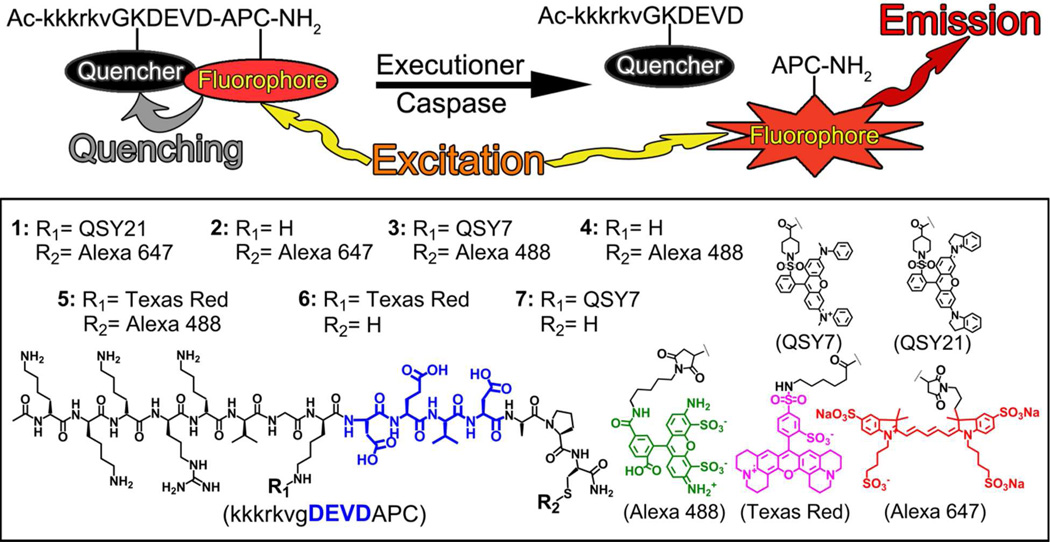
Abstract
Caspase-activatable cell-penetrating peptide (CPP) probes, designed for efficient cell uptake and specificity via cleavable intramolecular quenched-fluorophore strategies, show promise for identifying and imaging retinal ganglion cell apoptosis in vivo. However, initial cell uptake and trafficking events cannot be visualized because the probes are designed to be optically quenched in the intact state. To visualize subcellular activation events in real-time during apoptosis, a new series of matched quenched and non-quenched CPP probes were synthesized. In both native and staurosporine-differentiated RGC-5 cells, probe uptake was time- and concentration-dependent through clathrine-, caveolin- and pinocytosis-mediated endocytic mechanisms. During apoptosis, KcapTR488, a novel dual fluorophore CPP probe, revealed by multi-spectral imaging a temporal coupling of endosomal release and effector caspase activation in RGC-5 cells. The novel CPPs described herein provide new tools to study spatial and temporal regulation of endosomal permeability during apoptosis.
Introduction
Executioner caspases are central mediators of the apoptosis pathway, representing a final commitment to programmed cell death. Caspases are one of the most specific proteases, showing an absolute requirement for cleavage after aspartic acid1 and at least four amino acids N-terminal to the cleavage site for efficient catalysis. The preferred recognition motif differs significantly between caspases, thereby contributing to their biologically diverse functions.2 In addition to high specificity, caspases are also highly efficient, with kcat/Km values > 106 M−1s−1.3 When viewed from the perspective of a target for molecular imaging, these characteristics facilitate the rapid and specific detection of caspase activity in vitro and in vivo.
Previously, we reported the chemical and biochemical characterization of two cell-penetrating probes, TcapQ647 and KcapQ647 (1), designed for imaging effector caspases.4–6 These probes are modular in design, consisting of a cell-penetrating peptide (CPP) sequence conjugated to an effector recognition sequence (DEVD) that is flanked by a fluorophore-quencher pair (Alexa Fluor 647 and QSY 21, respectively) rendering the intact probe optically silent. The cleavable domain of this probe, DEVD, is a specific substrate for effector caspases (3 > 7).7,8 Upon recognition by effector caspases within cells, the probe is cleaved, separating the fluorophore from the quencher, thereby producing an intracellular fluorescent signal denoting activation of the apoptotic pathway (Figure 1). While the original probe, TcapQ647, showed promise both in vivo and in vitro,5,9,10 the HIV Tat sequence has shown evidence of systemic toxicity at high doses,11 prompting us to modify the cell-penetrating sequence. Replacement of the HIV-1 Tat sequence with the nuclear localization sequence of SV40 large T antigen (kkkrkv) yielded 1 (Figure 1).6 This peptide sequence retains cell-penetrating capabilities without the systemic toxicity of the HIV-1 Tat sequence.6
Figure 1.
Schematic of caspase-mediated activation of CPP probes (top). Structures of CPP probes (bottom).
Recently, we applied these caspase-activatable CPP probes to an in vivo rat model of retinal ganglion cell (RGC) degeneration as a demonstration of the potential for molecular imaging of RGC apoptosis in glaucoma.10,11 Glaucoma is an optic neuropathy characterized by RGC death and axon loss, resulting in an excavated appearance to the optic nerve head and associated loss of vision. A wide array of specific triggers of RGC apoptosis are implicated in glaucoma, including blockage of axonal transport with neurotrophin deprivation, antibodies to heat shock proteins, ischemia and oxidative stress, vasoactive regulators such as endothelins and nitric oxide, and glutamate excitotoxicity.12,13 Regardless of trigger, the apoptotic cascade results in cell suicide by invoking a series of cellular events leading to caspase activation, which is conserved in RGCs.14,15 Using TcapQ647 and 1 directly applied into the vitreous humor, we previously reported the capacity to detect RGC apoptosis in models of RGC degeneration in vivo.6,10
While CPP probes of this class are activated upon exposure to caspase 3 in vitro and in live animals, details of the mechanisms of subcellular transport, access to active caspases and the temporal relation to caspase activation remain unanswered. In particular, because it is not possible (by design) to visualize quenched probes prior to activation, the exact location and temporal events leading to intracellular activation of these and other quenched CPP probes are unresolved. Therefore, we synthesized a novel series of CPP probes based on this scaffold conjugated with either quenched fluorophore or non-quenched fluorophore combinations that afforded a variety of spectral properties. These probes enabled the multi-spectral resolution of subcellular trafficking events in relation to probe activation both spatially and temporally in RGC-5 cells, a cell line highly relevant to the study of glaucomatous neurodegeneration.16–19 Observing endosomal trafficking of these novel probes in healthy cells in relation to death-induced endosomal release and localization of caspase-cleaved probe fragments in dying cells provided direct evidence for temporal coupling of endosomal release and effector caspase activation in RGC-5 cells.
Experimental Procedures
Probe Synthesis
Peptides were synthesized as previously reported with revisions to improve probe yield and simplify reaction conditions.6 For 1, standard solid phase N-α-Fmoc chemistry was used to synthesize the peptide (Tufts University Peptide Synthesis Core, Boston, MA). Kcap, the core peptide, was composed of an all D cell-penetrating peptide comprising the SV40 TAg nuclear localization signal (kkkrkv) linked to a cleavage sequence consisting of L amino acids (KDEVDAPC). The N-terminus was acetylated and resin coupling afforded C-terminus amidation of the final product. For conjugation, in a small microwave reaction vial, 1 mL of 2% hydrazine in DMF was added to 25 mg of peptide on resin to selectively remove the single Dde group protecting the lysine N-terminal of the DEVD sequence. The reaction was stirred for 20 min at RT, the supernatant was then collected, and analyzed by UV-Vis spectroscopy to confirm removal of the Dde protecting group. This step was repeated until complete removal of the Dde group was confirmed (5 repetitions). Deprotected peptide on resin was then rinsed with methanol followed by an ether rinse and dried via a stream of argon. To deprotected peptide on resin, 5 mg of QSY21 succimidyl ester (Invitrogen) in a minimal volume (<2 mL) of 2% N,N-diisopropyethylamine/DMF were added, and microwaved in a microwave reactor (CEM Discover) at 10 W for 10 minutes (max temp 65°C). The reaction was then allowed to continue overnight at room temperature. The resin was then transferred to a 10 mL peptide reaction vessel, rinsed with methanol, followed by ether and dried via a stream of argon. Peptide was cleaved and deprotected from resin in 95% trifluoroacetic acid (TFA):5% water for 2 hrs at RT followed by precipitation in cold ether (dry ice acetone bath) to yield the intermediate product 7. 7 was then isolated by centrifuging the sample (2000 g) and the collected pellet dried under argon. 1 mg of Alexa Fluor 647 maleimide (Invitrogen) was dissolved in 1 mL of PBS buffer and added to 2 mL 7 in PBS. Conjugation to the C-terminal cysteine residue was allowed to proceed for 2 h. The final product, 1, was purified using reverse-phase HPLC using a flow rate of 1 mL/min and a 5%/95% to 40%/60% acetonitrile:0.1% TFA/water:0.1% TFA gradient over 10 minutes followed by a 42%/58% acetonitrile:0.1% TFA/water:0.1% TFA gradient over 20 minutes (tR = 17.2 min). 1 was collected by measuring absorbance at 214 nm and 605 nm (>98% purity) and characterized by MALDI mass spectrometry (1: m/z: 3381.0 (1 + 3Na+); calc: 3381.3).
3, 5 and D-5 were synthesized using the same procedure as 1, except that QSY7 succimidyl ester (Invitrogen) (in 3, 7), or Texas Red succimidyl ester (Invitrogen) (in 5, 6) replaced QSY21, and Alexa Fluor 488 maleimide (Invitrogen) replaced Alexa Fluor 647 maleimide. 3 was collected during HPLC purification by measuring absorbance at 214 nm and 595 nm, while 5 was monitored at 495 nm and 595 nm. Constitutively active probes (2 and 4) were isolated during purification of 1 or 3 (tR = 11.5 min 2, tR = 17.8 min for 3, tR = 12.2 min for 4). 5, 6 and 7 were isolated at tR = 17.7 min, tR = 16.3 min and tR = 16.1 min, respectively. Tcap488 and Tcap647 were synthesized and isolated (tR = 12.3 min Tcap488, tR = 11.7 min for Tcap647) using the same procedures as 1–7. Upon purification, probes were lyophilized and stored at -20 °C. All probes were characterized by MALDI mass spectrometry (2: m/z: 2717.2 (2 + 3Na+), calc: 2717.1; 3: m/z: 3082.3, calc: 3080.3; 4: m/z: 2441.4, calc: 2440.1; 5: m/z: 3139.5, calc: 3141.3; D-5: m/z: 3139.9, calc: 3141.3; 6: m/z: 2242.4, calc: 2443.2; 7: m/z: 2382.4, calc: 2382.2; Tcap488: m/z: 2979.4, calc: 2978.4; Tcap647: m/z: 3266.0 (Tcap647 + KCl), calc: 3264.4;). 1, 3 and 5 were characterized by amino acid analysis (Texas A&M Peptide Chemistry Lab). Working solutions of each probe were prepared in DI water and the concentration standardized using molecular extinction coefficients (112,000 M−1 cm−1 at 605 nm for 1; 239000 M−1 cm−1 at 650 nm for 2 and Tcap647; 69500 M−1 cm−1 at 496 nm for 3; 71000 M−1 cm−1 at 495 nm for 4 and Tcap488; 76586 M−1 cm−1 at 595 nm for 5 and D-5; 89000 M−1 cm−1 at 592 nm for 6, 90000 M−1 cm−1 at 565 nm for 7).
Cell Culture
RGC-5 cells were obtained from the ATCC, cultured in phenol-free DMEM supplemented with 10% FBS and incubated at 37°C in a humidified, 5% CO2 atmosphere. For high-throughput microscopy experiments, RGC-5 cells were plated onto 96-well clear bottom black-walled plates (Costar) and allowed to grow to ~70% confluence. For high-resolution microscopy experiments, RGC-5 cells were plated onto #1.5 borosilicate 8-well chambered cover slips (Nunc) and allowed to grow to ~70% confluence. For flow cytometry experiments, RGC-5 cells were plated onto 24-well clear plates and allowed to grow to ~70% confluence. RGC-5 cells were differentiated by incubating in growth medium supplemented with 0.5 µM staurosporine (SS) for 1 hour. Following SS exposure, growth media was removed and replaced with fresh media for experiments.
In Vitro Enzyme Assays
Caspase (human, recombinant from E. coli) assays were performed for caspases 2 (1.25 U; 8.3 × 10−5 mg), 3 (50 U; 1.4 × 10−6 mg), 6 (1.25 U; 9.6 × 10−5 mg), 7 (0.25 U; 1.0 × 10−5 mg) and 9 (1.25 U; 3.1 × 10−3 mg) (Calbiochem, La Jolla CA). All caspase assays were conducted in 50 mM HEPES, 1 mM EDTA, 100 mM NaCl, 10% glycerol, 0.1% CHAPS, pH 7.4 supplemented with 10 mM dithiothreitol (DTT) immediately prior to assay preparation. Granzyme B (human, recombinant from E. coli) and trypsin assays were conducted in TBS buffer (50 mM Tris, 100 mM NaCl, pH 7.5). Cathepsin L (human liver; Calbiochem, La Jolla CA) assays (0.06 mU; 5.0 × 10−5 mg) were conducted in 50 mM sodium acetate, 50 mM NaCl, 0.5 mM EDTA, pH 5.5 buffer supplemented with 10 mM DTT immediately prior to assay preparation. Enzymes were diluted in their respective assay buffer (50 µL final volume) and allowed to equilibrate at 37°C for 10 min. Assays were conducted in 96 well clear-bottomed black-walled plates (Costar) and initiated by adding 1 or 3 (2 µM in 50 µL of enzyme buffer) to enzyme solutions. Assays were performed with 640 nm Ex. and 680 nm Em. filters for 1 or 485 nm Ex. and 535 nm Em. filters for 3 in a fluorometric plate reader (EnVision, Perkin Elmer) at 37°C for 30 minutes. Data were then background subtracted, normalized by enzyme mass (mg) and plotted against time (min). For initial cleavage rates (0–5 min), plots were fitted against a linear regression analysis to obtain the slope. Km and kcat values were determined using nonlinear regression analysis of probe cleavage rates.
Cell Viability Assays
Cell viability was determined via Alamar Blue assays using a plate reader (EnVision, Perkin Elmer). Briefly, resazurin was added directly to cell culture wells (final concentration, 40 µM). Cells were incubated for 3 hours at 37°C in a humidified, 5% CO2 atmosphere and subsequently assayed for resorufin fluorescence at 560 nm Ex. and 590 nm Em. Viability of treated cells was expressed as a ratio by subtracting background signal from all values measured and normalizing against respective untreated cells.
Fluorescence Microscopy
High-resolution microscopy was conducted using a Nikon Ti-E PFS microscope equipped with a Nikon 40× 0.95 NA Plan APO objective, a Nikon 60× 1.4 NA Plan APO oil immersion objective, and Prior Lumen 200PRO illumination system with standard DAPI, FITC, Texas Red and Cy5 filter sets. Images were acquired using a Photometrics CoolSNAP HQ2 digital camera and images were processed and analyzed using the Image J software package (NIH). For co-localization experiments, the commercially available marker ER Tracker Green (Invitrogen) was used for the endoplasmic reticulum, Mito Tracker Green (Invitrogen) for mitochondria, C6-NBD ceramide (Invitrogen) for the golgi apparatus, LysoTracker Red (Invitrogen) for lysosomes and H33342 (Invitrogen) for nuclei.
High-throughput fluorescence microscopy was conducted using an InCell 1000 (GE Healthcare) equipped with DAPI, FITC, Cy3 and Cy5 filter sets using a Nikon 20× 0.45 NA S Plan Fluor objective. For time-lapse movies (Movies S1 and S2), a single representative field of view was selected and the time series acquisition option was utilized in the InCell software. All images were processed and analyzed using the Image J software package (NIH).
Immunohistochemistry
For immunohistochemistry, native and SS-treated RGC-5 cells were fixed using 4% paraformaldehyde solution in PBS buffer for 20 minutes. Cells were then rinsed 3 times with Tris-buffered saline (TBS; 100 mM Tris, 0.9% NaCl, pH 7.2). For Thy-1 antibody labeling, cells were blocked in a 5% normal goat serum in TBS for 30 minutes at room temperature. Cells were then exposed to a FITC-Thy-1 primary antibody (Santa Cruz, SC-18914FITC) in blocking buffer overnight at room temperature. For NMDAR1 antibody labeling, cells were blocked for 30 minutes using 5% normal goat serum solution in TBS. Cells were then exposed to 2.5 µg/mL NMDAR1 primary antibody (BD Biosciences clone 54.1) overnight at room temperature. Cells were then rinsed with TBS buffer and exposed to a 10 µg/mL solution of goat anti-mouse secondary antibody conjugated to Alexa Fluor 594 for 45 minutes at room temperature. For NF70 antibody labeling, cells were blocked for 30 minutes using 5% normal goat serum solution in TBS. Cells were then exposed to 2 µg/mL NF-L primary antibody (Santa Cruz, SC-58559) for 45 minutes at room temperature. Following primary antibody labeling, cells were then rinsed with TBS buffer and exposed to an 10 µg/mL solution of goat anti-mouse secondary antibody conjugated to Alexa Fluor 594 in a 1% normal goat serum solution for 45 minutes at room temperature. Following secondary antibody alone (control), labeled cells were rinsed 3 times with TBS buffer and immediately imaged using fluorescence microscopy.
Toxicity Models
For ionomycin toxicity, RGC-5 cells were exposed to 1, 3, 5, 10, or 20 µM ionomycin in cell growth media for up to 1 hour. For SS overdose, cells were exposed to 5 µM SS in cell growth media for 3 hours. Following toxin exposure, cells were co-incubated with up to 10 µM 3/Alexa Fluor 647-annexin V or 1/FITC-annexin V for 30 minutes and exposed to 500 nM propidium iodide for 5 min. Cells were then gently rinsed 3 times with phenol-free cell growth media and imaged using fluorescence microscopy.
Flow Cytometry
Following treatments, RGC-5 cell cultures were prepared for analysis by removing media, rinsing with PBS buffer, and treating with trypsin (0.25% in PBS) to detach cells from culture wells. Flow cytometry (FACSCalibur, BD Biosciences) was performed, and data were analyzed using the Cell Quest software package. Analysis was conducted for a minimum of 10,000 counting events and median values of histograms were used to quantify probe uptake.
Endocytosis Inhibition
To globally inhibit endocytosis, RGC-5 cells were incubated at 4 °C for 15 min, exposed to 4 (10 µM) in growth media, and incubated for another 3 h at 4 °C. Cells were then prepared for analysis by flow cytometry. To inhibit specific endocytosis mechanisms, cells were treated with chlorpromazine (30 µM), amiloride (3 mM), colchicine (10 µM) or genistein (200 µM) for 1 h prior to incubation with 4 in growth media. After incubation at 37 °C, 5% CO2 for 3 h, cells were prepared for analysis by flow cytometry. Probe co-localization experiments with CF-dextran (3000 Da average molecular weight, Sigma-Aldrich) and CF-albumin (Sigma-Aldrich) were conducted under standard cell culture conditions.
Results
Chemical and Biochemical Characterization of CPP Probes
This class of activatable CPP imaging probes is modular in design, comprising a CPP motif, a protease-cleavable peptide sequence, and a flanking quencher-fluorophore pair, each of which can be interchangeably mixed to yield the desired spectral properties.4 Near-infrared probes with optical properties most favorable for whole body applications in vivo have been characterized previously,5,6 but probes compatible with the wavelengths of fluorophores commonly utilized in clinical imaging of the retina (e.g., fluorescein) have not been reported. For this study, the following CPPs were synthesized: KcapQ647 (1), Kcap647 (2), KcapQ488 (3), Kcap488 (4), L- and D-KcapTR488 (5), KcapTR (6), and KcapQ (7), (Figure 1). 16 and 2, a quenched/non-quenched probe pair, emit in the near-infrared window. The non-quenched analog, 2, lacks a quencher moiety, affording fluorescence regardless of caspase activity. 3 and 4, a novel quenched/non-quenched probe pair that emit in the green optical window, incorporated a QSY7-Alexa Fluor 488 quencher-fluorophore pair instead of the QSY21-Alexa Fluor 647 pair. Alexa Fluor 488 is efficiently quenched when paired with QSY7. 4 was the non-quenched analog of 3. 5, a novel dual fluorophore CPP, which contained Texas Red (TR) and Alexa Fluor 488, 6, a non-quenched probe, which emitted in the red, and 7, which contained only a quencher moiety, were synthesized using analogous synthetic strategies. All orthogonal conjugation reactions of fluorophores and quenchers with the peptide backbone were monitored by UV-Vis spectroscopy, and confirmed by mass spectrometry and amino acid analysis.
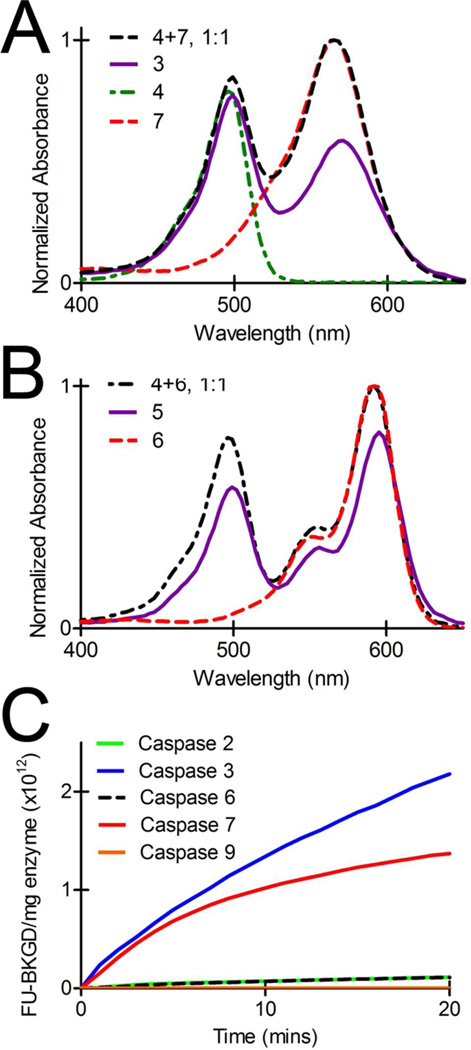
For 3, the quencher-fluorophore combination rendered a new probe potentially compatible with current clinical retinal imaging devices equipped for detection of fluorescent reporters in the visible spectrum of light, specifically bandwidths similar to carboxyfluorescein. The absorption spectrum of 3 revealed a substantial absorbance increase at 570 nm due to conjugation of QSY7 to the peptide backbone as compared to Alexa Fluor 488 alone conjugated to peptide (λmax = 495 nm) (Figure 2A). The extinction coefficient at λmax (496 nm) for 3 was 69500 M−1 cm−1 via UV-Vis spectroscopy coupled with amino acid analysis. The fluorescence quenching efficiency of 3 was high as determined by fluorometry following a 60 min incubation at 37°C in either deionized water, PBS buffer, phenol-free DMEM media with 10% heat-inactivated fetal bovine serum, or 100% heat inactivated fetal bovine serum (99%, 98%, 95% and 95%, respectively). 3 displayed a ratio of 1.3-fold between the absorbances at 496 nm and 570 nm, a significant blue-shift when compared to an equimolar solution of 4 and 7 wherein the ratio of the λmax for the individual probes was 0.8 (Figure 2A), providing evidence for an H-dimer-mediated quenching mechanism.20 The absorption spectrum of 5 revealed a substantial absorbance increase at 595 nm upon conjugation of Texas Red to the peptide backbone compared to Alexa Fluor 488 alone conjugated to peptide (Figure 2B). 5 displayed an extinction coefficient of 76586 M−1 cm−1 at λmax 595 nm with a ratio of 0.7-fold between the peak absorbances at 499 nm and 595 nm, compared to an equimolar solution of 4 and 6 wherein the ratio between the λmax of 4 and 6 (592 nm) was observed to be 0.8. This shift in λmax ratio and a 3 nm red shift in the absorbance spectra provided evidence for J-dimer formation between the Alexa Fluor 488 and Texas Red fluorophores of this intact probe.
Figure 2.
UV-Vis absorption spectra of an eqimolar solution of 4 + 7 ( ), 3 (
), 3 ( ), 4 (
), 4 ( ) and 7 (
) and 7 ( ) (A). UV-Vis absorption spectra of an equimolar solution of 4 + 6 (
) (A). UV-Vis absorption spectra of an equimolar solution of 4 + 6 ( ), 5 (
), 5 ( ) and 6 (
) and 6 ( ) (B). Enzyme assays demonstrating the selectivity of 3 (1 µM) cleavage by recombinant caspases. Assays were normalized to mg of caspase for each enzyme (C).
) (B). Enzyme assays demonstrating the selectivity of 3 (1 µM) cleavage by recombinant caspases. Assays were normalized to mg of caspase for each enzyme (C).
Recombinant caspase assays were performed using continuous fluorometric analysis. To compare initial cleavage rates of 3 by each of the caspases 2, 3, 6, 7 and 9, results were normalized to mg of enzyme (Figure 2C) and rates of 3 cleavage determined by linear fit. Caspase 3 cleaved 3 with an initial cleavage rate of 2 × 1011 FU min−1 mg−1, while caspases 7 and 6 cleaved 3 at 1 × 1011 FU min−1 mg−1 and 1 × 1010 FU min−1 mg−1, respectively. Cleavage of 3 by initiator caspases 2 and 9 were 9 × 109 FU min−1 mg−1 and 1 × 107 FU min−1 mg−1, respectively. Off-target proteases granzyme B (4 × 105 FU min−1 mg−1) and trypsin (1 × 103 FU min−1 mg−1) showed minimal fluorescence activation, providing further evidence of specificity. These data confirmed that 3 was specifically cleaved by effector caspases, preferentially caspase 3, similar to other probes of this class conjugated to different quencher-fluorophore pairs.5,6
Characterization and Differentiation of RGC-5 Cells
To determine the mechanism of cell penetration and subcellular localization kinetics, matched quenched/non-quenched probes were characterized in an in cellulo model of RGC neurodegeneration. RGC-5 cells are mouse retinal ganglion cells virally transformed by the ψ2 E1A virus for in cellulo experimental models of RGC degeneration.21 These cells were characterized by RT-PCR and IHC analysis of various markers representative of retinal ganglion cells, and were originally reported to be sensitive to glutamate-induced excitotoxicity upon differentiation with succinyl concanavalin A.22 Additionally, this cell line undergoes rapid differentiation into a more distinct neurite-like morphology with upregulation of neuronal markers upon low-dose treatment with the broad spectrum kinase inhibitor staurosporine (SS).23 As use of the RGC-5 cell line increased, greater resistance to glutamate toxicity was reported over time.24–26 To characterize the RGC-5 cells used for our studies, we subjected this cell line to an array of experiments to confirm differentiation upon SS exposure and to assess neuronal marker expression.
With 0.5 µM SS, changes in cell morphology consistent with differentiation were rapid, primarily occurring within 1 hour at 37°C. The distinct morphological alterations observed in RGC-5 cells, such as a change in soma from flat and polygonal to round and elevated as well as formation of multiple neurite projections from the soma, were consistent with previous findings.23 Differentiation was easily monitored via serial imaging by bright field microscopy, enabling production of time lapse movies of the differentiation process (Figure S1 and Movie S1). Differentiation progressed less rapidly at room temperature as completion of morphological changes required approximately 2 hours. Exposure to SS at concentrations exceeding 2 µM was toxic, causing blebbing and disruption of neurite morphology.
Immunohistochemistry documented the expression of several neuronal markers in native and SS-differentiated RGC-5 cells. Immunohistochemistry of NF70, Thy-1 and NMDAR1, markers of mature RGCs, was determined by fluorescence microscopy. NF70, Thy-1 and NMDAR1 antibody labeling in both native and SS-treated cells showed similar labeling intensity when judged qualitatively between cell states (Figures S2, S3). Semiquantitative ROI analysis with ImageJ software revealed approximately a 3-fold increase in fluorescence associated with NF70, 8-fold increase with Thy-1, and 2-fold with NMDAR1 for both cell states when compared to control cells (secondary antibody exposure alone for NF70 and NMDAR1; no antibody exposure for Thy1). Labeling patterns were diffuse throughout the cell body, consistent with previous findings.23 Thus, we concluded that our RGC-5 cells retained a neuronal phenotype appropriate for further analysis.
Characterization of Probe Uptake in RGC-5 Cell Culture Models
The optimal concentration range of CPP probe for use in RGC-5 cells was determined by Alamar Blue assays to monitor cell viability after prolonged exposure to the quenched probe 1. Increasing concentrations of 1 were added to a 96-well plate containing RGC-5 cells at 70% confluence. Under these conditions, 1 showed only a 7% loss of viability after a 24 hour exposure at 19 µM and a 39% loss of cell viability at 19 µM after a 72 h exposure. For comparison, a first generation probe, TcapQ647, which differs only in the CPP sequence, was tested under similar conditions. Cells exposed to TcapQ647showed a loss of 40% viability at 20 µM after 24 hours, and >90% loss of viability at 10 µM after 72 hours (Figure S4). Therefore, probes were typically used at 10 µM or less.
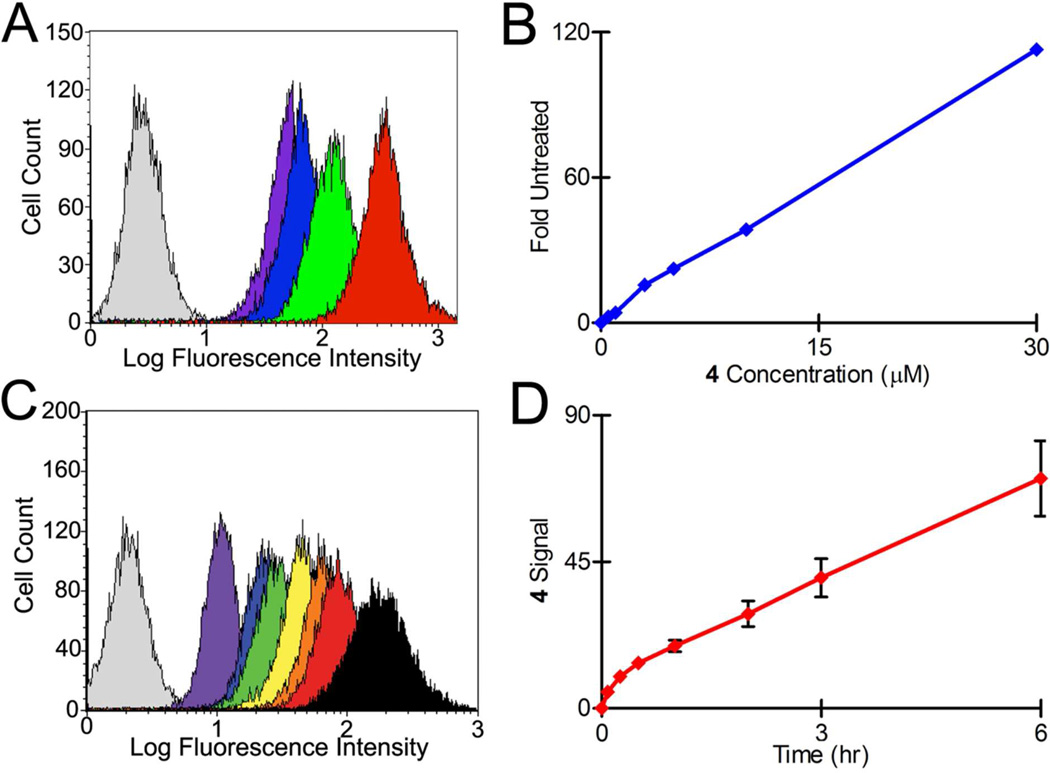
First, to investigate the trafficking pathways underlying probe uptake, we utilized non-quenched probes designed to be constantly fluorescent regardless of caspase activity. Flow cytometry of RGC-5 cells exposed to non-quenched 4 (10 µM) showed a concentration-dependent increase in probe uptake (Figure 3A–B) as well as a time-dependent increase in probe uptake over a 6 hour interval (Figure 3C–D). When compared to Tcap488, a non-quenched version of our previous generation of CPP probes, 4 showed a 25-fold lower uptake under equivalent conditions of exposure (10 µM for 3 hrs). Imaging by fluorescence microscopy under identical conditions confirmed localization of probe within punctate foci localized to the cytosol of RGC-5 cells. To further characterize CPP probes in RGC-5 cells, native and SS-differentiated RGC5 cells were exposed to the non-quenched probes 2, 4 or co-exposure of each, and examined by multi-spectral high-resolution fluorescence microscopy. The probes were readily imaged. Both native (Figure 4) and differentiated RGC-5 cells demonstrated rapid and distinct probe uptake with a punctate distribution pattern. Cells simultaneously exposed to 2 and 4 displayed the identical subcellular localization pattern demonstrating that different fluorophores had no observable effect on the initial subcellular distribution of this class of CPP probes (Figure 4C). Similarly, cells simultaneously exposed to 4 and Tcap647 also displayed the same subcellular localization, providing evidence that these fluorophores, when appended to either the HIV-Tat cell penetrating sequence or the SV40 NLS, internalized into RGC-5 cells via the same trafficking pathways (Figure S5).
Figure 3.
Flow cytometry histograms of native RGC-5 cells exposed to 0  , 1
, 1  , 5
, 5  , 10
, 10  , and 30 µM
, and 30 µM  4 for 3 hours (A). Plot of 4 signal versus concentration in native RGC-5 cells (B). Flow cytometry histograms of native RGC-5 cells exposed to 10 µM 4 for 0
4 for 3 hours (A). Plot of 4 signal versus concentration in native RGC-5 cells (B). Flow cytometry histograms of native RGC-5 cells exposed to 10 µM 4 for 0  , 0.08
, 0.08  , 0.25
, 0.25  , 0.5
, 0.5  , 1
, 1  , 2
, 2  , 3
, 3  , and 6
, and 6  hours (C). Plot of 4 signal versus time in native RGC-5 cells (D).
hours (C). Plot of 4 signal versus time in native RGC-5 cells (D).
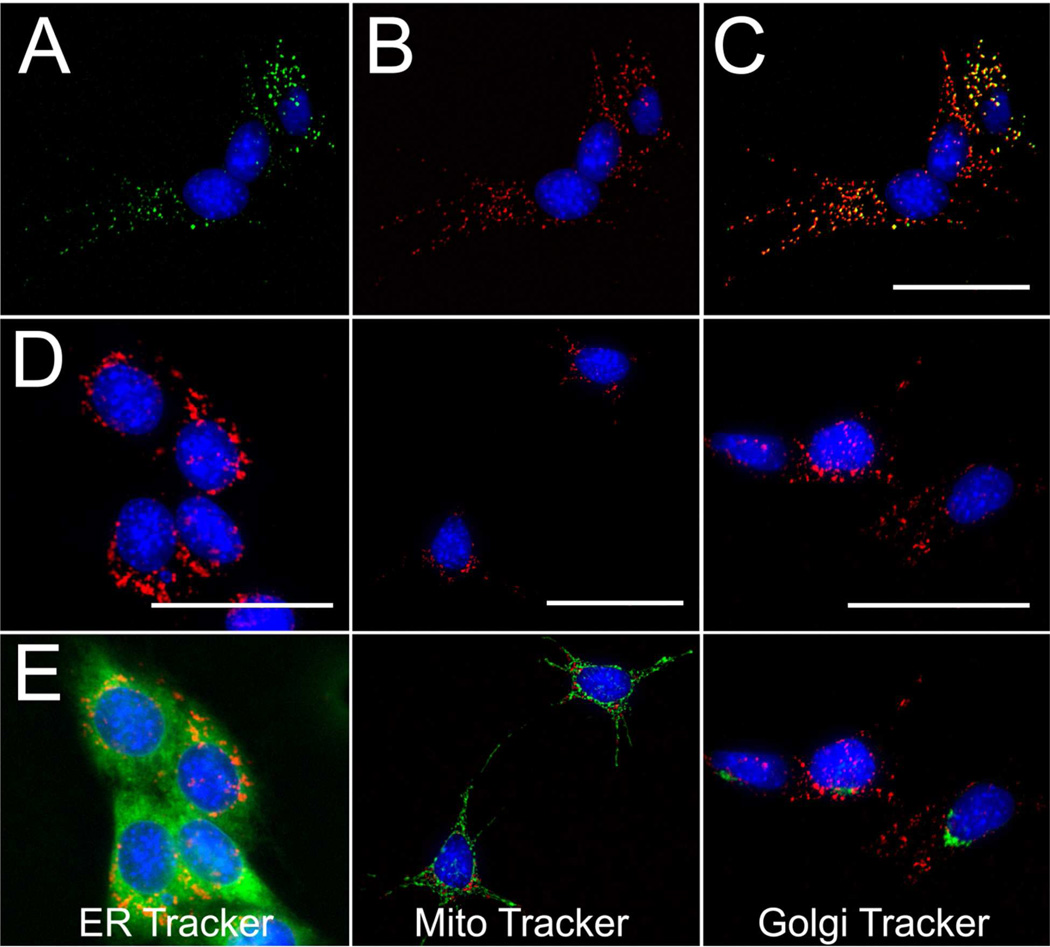
Figure 4.
Fluorescence microscopy images of native RGC-5 cells following a 3 hour co-incubation with 10 µM 4 (A) and 10 µM 2 (B). Overlay of panels A and B (C) showing co-localization of each probe. Images of native RGC-5 cells following co-exposure to 2 (10 µM, 3hrs) (row D) and ER Tracker Green (1 µM, 30 min) (E, left), Mito Tracker Green (50 nM, 30 min) (E, middle), or C6-ceramide-NBD (Golgi Tracker, 2.5 µM, 30 min) (E, right); overlay image of fluorescence fields (row E). Cells were counterstained with H33342 (pseudocolor blue) to show subcellular distribution relative to nuclei (50 µm scale bars).
Co-localization experiments were then conducted with 2 and carboxyfluorescein (CF)-labeled albumin or dextran, markers of cell surface protein binding and fluid phase-mediated endocytosis, respectively.27–29 High-resolution fluorescence microscopy of RGC-5 cells revealed strong co-localization of 2 with CF-albumin (Figure S6) and nearly identical co-localization with CF-dextran. High thorughput fluorescence microscopy of RGC-5 cells co-treated with 2 and these endocytic markers followed by co-localization analysis via Image J produced Pearson’s coefficients30 of 0.732 and 0.873 for CF-dextran and CF-albumin, respectively, in native RGC-5 cells, while Pearson’s coefficients of 0.639 and 0.831 were obtained for differentiated RGC-5 cells. In cells exposed to 2, additional co-staining experiments for endoplasmic reticulum, mitochondria and golgi apparatus showed little if any co-localization of probe distribution with these subcellular organalles (Figure 4D, E). Similar results were obtained with SS-differentiated RGC-5 cells (Figure S7). Overall, these experiments provided evidence that the CPP probes were internalized into RGC-5 cells via endocytic mechanisms and retained for in endosomal endosomes compartments.
The contribution of various endocytic pathways to probe uptake was further characterized by exposing RGC-5 cells to 4 in the presence of known inhibitors of endocytic processes31 followed by quantification with flow cytometry (Table 1). The first method involved incubation of cell cultures at 4°C, which blocked over 90% of the uptake of 4. More detailed mechanistic insight was gained by exposing RGC-5 cells for 1 hour to compounds that inhibit specific endocytic processes prior to addition of 4. Chlorpromazine inhibits clathrin-mediated endocytosis, while genistein inhibits caveolin-mediated mechanisms.32 Cells were also exposed to amiloride33 and colchicine34, each an inhibitor of pinocytosis. All compounds showed partial inhibitory effects on uptake, but taken together, the data in Table 1 indicated that 4 was primarily transported by caveolin-mediated endocytosis in native cells, while probe uptake was more dependent on pinocytosis in SS-differentiated cells.
Table 1.
Relative uptake of Kcap488 (4) into native and SS-differentiated RGC-5 cells upon treatment with inhibitors of endocytosis.
| Treatment | Inhibition Target | RGC-5 Cell State |
|
|---|---|---|---|
| Native | SS-Differentiated | ||
| 4 (10 µM) alone | - | 1 | 1 |
| 4°C incubation | General Endocytosis | 0.08 ± 0.03 | - |
| Chloropromazine (30 µM) | Clathrin-Mediated | 0.71 ± 0.05 | 0.74 ± 0.06 |
| Amiloride (3 mM) | Pinocytosis | 0.58 ± 0.12 | 0.59 ± 0.12 |
| Colchicine (10 µM) | Pinocytosis | 0.54 ± 0.05 | 0.55 ± 0 .9 |
| Genestein (200 µM) | Caveolin-Mediated | 0.43 ± 0.03 | 0.65 ± 0.04 |
Cells were incubated with inhibitors for 1 hour prior to, and for the duration of probe exposure (10 µM 4 for 3h), and probe uptake quantified by flow cytometry. Values are the median ± standard deviation obtained from three independent experiments normalized to 4 alone; n = 3 samples per treatment.
As would be expected for quenched probes prior to activation, imaging healthy RGC-5 cells by fluorescence microscopy following a 2 hr exposure to 1 yielded no significant probe signal. However, non-target activation of CPP-based probes has been previously reported with evidence of caspase-activatable probes undergoing lysosomal degradation via proteases such as cathepsins and legumains.35–37 Degradation of the probe backbone could disrupt quencher-fluorophore interactions leading to non-target probe signal. To evaluate potential lysosome-mediated degradation, fluorescence microscopy co-localization experiments after prolonged exposure of healthy RGC-5 cells to probe 1 (3 hrs) as well as biochemical characterization of cathepsin-mediated activation of 1 were conducted. High-resolution fluorescence microscopy of healthy native and SS-differentiated RGC-5 cells showed the delayed appearance of punctate signal with co-localization between LysoTracker-labeled compartments and activated 1 at 3 hrs (Pearson’s coefficients of 0.566 and 0.612 for native and differentiated cells, respectively; Figure S8). To further characterize this delayed activation, cathepsin-mediated cleavage of 1 was studied using human cathepsin L and compared with cleavage via caspase 3 in vitro. Using nonlinear regression analysis of probe cleavage rates, Km and kcat values were determined (Table 2). These data confirmed that while executioner caspases were preferred for activation of our probes, compartmentalized non-target signal was possible due to lysosomal degradation with prolonged probe exposure to healthy cells.
Table 2.
Kinetic parameters of KcapQ647 (1) cleavage.
| Enzyme | Km (nM) |
kcat (FU min−1 µM−1) |
kcat/Km ((FU min−1 µM−1)/nM) |
|---|---|---|---|
| Caspase 3 | 2,404 | 1,046,000 | 435 |
| Cathepsin L | 3,872 | 375,000 | 97 |
Characterization of Probe Activation in RGC-5 Cells
Excitotoxicity initiates apoptosis via excessive cellular calcium influx mediated by overstimulation of specific receptors, such as the NMDA or AMPA receptors.38 Using the quenched probe, 1-mediated detection of apoptosis was first attempted via NMDA and glutamate excitotoxicity models; however, both native and SS-differentiated RGC-5 cells displayed no loss in viability or probe activation at concentrations up to 5 mM and exposure times up to 48 hrs (data not shown). As an alternative to NMDA and glutamate excitotoxicity, we evaluated 1- and 3-mediated detection of apoptosis via an ionomycin toxicity model.39 Ionomycin is a calcium ionophore commonly used to raise intracellular calcium levels and can be used to trigger cell death in a manner similar to excitotoxicity. Native and SS-differentiated RGC-5 cells were treated with 3 µM ionomycin followed by exposure to 10 µM 3 for 30 minutes. High-resolution fluorescence microscopy revealed distinct probe activation in dying cells within this time frame. CPP probe activation was confined to the cytosolic compartment and at no time after ionomycin treatment were endosomes observed (Figures 5 and S9). Thus, quenched probe escaped from endosomes into the cytosol during the course of apoptosis rather than being activated within endosomes prior to escaping. Additionally, apoptosis was induced in RGC-5 cells by an extended exposure to SS. Herein, cells were incubated with SS for 3 hours at 5 µM, exposed to 3 for 30 minutes and then imaged (Figure 6). Importantly, independent confirmation of apoptosis was obtained with annexin V co-staining. Most cells displaying probe activation were also bound by Alexa Fluor 647-annexin V, with occasional cells showing only annexin V, a marker of phosphatidylserine externalization, an event generally found to be earlier in the apoptotic cascade than effector caspase activation (Figure 5B). Identical results were obtained in this model with 1 and FITC-annexin V staining (data not shown). Notably, in both models of cell death, 1 and 3 were also activated in apoptotic bodies. These cell fragments were readily visualized using combined high-resolution fluorescence and bright field microscopy following co-staining with fluorescent annexin V conjugates and propidium iodide (Figure 6). Furthermore, when cells were preloaded with 3 (5 µM) for 30 minutes and then, in the continued presence of quenched probe, treated with ionomycin (20 µM) and monitored for 1 hour using real-time fluorescence microscopy, distinct intracellular activation of the probe was observed within the cytosol of individual cells starting at ~15 minutes. Sequential image acquisition enabled generation of time-lapse movies showing both morphological changes of the cells undergoing apoptosis and concurrent activation of 3 (Figure 6, bottom row and Movie S2).
Figure 5.
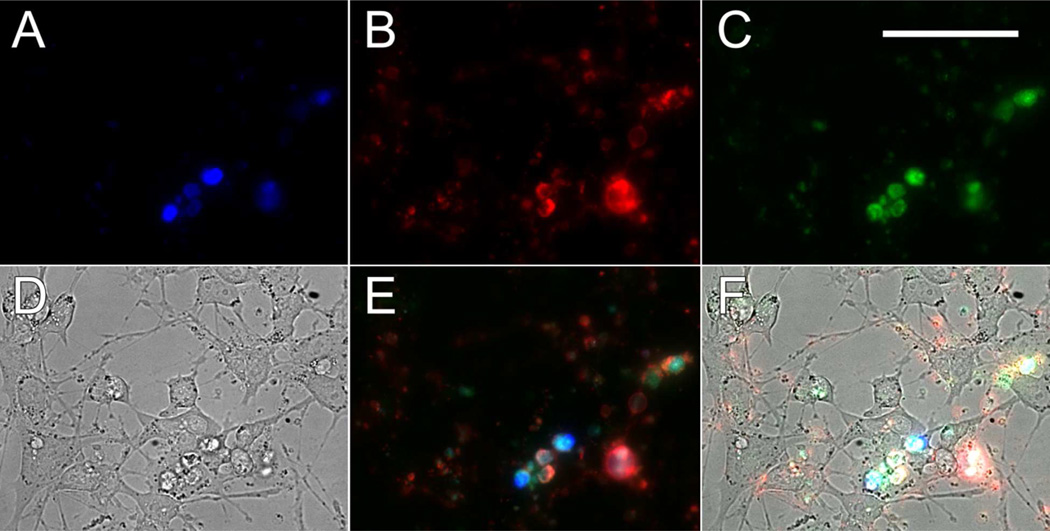
Fluorescence microscopy images of propidium iodide (500 nm) permeable (A), Alexa Fluor 647-annexin V-bound (B) and 3-labeled (C) RGC-5 cells following a 1 hour exposure to 10 µM ionomycin. Bright-field microscopy image of ionomycin-treated RGC-5 cells (D). Overlay of panels A–C showing KcapQ488 (pseudocolor green) activation and co-localization with propidium iodide (pseudocolor blue) and Alexa Fluor 647-annexin V (pseudocolor red) stained cells (E). Overlay of panels D and E (F) (50 µm scale bar).
Figure 6.
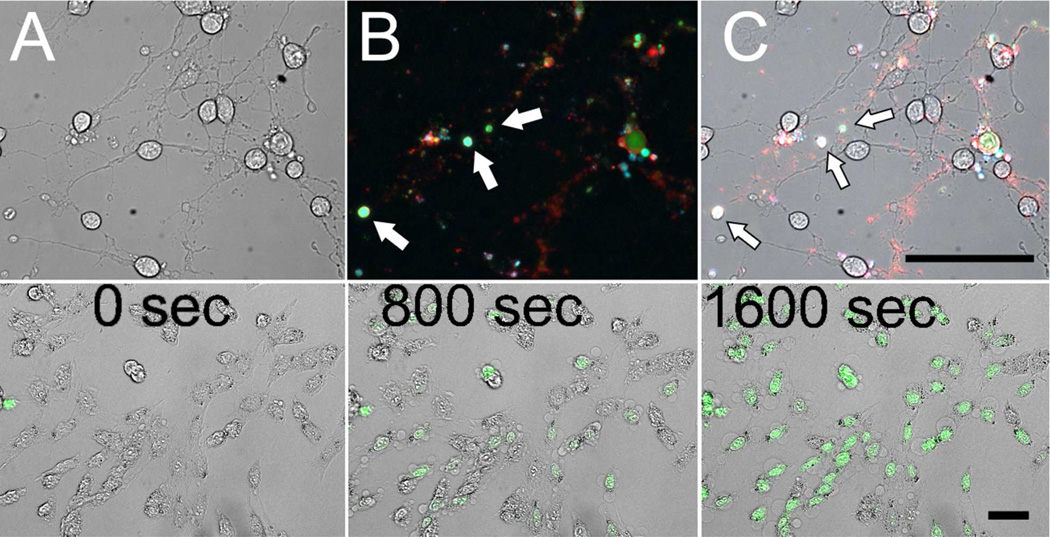
Transmitted light image of RGC-5 cells undergoing SS-induced apoptosis (5 µM SS, 3 hrs) (A). Overlay of fluorescence microscopy images of RGC-5 cells undergoing SS-induced apoptosis with concurrent propidium iodide staining (pseudocolor blue), 3 (10 µM) activation (pseudocolor green) and Alexa Fluor 647-annexin V staining (pseudocolor red) (B). Overlay of panels A and B (C). Arrows denote apoptotic bodies (B and C). Transmitted light and fluorescence microscopy overlay images taken from a time lapse movie of 3 (5 µM) activation in RGC-5 cells following exposure to ionomycin (20 µM) (bottom row). Times correspond to movie acquisition time (sec). Image acquisition began 1 minute following ionomycin exposure (50 µm scale bars).
Our data thus far demonstrate that CPP probes were internalized and retained in endocytic compartments within healthy cells. However, effector caspases are thought to reside entirely within the cytosol.40–42 Thus, in the course of the apoptotic cascade, endosomal or endosomally-associated release mechanisms43–47 must enable probe access to the active pool of caspases. To temporally monitor CPP probe released into the cytosol, and the fate of CPP fragments after cleavage, 5 was designed and synthesized to study probe trafficking in both healthy and apoptotic RGC-5 cells by multi-spectral fluorescence microscopy. This novel probe, which incorporated a Texas Red fluorophore in place of the QSY7 quencher, allowed concurrent multi-spectral tracking of the probe in both its intact and cleaved states. Live RGC-5 cells were exposed to 5 for 3 hours, rinsed to remove exogenous probe, and examined. Healthy cells again displayed punctate, endosomal distribution patterns of the probe concurrently observable with both spectral windows, as expected for the intact probe and consistent with single channel analysis of RGC-5 cells exposed to either 2 or 4 alone (Figure 7, row A). Subsequently, cells were exposed to 20 µM ionomycin for 1 hr. Examination by real-time fluorescence microscopy revealed a redistribution of 5 from endosomes to the cytosol wherein both C- and N-terminal fragments were observed. As expected by the conjugation design, the nucleus contained only the N-terminus with the progression of cell death (Figure 7, row C). Localization of the N-terminal fragment to the nucleus was expected as this fragment incorporated the SV40 large T antigen NLS. Importantly, there was no evidence for the asymmetric loss of fragments within endosomes, such as differential localization of probe signal between spectral windows, or transport into non-vesicular compartments prior to the appearance of cytosolic activation events during the apoptotic cascade (Figure 7, row C, right). Additionally, distribution of an all D-amino acid (cleavage resistant) peptide, D-5, displayed punctuate co-localization of signal in healthy cells (identical to 5) with a cytosolic and concurrent nuclear distribution in both spectral windows in ionomycin-exposed cells (Figure 7, rows B, D). Following endosomal release, intact D-5 was transported predominantly to the nucleus by the NLS. Similar results were obtained with SS-differentiated RGC-5 cells (Figure S10). Together, these data demonstrated that, during the cascade of apoptosis, CPP probes were first released from the endosomes to gain access to the cytosol and in turn were accessible to activated caspases as apoptosis progressed. Thus, endosomal release appears temporally coupled to effector caspase activation. Furthermore, the dual-label cleavable probe 5 also afforded a novel multi-spectral method to detect apoptosis by analysis of differential red/green ratios between the cytosol and nucleus.
Figure 7.
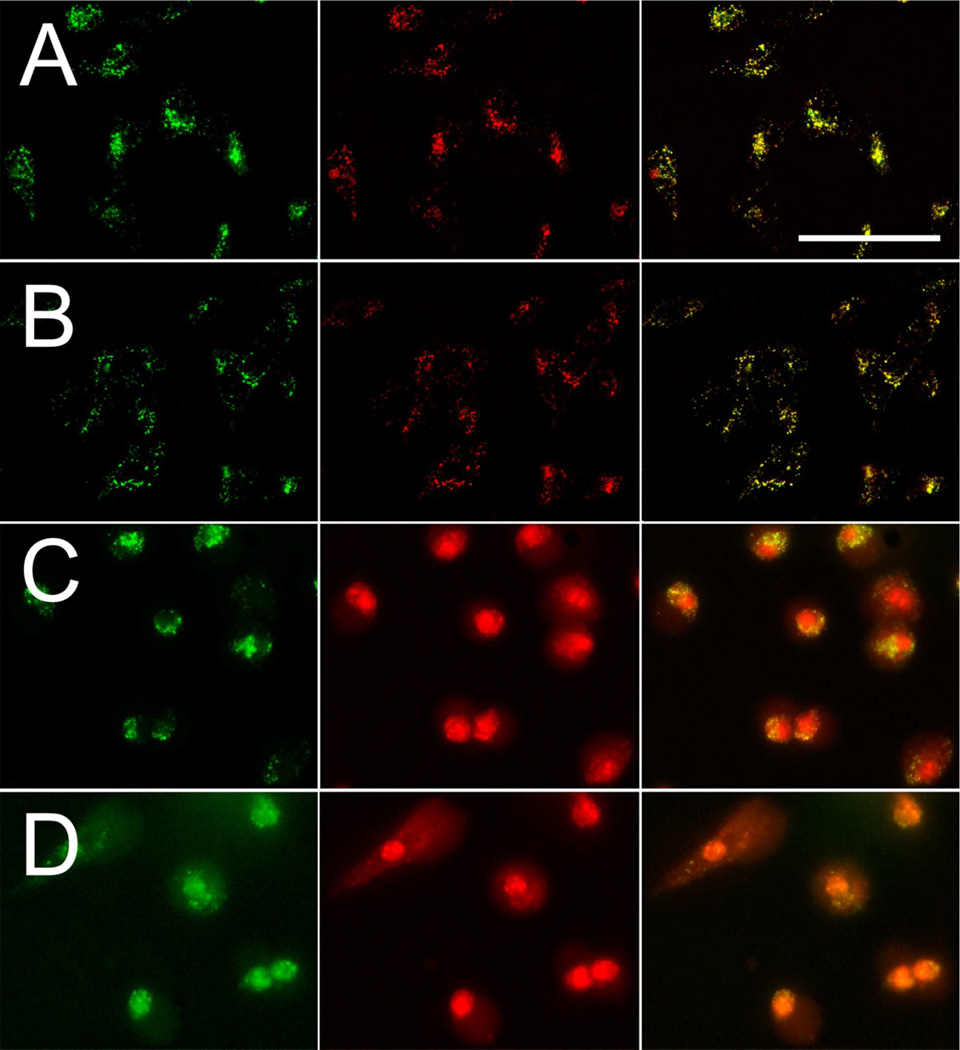
Fluorescence microscopy images of 5 (12 µM) distribution in native RGC-5 cells (row A); D-5 (12 µM) distribution in native RGC-5 cells (row B); 5 (12 µM) distribution in ionomycin-treated (10 µM) native RGC-5 cells (row C); D-5 (12 µM) distribution in ionomycin-treated (10 µM) native RGC-5 cells (row D). Left, fluorescence field for the Alexa Fluor 488-labeled C-terminus (pseudocolored green); middle, fluorescence for the Texas Red-labeled N-terminus (pseudocolored red); right, overlays of Alexa Fluor 488 and Texas Red images (50 µm scale bar).
Discussion
These experiments provided validation of intramolecular-quenched, caspase-activatable optical imaging peptides as apoptosis biosensors for use in live cell models of RGC degeneration. Apoptosis assays are routinely used in cell biology research and in the drug discovery process. These typically sense key events in the apoptotic cascade, such as externalization of phosphatidylserine,48–50 activation of caspases,4,6,35–37,51 DNA fragmentation52 and depolarization of mitochondrial membrane potential.53 While each of these methods have application in a variety of studies, each has its own limitations, such as cumbersome labeling conditions (annexin V), limited selectivity between necrotic and apoptotic cells (annexin V, depolarization probes, DNA fragmentation), limited access to active caspases within intact cells (non-permeant caspase substrates), or poor optical properties for analysis in real-time or in vivo (commercial caspase substrates, e.g., Ac-DEVD-AMC and Z-DEVD-R110). 1 and 3 facilitated detection of cells undergoing apoptosis in real-time using high-throughput imaging methods because the quenched probes could be introduced directly onto cells in cell culture media, readily internalized into cells, selectively activated by executioner caspases and imaged with minimal sample preparation.
Delivery of optically silent, activatable probes to the interior of living cells facilitated analysis by providing enhanced signal-to-noise and retention of the spatial distribution of probe signal. The strategy was scalable, enabling apoptosis detection in both population- and single cell-based assays in real-time. The new quenched CPP probe, 3, which incorporated a QSY7-Alexa Fluor 488 quencher-fluorophore pair, showed evidence of blue-shifted spectral changes similar to previously characterized probes containing QSY21-Alexa Fluor 647 pairs.5,6 3 also displayed quenching efficiencies of 99% in deionized water and 95% in 10% serum, while previously reported nucleotide-based probes incorporating QSY7-Alexa Fluor488 pairs optimized for FRET and static/contact-mediated quenching demonstrated 86% and 95% quenching efficiencies, respectively.54 Overall, the blue-shifted spectral properties and high quenching efficiency observed with the QSY7-Alexa Fluor 488 pair in 3 are characteristic of an H-dimer mediated quenching mechanism.54,55
The data showing the preferential cleavage of 3 by caspase 3 > caspase 7 > caspase 6 ≫ initiator caspases, and the lack of activity in the presence of granzyme B or trypsin were consistent with previous probes of this class, indicating that the identity of the quencher-fluorophore pair did not impact biochemical specificity. We previously published that all D-amino acid probes of this class as well as addition of caspase inhibitors blocked activation,5,6 providing further evidence of specificity. Recent studies have demonstrated cleavage of activatable peptide probes via cathepsins and legumains,35–37 as observed with the present CPP probes. While this can generate non-target signal, short exposure times can circumvent undesired activation. Changes in probe architecture such as lysosomally-inactivated fluorophores and alternative caspase cleavage sequences can be explored to further diminish non-target signal. Additionally, non-target-mediated activation of these CPP probes in vivo remains to be observed.
Alexa Fluor 488 has similar excitation and emission profiles as carboxyfluorescein and thus, 3, 4 and 5 are more compatible with instrumentation such as microscopes, plate readers, and flow cytometers that are not equipped with NIR optical systems. These wavelengths are also commonly utilized in clinical retinal imaging devices, an area of potential application of these probes. Broadly, the availability of a variety of caspase-activatable CPP probes in various emission windows may facilitate multi-spectral analysis when imaged in the presence of other reagents and labeled antibodies.
Live cell cytotoxicity assays demonstrated that 1 and 3 were specifically activated in cells undergoing apoptosis and enabled real-time fluorescence imaging of programmed cell death. Neuronal excitotoxicity models were attempted with NMDA and glutamate toxicity; however, RGC-5 cells were unresponsive to these treatments, consistent with literature reports of loss of sensitivity of RGC-5 cells to these compounds.21 Additionally, NMDA and glutamate toxicity models (up to 5 mM each) were attempted with combinations of BSO (up to 200 µM),26 D-serine (up to 100 mM),56,57 and glycine (up to 100 mM)58 with no significant loss in cell viability (data not shown). However, apoptosis and probe activation were readily observed upon treatment with ionomycin (Figures 5–7, S9, and S10), SS (Figure 6) or doxorubicin (data not shown). The capacity for this probe to detect apoptosis in single cells in real-time using a multi-well plate format will be useful for high-throughput assays as probe activation is rapid and does not require rinse steps to remove exogenous or non-specific signal. Additionally, our non-quenched probes 2, 4 and 5 allowed for detailed examination of probe behavior in cellulo. These probes were amenable to live cell studies as they could be introduced directly into cell culture media without any special preparation and performance was not hindered by media components, such as serum, simplifying experimental conditions and sample preparation.
The pharmacological inhibition of endocytic mechanisms which impacted 4 accumulation levels lends further understanding of how these probes were internalized by cells, while ionomycin- and SS-induced apoptosis models with 5 demonstrated how CPP probes were released and gained access to active caspases during cell death processes. Mechanisms underlying endosomal permeability during the progression of apoptosis are not fully understood, but may involve several distinct mechanisms that are likely engaged in a death stimulus- and cell-type-dependent fashion as with other organelles.43–47 Thus, the dual-labeled probe 5 may serve as a valuable tool for further investigations into mechanisms of endosomal release. The multi-spectral characteristics of 5 could be used in conjunction with other molecular markers and imaging methodologies to further characterize distinct steps of endosomal release associated with apoptosis during organ development and disease.
In summary, we have demonstrated the utility of caspase-activatable cell-penetrating optical imaging peptides using high-throughput and high-resolution live cell microscopy techniques. These probes can selectively identify cells undergoing apoptosis in real-time allowing for detailed examination of cell death processes in cellulo. Future experiments with this toolbox of probes will focus on further understanding endosomal permeability and trafficking behavior during cell death processes as well as optimization of probe use for monitoring apoptosis in vivo.
Supplementary Material
Acknowledgements
Special thanks to Seth Gammon, Vijay Sharma, Jothilingam Sivapackiam, and Sundaram Guruswami for valuable discussions, as well as the High-Throughput Screening Core of the Molecular Imaging Center and Siteman Cancer Center at Washington University for microscopy use and support (P30 CA091842). This work was supported by NIH grants F32 EY20051-01 (J.R.J.), NIH R01 EY019587 (D.P.-W.) and P50 CA94056 (D.P.-W.). J.R.J., E.M.B., and D.P.-W. designed research; J.R.J., B.K., and J.M. performed research; J.R.J and D.P.-W. contributed new reagents/analytic tools; J.R.J., J.M., and D.P.-W. analyzed data; J.R.J. and D.P.-W. wrote the paper; and J.R.J., B.K., E.M.B. and D.P.-W. edited the paper.
Abbreviations
- SS
Staurosporine
- CPP
cell-penetrating peptide
- RGC
retinal ganglion cell
Footnotes
Supporting Information
Supplementary figures and movies further document RGC-5 cell line characterization, CPP probe co-localization, and caspase-activatable CPP probes in an ionomycin-induced cell death model. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thornberry N, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 2.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS, Riedl SJ. Caspase mechanisms. In: Khosravi-Far R, White E, editors. Programmed Cell Death in Cancer Progression and Therapy. New York: Springer; 2008. pp. 13–23. [Google Scholar]
- 4.Bullok K, Piwnica-Worms D. Synthesis and characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. J Med Chem. 2005;48:5404–5407. doi: 10.1021/jm050008p. [DOI] [PubMed] [Google Scholar]
- 5.Bullok KE, Maxwell D, Kesarwala AH, Gammon S, Prior JL, Snow M, Stanley S, Piwnica-Worms D. Biochemical and in vivo characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. Biochemistry. 2007;46:4055–4065. doi: 10.1021/bi061959n. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell D, Chang Q, Zhang X, Barnett E, Piwnica-Worms D. An improved cell-penetrating, caspase-activatable, near-infrared fluorescent peptide for apoptosis imaging. Bioconjug Chem. 2009;20:702–709. doi: 10.1021/bc800516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thornberry NA, Rano TA, Peterson EP, Rasperi DM, Timkey T, Garcia-Calvo M, Houtzageri VM, Nordstromi PA, Royi S, Vaillancourti JP, Chapman KT, Nicholsoni DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B: functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 8.Lien S, Pastor R, Sutherlin R, Lowman H. A substrate-phage approach for investigating caspase specificity. The Protein J. 2004;23:413–425. doi: 10.1023/b:jopc.0000039555.92058.51. [DOI] [PubMed] [Google Scholar]
- 9.Barnett EM, Elangovan B, Bullok KE, Piwnica-Worms D. Selective cell uptake of modified Tat peptide-fluorophore conjugates in rat retina in ex vivo and in vivo models. Invest Ophthalmol Vis Sci. 2006;47:2589–2595. doi: 10.1167/iovs.05-1470. [DOI] [PubMed] [Google Scholar]
- 10.Barnett E, Zhang X, Maxwell D, Chang Q, Piwnica-Worms D. Single-cell imaging of retinal ganglion cell apoptosis with a cell-penetrating, activatable peptide probe in an in vivo glaucoma model. Proc Natl Acad Sci USA. 2009;106:9391–9396. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDunn J, Muenzer J, Dunne B, Zhou A, Yuan K, Hoekzema A, Hilliard C, Chang K, Davis C, McDonough J, Hunt C, Grigsby P, Piwnica-Worms D, Hotchkiss R. An anti-apoptotic peptide improves survival in lethal total body irradiation. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.03.080. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinnon SJ. Glaucoma: ocular Alzheimer's disease? Frontiers Bioscience. 2003;8:s1140–s1156. doi: 10.2741/1172. [DOI] [PubMed] [Google Scholar]
- 13.Qu J, Wang D, Grosskreutz CL. Mechanisms of retinal ganglion cell injury and defense in glaucoma. Exp Eye Res. 2010;91:48–53. doi: 10.1016/j.exer.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Valenzuela E, Shareef S, Walsh J, Sharma S. Programmed cell death of retinal ganglion cells during expreimental glaucoma. Exp Eye Res. 1995;61:33–44. doi: 10.1016/s0014-4835(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 15.Nickells R. Apoptosis of retinal ganglion cells in glaucoma: an update of the molecular pathways involved in cell death. Surv Ophthalmol. 1999;43:S151–S161. doi: 10.1016/s0039-6257(99)00029-6. [DOI] [PubMed] [Google Scholar]
- 16.Kalapesi FB, Coroneo MT, Hill MA. Human ganglion cells express the alpha-2 adrenergic receptor: relevance to neuroprotection. Br J Ophthalmol. 2005;89:758–763. doi: 10.1136/bjo.2004.053025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, Rotiroti D, Morrone LA, Bagetta G, Corasaniti MT. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju WK, Kim KY, Duong-Polk KX, Lindsey JD, Ellisman MH, Weinreb RN. Increased optic atrophy type 1 expression protects retinal ganglion cells in a mouse model of glaucoma. Mol Vis. 2010;16:1331–1342. [PMC free article] [PubMed] [Google Scholar]
- 19.Agar A, Li S, Agarwal N, Coroneo MT, Hill MA. Retinal ganglion cell line apoptosis induced by hydrostatic pressure. Brain Res. 2006;1086:191–200. doi: 10.1016/j.brainres.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa M, Kosaka N, Choyke PL, Kobayashi H. H-type dimer formation of fluorophores: a mechanism for activatable, in vivo optical molecular imaging. ACS Chem Biol. 2009;4:535–546. doi: 10.1021/cb900089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Bergen NJ, Wood JP, Chidlow G, Trounce IA, Casson RJ, Ju WK, Weinreb RN, Crowston JG. Recharacterization of the RGC-5 retinal ganglion cell line. Invest Ophthalmol Vis Sci. 2009;50:4267–4272. doi: 10.1167/iovs.09-3484. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamoorthy R, Agarwal P, Prasanna G, Vopat K, Lambert W, Sheedlo H, Pang I, Shade D, Wordinger R, Yorio T, Clark A, Agarwal N. Characterization of a transformed rat retinal ganglion cell line. Brain Res Mol Brain Res. 2001;86:1–12. doi: 10.1016/s0169-328x(00)00224-2. [DOI] [PubMed] [Google Scholar]
- 23.Frassetto L, Schlieve C, Lieven C, Utter A, Jones M, Agarwal N, Levin L. Kinase-dependent differentiation of a retinal ganglion cell precursor. Invest Ophthalmol Vis Sci. 2006;47:427–438. doi: 10.1167/iovs.05-0340. [DOI] [PubMed] [Google Scholar]
- 24.Aoun P, Simpkins JW, Agarwal N. Role of PPAR-gamma ligands in neuroprotection against glutamate-induced cytotoxicity in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2003;44:2999–3004. doi: 10.1167/iovs.02-1060. [DOI] [PubMed] [Google Scholar]
- 25.Fan W, Agarwal N, Kumar MD, Cooper NG. Retinal ganglion cell death and neuroprotection: Involvement of the CaMKIIalpha gene. Brain Res Mol Brain Res. 2005;139:306–316. doi: 10.1016/j.molbrainres.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Maher P, Hanneken A. The molecular basis of oxidative stressinduced cell death in an immortalized retinal ganglion cell line. Invest Ophthalmol Vis Sci. 2005;46:749–757. doi: 10.1167/iovs.04-0883. [DOI] [PubMed] [Google Scholar]
- 27.Cheng ZJ, Singh RD, Sharma DK, Holicky EL, Hanada K, Marks DL, Pagano RE. Distinct mechanisms of clathrin-independent endocytosis have unique sphingolipid requirements. Mol Biol Cell. 2006;17:3197–3210. doi: 10.1091/mbc.E05-12-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnitzer JE, Oh P. Albondin-mediated capillary permeability to albumin. Differential role of receptors in endothelial transcytosis and endocytosis of native and modified albumins. J Biol Chem. 1994;269:6072–6082. [PubMed] [Google Scholar]
- 29.Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrinindependent pinocytic pathway. Dev Cell. 2002;2:411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 30.Manders EEM, Verbeek FJ, Aten JA. Measurement of co-localisation of objects in dual-colour confocal images. J Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 32.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 33.Dharmawardhane S, Schurmann A, Sells MA, Chernoff J, Schmid SL, Bokoch GM. Regulation of macropinocytosis by p21-activated kinase-1. Mol Biol Cell. 2000;11:3341–3352. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krakauer KA, Zurier RB. Pinocytosis in human synovial cells in vitro. Evidence for enhanced activity in rheumatoid arthritis. J Clin Invest. 1980;66:592–598. doi: 10.1172/JCI109891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, Bogyo M. An optimized activity-based probe for the study of caspase-6 activation. Chem Biol. 2012;19:340–352. doi: 10.1016/j.chembiol.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Curr Opin Chem Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manev H, Favaron M, Guidotti A, Costa E. Delayed increase of Ca2+ influx elicited by glutamate: role in neuronal death. Mol Pharmacol. 1989;36:106–112. [PubMed] [Google Scholar]
- 39.Trump BF, Berezesky IK. Calcium-mediated cell injury and cell death. FASEB J. 1995;9:219–228. doi: 10.1096/fasebj.9.2.7781924. [DOI] [PubMed] [Google Scholar]
- 40.Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 41.Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvesen GS, Riedl SJ. Caspase mechanisms. Adv Exp Med Biol. 2008;615:13–23. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- 43.Repnik U, Turk B. Lysosomal-mitochondrial cross-talk during cell death. Mitochondrion. 2010;10:662–669. doi: 10.1016/j.mito.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Matarrese P, Manganelli V, Garofalo T, Tinari A, Gambardella L, Ndebele K, Khosravi-Far R, Sorice M, Esposti MD, Malorni W. Endosomal compartment contributes to the propagation of CD95/Fas-mediated signals in type II cells. Biochem J. 2008;413:467–478. doi: 10.1042/BJ20071704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouasti S, Matarrese P, Paddon R, Khosravi-Far R, Sorice M, Tinari A, Malorni W, Degli Esposti M. Death receptor ligation triggers membrane scrambling between Golgi and mitochondria. Cell Death Differ. 2007;14:453–461. doi: 10.1038/sj.cdd.4402043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiraishi H, Okamoto H, Yoshimura A, Yoshida H. ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J Cell Sci. 2006;119:3958–3966. doi: 10.1242/jcs.03160. [DOI] [PubMed] [Google Scholar]
- 47.Johansson AC, Appelqvist H, Nilsson C, Kagedal K, Roberg K, Ollinger K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15:527–540. doi: 10.1007/s10495-009-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanshaw RG, Lakshmi C, Lambert TN, Johnson JR, Smith BD. Fluorescent detection of apoptotic cells by using zinc coordination complexes with a selective affinity for membrane surfaces enriched with phosphatidylserine. Chembiochem. 2005;6:2214–2220. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 49.Smith BA, Akers WJ, Leevy WM, Lampkins AJ, Xiao S, Wolter W, Suckow MA, Achilefu S, Smith BD. Optical imaging of mammary and prostate tumors in living animals using a synthetic near infrared zinc(II)-dipicolylamine probe for anionic cell surfaces. J Am Chem Soc. 2010;132:67–69. doi: 10.1021/ja908467y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordeiro M, Guo L, Luong V, Harding G, Wang W, Jones H, Moss S, Sillito A, Fitzke F. Real-time imaging of single nerve cell apoptosis in retinal neurodegeneration. Proc Natl Acad Sci USA. 2004;101:13352–13356. doi: 10.1073/pnas.0405479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizukami S, Kikuchi K, Higuchi T, Urano Y, Mashima T, Tsuruo T, Nagano T. Imaging of caspase-3 activation in HeLa cells stimulated with etoposide using a novel fluorescent probe. FEBS Lett. 1999;453:356–360. doi: 10.1016/s0014-5793(99)00755-3. [DOI] [PubMed] [Google Scholar]
- 52.Negoescu A, Lorimier P, Labat-Moleur F, Drouet C, Robert C, Guillermet C, Brambilla C, Brambilla E. In situ apoptotic cell labeling by the TUNEL method: improvement and evaluation on cell preparations. J Histochem Cytochem. 1996;44:959–968. doi: 10.1177/44.9.8773561. [DOI] [PubMed] [Google Scholar]
- 53.Del Vecchio S, Zannetti A, Aloj L, Salvatore M. MIBI as prognostic factor in breast cancer. Q J Nucl Med. 2003;47:46–50. [PubMed] [Google Scholar]
- 54.Marras S, Kramer F, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucl Acids Res. 1999;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansson M, Fidder H, Dick D, Cook R. Intramolecular dimers: a new strategy to fluorescence quenching in dual-labeled oligonucleotide probes. J Am Chem Soc. 2002;124:6950–6956. doi: 10.1021/ja025678o. [DOI] [PubMed] [Google Scholar]
- 56.Matsui T, Sekiguchi M, Hashimoto A, Tomita U, Nishikawa T, Wada K. Functional comparison of D-serine and glycine in rodents: the effect on cloned NMDA receptors and the extracellular concentration. J Neurochem. 1995;65:454–458. doi: 10.1046/j.1471-4159.1995.65010454.x. [DOI] [PubMed] [Google Scholar]
- 57.Stevens ER, Esguerra M, Kim PM, Newman EA, Snyder SH, Zahs KR, Miller RF. D-serine and serine racemase are present in the vertebrate retina and contribute to the physiological activation of NMDA receptors. Proc Natl Acad Sci U S A. 2003;100:6789–6794. doi: 10.1073/pnas.1237052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.