ATR-CHK1 pathway in J45.01 cells is responsive to the individual drugs but is not further activated by combinations of the drugs.
[B+M+G+S] combinations trigger extensive apoptosis
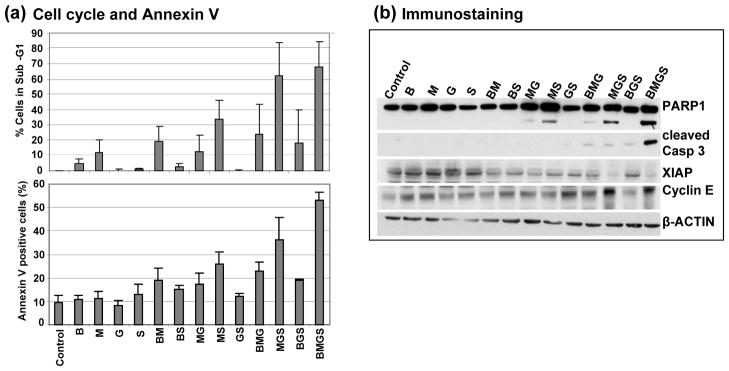
In general, cells of hematological origin undergo apoptosis in response to an overwhelming DDR. The observed activation of the ATM-CHK2 and ATR-CHK1 pathways by these drugs (Figure 3) is strongly suggestive of such a substantial DDR that may, among other responses, trigger the apoptotic pathway. Indeed, flow cytometric analysis of J45.01 cultures exposed to BMGS showed almost 70% of the cells with sub-G1 DNA content and ~56% cells positive for Annexin V staining, both hallmarks of apoptosis pathway activation (Figure 4a). These results parallel the elevated cleavage of PARP1 and caspase 3 in cells exposed to the four-drug combination compared with any other drug combination (Figure 4b). The BMGS-mediated activation of the apoptotic pathway is further suggested by the observed decrease in the level of XIAP1, an inhibitor of apoptosis [33], and increase in cyclin E, which may cause S-phase blockade in response to replication fork stress [34].
Figure 4.
Activation of apoptosis in J45.01 cells exposed to various combinations of busulfan-melphalan-gemcitabine-SAHA (B±M±G±S). Cells were exposed to the indicated drugs for 48 hr and analyzed by flow cytometry for two markers of apoptosis: cells with sub-G1 DNA content (a, upper panel) and cells positive for Annexin V staining (a, lower panel). Cell extracts were analyzed by Western blot for proteins involved in apoptosis and cell survival (b). B: 20 μM busulfan; M: 0.7 μM melphalan; G: 0.01 μM gemcitabine; S: 0.6 μM suberoylanilide hydroxamic acid.
[B+M+G+S] combinations mediate structural alterations in chromatin
We previously suggested a model for the mechanism of enhanced cytotoxicity of combined nucleoside analogs and busulfan [15, 17]. We proposed that nucleoside analogs cause chromatin remodeling and make the genomic DNA more susceptible to alkylation by busulfan. In the present study, we similarly hypothesize that gemcitabine and SAHA synergize in altering chromatin structure and make the DNA more accessible to alkylation by busulfan and melphalan.
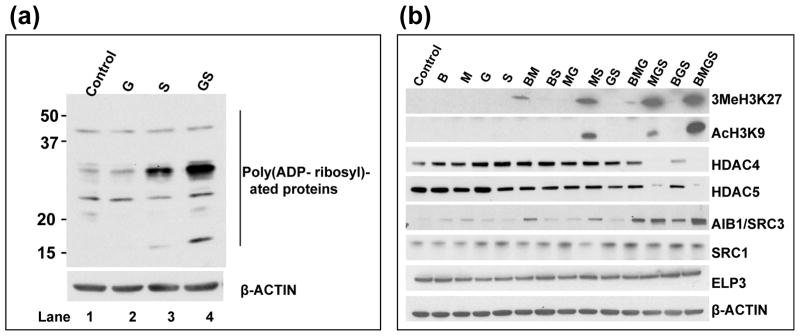
Among the various post-translational modification of histones and other chromatin proteins known to contribute to chromatin remodeling is poly(ADP-ribosyl)ation, which is catalyzed by PARP enzymes [35]. Such modification apparently facilitates access of repair factors to DNA damage sites. We therefore examined changes in the status of poly(ADP-ribosyl)ation of proteins after exposure of J45.01 cells to gemcitabine and SAHA for 24 hr. Gemcitabine (30 nM) alone did not cause significant changes in the poly(ADP-ribosyl)ation of whole-cell proteins relative to the control (Figure 5a, lane 2 versus 1). SAHA alone (0.6 μM) increased the level of poly(ADP-ribosyl)ation of at least two proteins with apparent molecular weights of ~15 kDa and ~30 kDa (Figure 5a, lane 3). A more dramatic increase was observed when SAHA was combined with gemcitabine (Figure 5a, lane 4), suggesting synergistic effects and a possibility of consequential increased chromatin remodeling. These changes in poly(ADP-ribosyl)ation were not apparent in cells exposed to the drugs for 48 hr (data not shown), possibly because of activation of apoptosis and associated inactivation of PARP.
Figure 5.
Epigenetic changes in cells exposed to various combinations of busulfan-melphalan-gemcitabine-SAHA (B±M±G±S). (a) J45.01 cells were continuously exposed for 24 hr to the indicated drugs and total cell extracts were analyzed for early changes in the status of protein poly(ADP-ribosyl)ation by Western blot. G: 0.03 μM gemcitabine; S: 0.6 μM suberoylanilide hydroxamic acid. (b) J45.01 cells were exposed to drugs for 48 hr and the levels of expression and modification status of proteins involved in chromatin remodeling were determined by Western blot. B: 20 μM busulfan; M: 0.7 μM melphalan; G: 0.01 μM gemcitabine; S: 0.6 μM suberoylanilide hydroxamic acid.
Covalent modifications of histones provide additional markers of chromatin remodeling. We, therefore, examined the effects of drug exposure on the methylation and acetylation of histone 3. Exposure of J45.01 cells to the individual drugs showed negligible trimethylation of histone 3 at Lys27. Exposure to BM, MS, MGS and BMGS showed a trend of increasing methylation, with the four-drug combination causing the highest increase in the level of 3MeH3K27 (Figure 5b). Although 0.6 μM SAHA, a known histone deacetylase inhibitor [36], by itself did not increase the level of acetylated histone 3 at Lys9 (AcH3K9), MS and MGS slightly increased the level of AcH3K9. The greatest increase in the level of AcH3K9 was seen in cells exposed to BMGS (Figure 5b). This observed increase in histone 3 acetylation (AcH3K9) correlates with an increase in the level of histone acetyltransferase AIB1/SRC3 and a decrease in the level of histone deacetylases HDAC4 and HDAC5 (Figure 5b). Two other histone acetyltransferases – SRC1 and ELP3 – did not show significant changes in their protein level (Figure 5b). This decrease in the levels of HDAC4 and HDAC5 proteins in response to certain drug combinations might be due to their sumoylation and subsequent proteosomal degradation, as was previously reported for breast cancer and osteosarcoma cell lines treated with SAHA and other HDAC inhibitors [37]. Although the mechanism of SAHA-mediated increase in the level of AIB1/SRC3 is not known, this effect possibly contributed to the increased levels of AcH3K9. The observed similarity in the pattern of increased methylation and acetylation of histone 3 in Figure 5b is consistent with the previously reported interaction between methyltransferases and histone lysine deacetylases [38].
[B+M+G+S] combinations induce expression of DNA methyltransferases
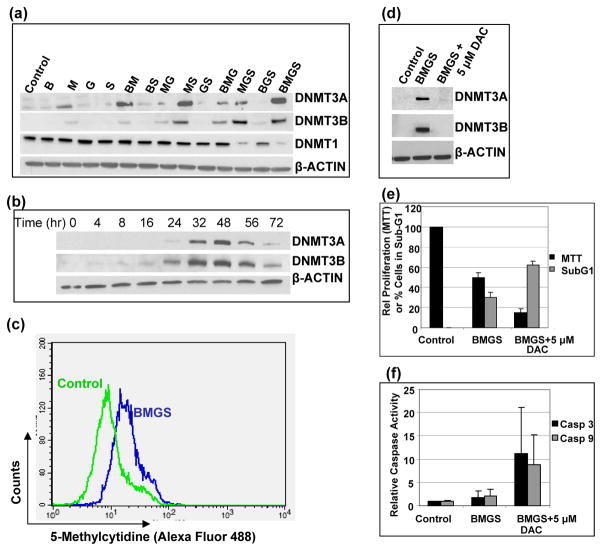
Previous reports suggest functional interactions between DNA methyltransferases and histone deacetylases in gene silencing [39, 40]. We therefore sought to determine the effects of these drugs on the DNA methyltransferases to further decipher the possible underlying role of epigenetic alteration in the combined efficacy of BMGS. Western blot analysis showed increased protein levels of DNA methyltransferases DNMT3A and DNMT3B in cells exposed to BMGS, whereas the level of DNMT1 decreased (Figure 6a). DNMT3A and DNMT3B enzymes are known to be responsible for de novo DNA methylation [41] and might also catalyze drug-induced methylation of DNA.
Figure 6.
Induction of DNA methyltransferases by various combinations of busulfan-melphalan-gemcitabine-SAHA (B±M±G±S). (a) J45.01 cells were exposed to drugs for 48 hr and analyzed for the levels of DNA methyltransferases. (b) The kinetics of the expression of the DNMT3A and -3B proteins was determined in cells exposed to BMGS for up to 72 hr. (c) The level of 5-methylcytidine, indicative of global DNA methylation, was determined by flow cytometry in cells exposed to solvent (Control) or four-drug combination BMGS for 48 hr. The effects of 5-aza-2′-deoxycytidine (DAC) on the drug-induced expression of DNA methyltransferases (d), cell proliferation (e) and caspase activities (f) were determined after a 48-hr drug exposure. B: 20 μM busulfan; M: 0.7 μM melphalan; G: 0.01 μM gemcitabine; S: 0.6 μM suberoylanilide hydroxamic acid.
The induction of DNMT3A and DNMT3B observed in this study peaked at approximately 48 hr after drug exposure (Figure 6b). We therefore exposed J45.01 cells to BMGS for 48 hr and analyzed the level of global DNA methylation by flow cytometry using an anti-5-methylcytidine antibody. Compared with the solvent-treated cells, exposure of J45.01 cells to the four-drug combination increased the level of methylated DNA two-fold as shown by the shifting of the histogram to the right (Figure 6c).
This increased global DNA methylation following BMGS exposure suggests that the ability of SAHA to sensitize cells to DNA-damaging agents mediated by chromatin remodeling might not have been maximized. This leads to the possibility that inhibition of DNA methyltransferases by 5-aza-2′-deoxycytidine (DAC) could further enhance the cytotoxicity of BMGS. Again, exposure of cells to BMGS for 48 hr increased the levels of DNMT3A and DNMT3B proteins, and addition of 5 μM DAC abrogated such induction (Figure 6d). Analysis by the MTT assay indicated a decrease in cell proliferation to 50% of control in the presence of BMGS which further decreased to 13% of control in the presence of DAC; cell cycle analysis showed that the proportion of cells with sub-G1 DNA content increased from 28% to 62% after similar exposures (Figure 6e), suggesting enhanced activation of apoptosis by DAC. This observation is further supported by an increase in the enzymatic activities of caspases 3 and 9 in cells exposed to [BMGS+DAC] (Figure 6f). These results suggest that the efficacy of BMGS may be maximized by the inclusion of DAC as part of the formulation.
DISCUSSION
In our continued search for more efficacious and safer combinations of DNA alkylators and nucleoside analogs for hematologic malignancies, we report in this study the cytotoxicity of busulfan-melphalan-gemcitabine (BMG) in lymphoma cells. The synergistic cytotoxicity seen with the BMG combination is further enhanced by the addition of SAHA, an HDAC inhibitor. These effects are broadly paralleled by the levels of apoptosis seen in the various treatment groups.
Consistent with our previous reports with other nucleoside analog/DNA alkylator combinations [15–18], the BMG±S combinations invoke an extensive DDR in J45.01 cells. However, the patterns of ATM-CHK2 and ATR-CHK1 pathway activation were quite distinct. The extent of activation of the ATM-CHK2 pathway, based on both the auto-phosphorylation of ATM at Ser1981 and the phosphorylation status of selected downstream targets of ATM, notably histone 2AX, CHK2, SMC1 and KAP1, closely parallels the observed degree of cytotoxicity and apoptosis, with the largest response being seen in cells exposed to either BMG or BMGS (Figure 3).
As regards an underlying mechanism for these effects, we previously suggested that the synergistic cytotoxicity of nucleoside analog-alkylating agent combinations may relate to the ability of the former drugs to induce chromatin remodeling and make the genomic DNA transiently more accessible to DNA alkylation [15, 17]. By extension, a similar mechanism might underlie the synergistic effects seen with the BMG combinations. Addition of SAHA presumably sensitizes cells to the combination of DNA-damaging agents by exacerbating these modifications of chromatin structure. Thus, the combination of gemcitabine and SAHA may initiate histone modifications that enhance chromatin relaxation and thus its susceptibility to busulfan- and melphalan-mediated DNA alkylation, leading to excessive complex damage to DNA and subsequent DDR that commits the cells to apoptosis. Consistent with this hypothesis we did observe increased protein poly(ADP-ribosyl)ation of cellular proteins after a 24-hr exposure of J45.01 cells to SAHA and even more so after the combination of gemcitabine and SAHA (S and GS in Figure 5a).
The profound effects of SAHA on chromatin remodeling in cancer cells are well known. SAHA induces replication-mediated DNA damage and activates dormant origins of replication [42]; it also stalls or slows down replication forks, which inadvertently results in recombination and genomic instability [42, 43]. Among the known biological effects of gemcitabine is the depletion of deoxynucleotide pools due to inhibition of ribonucleotide reductase [44], which results in activation of dormant replicons and increased incorporation of gemcitabine-triphosphate into nascent DNA strands, leading to the potentiation of drug effects. The effect of these two agents when given in combination with DNA-alkylating agents is clearly going to be even more complex, but the overall outcome appears to be an intensification of the DNA injury and activation of the apoptosis pathway in lymphoma cells.
SAHA is a pan-HDAC inhibitor and may affect the acetylation of many proteins [45]. Nonetheless, the observed increase in the acetylation of histone 3 in the presence of BMGS does correlate with a decrease in histone deacetelyases HDAC4 and HDAC5 and an increase in histone acetylase AIB1/SRC3 (Figure 5b) and thus may, at least in part, underlie the observed sensitization of lymphoma cells to the BMG combination. Moreover, the increase in AcH3K9 levels (Figure 5b) also correlates with the observed cytotoxicity of this drug combination (Figure 2).
Because these histone modifications reflect not only on early chromatin modifications but also on the overall extent of the DDR activation, the data are difficult to interpret in the context of the chromatin-relaxation hypothesis. The same caveat applies to read-outs such as the phosphorylation of KAP1, a heterochromatin binding protein that maintains the densely packed structure of chromatin and contributes to gene silencing [30]. Phosphorylation of KAP1 at Ser824 by ATM in response to DNA damage releases KAP1 from the DNA complex and causes chromatin relaxation to promote DNA repair [30, 46]. The dramatic increase in phosphorylation of KAP1 by BMGS (Figure 3) does, however, clearly indicate the extensive chromatin remodeling that occurs during the processing of these complex events by the ATM-CHK2 pathway.
An interesting observation in this study is the increase in levels of DNA methyltransferases DNMT3A and DNMT3B, especially in cells exposed to drug combinations that include melphalan, such as MS, MGS and BMGS (Figure 6A). The pattern of this effect suggests that melphalan-SAHA combinations are particularly potent inducers of DNMTs and that melphalan has a greater impact than busulfan in this regard; indeed, a similar pattern was seen for histone H3 modifications (Figure 5b), further indicating some dependence of these events on the specific types of DNA alkylation events. A similar increase in DNMT3A and DNMT3B protein levels was observed in KBM7/B5/Bu2506 (CML) and OCI-AML3 cells (data not shown). The increase in DNMT3A and DNMT3B protein levels in J45.01 cells is associated with an increase in global DNA methylation as indicated by an increased level of 5-methylcytidine (Figure 6c). These results raise the possibility that down-regulation or inhibition of these DNA methyltransferases might further enhance the cytotoxicity of BMGS. Indeed, addition of DAC, a DNA methyltransferase inhibitor, to the 4-drug combination further activates caspases 3 and 9 (Figure 6f) and inhibits the proliferation of J45.01 cells by almost 90% (Figure 6e). Inclusion of DAC in the BMGS formulation, however, may be advantageous only in tumor cell types with low constitutive levels of DNMT3A and DNMT3B proteins. The Daudi cell line (B-cell lymphoma) has a high constitutive level of these enzymes and addition of SAHA to the BMG combination actually decreases their DNMT3B protein levels (data not shown). Our findings underscore the likely importance of determining the DNMT status of lymphoma patients prior to inclusion of DAC in the BMGS combination.
In summary, these overall findings suggest that the enhanced cytotoxic effects of BMGS ± DAC combinations towards lymphoma cells are partly due to epigenetic changes with consequential alteration in chromatin structure, which may facilitate DNA adduct formation by busulfan and melphalan. This study provides the preclinical basis for an ongoing clinical trial using BMGS as part of pre-transplant conditioning therapy for lymphoma patients undergoing HSCT in our institution.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (P01 CA055164 and CCSG Core CA16672), and The Stephen L. and Lavinia Boyd Fund for Leukemia Research.
Footnotes
DISCLOSURES FOR POTENTIAL CONFLICT OF INTEREST:
B. Valdez – None
Y. Nieto – None
D. Murray – None
Y. Li – None
G. Wang – None
R. Champlin – None
B. S. Andersson - None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murawski N, Pfreundschuh M. New drugs for aggressive B-cell and T-cell lymphomas. Lancet Oncol. 2010;11:1074–1085. doi: 10.1016/S1470-2045(10)70210-2. [DOI] [PubMed] [Google Scholar]
- 2.Klyuchnikov E, Bacher U, Kröger N, Kazantsev I, Zabelina T, Ayuk F, Zander AR. The role of allogeneic stem cell transplantation in relapsed/refractory Hodgkin’s lymphoma patients. Adv Hematol. 2012 doi: 10.1155/2011/974658. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salit RB, Bishop MR, Pavletic SZ. Allogeneic hematopoietic stem cell transplantation: Does it have a place in treating Hodgkin lymphoma? Curr Hematol Malig Rep. 2010;5:229–238. doi: 10.1007/s11899-010-0065-7. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava S, Jones D, Wood LL, et al. A Phase I Trial of high-dose clofarabine, etoposide, and cyclophosphamide and autologous peripheral blood stem cell transplantation in patients with primary refractory and relapsed and refractory non-Hodgkin lymphoma. Biol Blood Marrow Transplant. 2011;17:987–994. doi: 10.1016/j.bbmt.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Suyani E, Sucak GT, Aki SZ, Yeğin ZA, Ozkurt ZN, Yağci M. Gemcitabine and vinorelbine combination is effective in both as a salvage and mobilization regimen in relapsed or refractory Hodgkin lymphoma prior to ASCT. Ann Hematol. 2011;90:685–691. doi: 10.1007/s00277-010-1113-z. [DOI] [PubMed] [Google Scholar]
- 6.Russell JA, Tran HT, Quinlan D, et al. Once-daily intravenous busulfan given with fludarabine as conditioning for allogeneic stem cell transplantation: study of pharmacokinetics and early clinical outcomes. Biol Blood Marrow Transplant. 2002;8:468–476. doi: 10.1053/bbmt.2002.v8.pm12374451. [DOI] [PubMed] [Google Scholar]
- 7.de Lima M, Couriel D, Thall PF, et al. Once-daily intravenous busulfan and fludarabine: clinical and pharmacokinetic results of a myeloablative, reduced-toxicity conditioning regimen for allogeneic stem cell transplantation in AML and MDS. Blood. 2004;104:857–864. doi: 10.1182/blood-2004-02-0414. [DOI] [PubMed] [Google Scholar]
- 8.Andersson BS, de Lima M, Thall PF, et al. Once daily i.v. busulfan and fludarabine (i.v. Bu-Flu) compares favorably with i.v. busulfan and cyclophosphamide (i.v. BuCy2) as pretransplant conditioning therapy in AML/MDS. Biol Blood Marrow Transplant. 2008;4:672–684. doi: 10.1016/j.bbmt.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddow A, Timmis GM. Myleran in chronic myeloid leukaemia; chemical constitution and biological action. Lancet. 1953;264:207–208. doi: 10.1016/s0140-6736(53)90884-8. [DOI] [PubMed] [Google Scholar]
- 10.Knock FE. Newer anticancer agents. Med Clin North Am. 1964;48:501–527. doi: 10.1016/s0025-7125(16)33478-2. [DOI] [PubMed] [Google Scholar]
- 11.Hanada K, Budzowska M, Modesti M, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plunkett W, Gandhi V. Purine and pyrimidine nucleoside analogs. Cancer Chemother Biol Response Modif. 2001;19:21–45. [PubMed] [Google Scholar]
- 13.Parker WB, Shaddix SC, Chang CH, et al. Effects of 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl) adenine on K562 cellular metabolism and the inhibition of human ribonucleotide reductase and DNA polymerases by its 5’-triphosphate. Cancer Res. 1991;51:2386–2394. [PubMed] [Google Scholar]
- 14.Yamauchi T, Nowak BJ, Keating MJ, Plunkett W. DNA repair initiated in chronic lymphocytic leukemia lymphocytes by 4-hydroperoxycyclophosphamide is inhibited by fludarabine and clofarabine. Clin Cancer Res. 2001;11:3580–3589. [PubMed] [Google Scholar]
- 15.Valdez BC, Li Y, Murray D, Champlin RE, Andersson BS. The synergistic cytotoxicity of clofarabine, fludarabine and busulfan in AML cells involves ATM pathway activation and chromatin remodeling. Biochem Pharmacol. 2011;81:222–232. doi: 10.1016/j.bcp.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson BS, Valdez BC, de Lima M, et al. Clofarabine ± Fludarabine with Once Daily i.v. Busulfan as Pretransplant Conditioning Therapy for Advanced Myeloid Leukemia and MDS. Biol Blood Marrow Transplant. 2011;17:893–900. doi: 10.1016/j.bbmt.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdez BC, Andersson BS. Interstrand crosslink inducing agents in pretransplant conditioning therapy for hematologic malignancies. Environ Mol Mutagen. 2010;51:659–668. doi: 10.1002/em.20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdez BC, Murray D, Nieto Y, Li Y, Wang G, Champlin RE, Andersson BS. Synergistic cytotoxicity of the DNA alkylating agent busulfan, nucleoside analogs and SAHA in lymphoma cell lines. Leuk Lymphoma. 2012;53:973–981. doi: 10.3109/10428194.2011.634043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdez BC, Murray D, Ramdas L, et al. Altered gene expression in busulfan-resistant human myeloid leukemia. Leuk Res. 2008;32:1684–1697. doi: 10.1016/j.leukres.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Hassan Z, Hassan M, Hellström-Lindberg E. The pharmacodynamic effect of busulfan in the P39 myeloid cell line in vitro. Leukemia. 2001;15:1240–1247. doi: 10.1038/sj.leu.2402193. [DOI] [PubMed] [Google Scholar]
- 22.Bosanquet AG, Bird MC. Degradation of melphalan in vitro: rationale for the use of continuous exposure in chemosensitivity assays. Cancer Chemother Pharmacol. 1988;21:211–215. doi: 10.1007/BF00262772. [DOI] [PubMed] [Google Scholar]
- 23.Martin SJ, Reutelingsperger CP, McGahon AJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exptl Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib M, Fares F, Bourgeois CA, et al. DNA global hypomethylation in EBV-transformed interphase nuclei. Exptl Cell Res. 1999;249:46–53. doi: 10.1006/excr.1999.4434. [DOI] [PubMed] [Google Scholar]
- 25.Drabløs F, Feyzi E, Aas PA, et al. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Ewald B, Sampath D, Plunkett W. Nucleoside analogs: molecular mechanisms signaling cell death. Oncogene. 2008;27:6522–6537. doi: 10.1038/onc.2008.316. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 28.Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 29.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 30.Ziv Y, Bielopolski D, Galanty Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 31.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7:1036–1046. doi: 10.4161/cc.7.8.5783. [DOI] [PubMed] [Google Scholar]
- 34.Lu X, Liu J, Legerski RJ. Cyclin E is stabilized in response to replication fork barriers leading to prolonged S phase arrest. J Biol Chem. 2009;284:35325–35337. doi: 10.1074/jbc.M109.035949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Science. 2004;117:815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- 36.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 37.Scognamiglio A, Nebbioso A, Manzo F, Valente S, Mai A, Altucci L. HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochim Biophys Acta. 2008;1783:2030–2038. doi: 10.1016/j.bbamcr.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodeling and histone deacetylation. Nat Gen. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 40.Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Develop. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 42.Conti C, Leo E, Eichler GS, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 44.Cerqueira NM, Fernandes PA, Ramos MJ. Understanding ribonucleotide reductase inactivation by gemcitabine. Chemistry. 2007;13:8507–8515. doi: 10.1002/chem.200700260. [DOI] [PubMed] [Google Scholar]
- 45.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 46.Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]