Abstract
Increasing evidence points to endoglin (Eng), an accessory receptor for the TGF-beta superfamily commonly associated with the endothelial lineage, as an important regulator of the hematopoietic lineage. We have shown that lack of Eng results in reduced numbers of primitive erythroid (EryP) colonies as well as down-regulation of key hematopoietic genes. To determine the effect of endoglin overexpression in hematopoietic development, we generated a doxycycline-inducible ES cell line. Our results demonstrate that induction of endoglin during embryoid body (EB) differentiation leads to a significant increase in the frequency of hematopoietic progenitors, in particular the erythroid lineage, which correlated with up-regulation of Scl, Gata1, Runx1 and embryonic globin. Interestingly activation of the hematopoietic program happened at the expense of endothelial and cardiac cells, as differentiation into these mesoderm lineages was compromised. Endoglin-induced enhanced erythroid activity was accompanied by high levels of Smad1 phosphorylation. This effect was attenuated by addition of a BMP signaling inhibitor to these cultures. Among the BMPs, BMP4 is well-known for its role in hematopoietic specification from mesoderm by promoting expression of several hematopoietic genes, including Scl. Since Scl is considered the master regulator of the hematopoietic program, we investigated whether Scl would be capable of rescuing the defective hematopoietic phenotype observed in Eng−/− ES cells. Scl expression in endoglin-deficient ES cells resulted in increased erythroid colony-forming activity and up-regulation of Gata1 and Gata2, positioning endoglin upstream of Scl. Taken together, these findings support the premise that endoglin modulates the hematopoietic transcriptional network, most likely through regulation of BMP4 signaling.
Keywords: Endoglin, overexpression, hematopoiesis, endothelial lineage, cardiac lineage, BMP signaling, Scl
INTRODUCTION
Endoglin (Eng) is a type III receptor for the TGFβ superfamily, meaning that it functions as an accessory molecule associated with a type I/type II receptor complex (1, 2). In endothelial cells, where endoglin has been most studied (3), this receptor has been proposed to function as a regulator of TGFβ-dependent responses by balancing activating (ALK-1) and inhibitory (ALK-5) TGFβ signals to bias signaling through the Smad1/5 or Smad2/3 signaling pathways, respectively (4, 5). High levels of endoglin are commonly associated with conditions involving alteration in vascular structure, such as angiogenesis, wound healing, and inflammation. Interestingly, mutations in both endoglin and ALK-1 (type I receptor) have been linked to hereditary hemorrhagic telangiectasia type I (6, 7), a disease characterized by hemorrhagic bleedings due to vascular malformations. These vascular defects are likely caused by a loss of endothelial cell (EC) function, which indirectly affects proper EC-smooth muscle cell/pericyte interactions, resulting in poor vascular integrity (8, 9).
Eng-null (Eng−/−) mice die around E10.5 due primarily to cardiovascular defects (10, 11). However, in addition to abnormal vasculature, Arthur and colleagues reported that the YS of 9.5 dpc Eng−/− embryos displayed anemia, which was assumed for several years to be an indirect result of insufficient blood flow. We have provided evidence, using Eng−/− differentiating ES cells, that endoglin is an important regulator of early stages of hemangioblast specification and hematopoietic commitment (12, 13).
Here we investigated whether induction of endoglin affects hematopoietic cell fate from mesoderm, and pinpoint the mechanism behind endoglin’s effects.
MATERIALS AND METHODS
Generation of inducible ES cell lines
Endoglin cDNA was obtained from Open Biosystems (clone MMM1013-7510850) and subcloned into the p2Lox targeting vector, which allows Cre-mediated recombination in the A2lox.cre mouse embryonic stem cell line (14). A2lox cells carry a tetracycline response element (TRE) driving a Cre transgene flanked by heterologous LoxP sites. p2Lox-Eng was electroporated into A2Lox.cre-targeting cells, which were sub-grown on 300 μg/ml of G418 to select for replacement of Cre by Eng. To generate iScl:Eng−/− ES cells, ES cells were transduced with a lentiviral vector expressing the reverse tet-transactivator (rtTA), followed by a vector enabling inducible Scl expression. Scl cDNA was sub-cloned into a lentiviral construct containing the transactivator TRE, which allows the expression of the target gene upon doxycycline (dox) induction, and IRES-EGFP, which allows confirmation of integration and inducible expression. Viruses for these plasmids, rtTA and TRE/Scl/ires.GFP, were generated through transfection in 293T cells using Fugene (Roche). Eng−/− ES cells were initially transduced with rtTA, then with TRE/Scl/Ires.GFP. Eng−/− ES cells containing the Scl insert were purified by FACS based on GFP expression following an overnight incubation with dox at 1μg/ml. After three rounds of purification, inducible expression of SCL was confirmed by western blot using the BTL-73 mouse monoclonal anti-SCL antibody (generously provided by Dr. Karen Pulford, Oxford, U.K.).
Growth and differentiation of ES cells
Inducible Eng (iEng), Eng−/−, and iScl;Eng−/− ES cell lines were used in this study. ES cells were maintained on irradiated mouse embryonic fibroblasts (MEFs) in DMEM (Gibco) supplemented with 1000U/ml LIF (leukemia inhibitory factor; Chemicon), 15% inactivated fetal bovine serum (Gibco), 0.1 mM non-essential amino acids (Gibco), and 0.1 mM of beta-mercaptoethanol (Sigma). For embryoid body (EB) differentiation, ES cells were plated as hanging drops (100 cells/10μl drop) in EB differentiation medium, which consisted of IMDM supplemented with 15% FBS (Gibco), 4.5 mM monothioglycerol (Sigma), 100 μg/ml ascorbic acid (Sigma), and 200 μg/ml iron-saturated transferrin (Sigma) in 150 mm Petri dishes. After 48 hours in culture, EBs were collected and transferred into 10 cm Petri dishes in 10ml of EB differentiation medium. These dishes were cultured on slowly swirling table rotator (set up inside of the tissue culture incubator). To induce appropriate endoglin or Scl expression during EB differentiation, doxycycline (Sigma) was added to the cultures at 1 μg/ml. Dorsomorphin (Stemgent) was added at 2μM.
Blast colony-forming cell (BL-CFC) assay
Embryoid bodies were disaggregated using trypsin, and plated at 5×104 cells in 1.5 ml of methylcellulose medium (M3120, StemCell Technologies) supplemented with 15% FBS, 50 μg/ml ascorbic acid, 200 μg/ml iron-saturated transferrin, 4.5×10−4 M MTG, TPO (25 ng/ml; Peprotech), VEGF (5 ng/ml; Peprotech) and SCF (100 ng/ml; Peprotech), as previously described (15). Plated cells were cultured in a humidified incubator at 37°C in an environment of 5% CO2. Blast colonies were enumerated 5 days later.
Hematopoietic CFC assays
Cells from day 4.25, 4.5, or 6 EBs were plated at 5×104 cells into 1.5 ml of methylcellulose medium containing interleukin 3 (IL3), interleukin 6 (IL6), erythropoietin (EPO), and SCF (M3434; StemCell Technologies). EryP and definitive hematopoietic colonies were scored 5 and 10 days after plating, respectively.
Flow cytometry
EB cells were collected after a short incubation with 0.25% trypsin-EDTA, washed twice with blocking buffer (PBS with 2% FBS), re-suspended in the blocking buffer containing 0.25 μg/106 cells of Fc block (Pharmingen), and incubated on ice for 5 minutes. Staining antibody was added at 1 μg/106 cells and incubated at 4°C for 20 minutes before washing with blocking buffer. We analyzed stained cells on a FACS Aria instrument (Becton-Dickinson) after adding propidium iodide (Pharmingen) to exclude dead cells. For FACS analysis, the following antibodies were used: PE-Cy7-conjugated anti-mouse CD105 (BioLegend), PE- and FITC-conjugated anti-mouse CD41, APC-conjugated anti-mouse c-Kit, APC-conjugated anti-mouse Flk-1, PE-conjugated anti-mouse PDGFR3, PE-conjugated anti-mouse Tie2 (all from eBioscience).
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated using Trizol (Invitrogen) as described by the manufacturer. cDNA was synthesized using Thermoscript reverse transcriptase (Invitrogen) with Oligo dT priming. For real-time PCR, all probe sets were from Applied Biosystems. For globins, we designed customized primer/probe sets (all shown 5′-3′), as follows: Beta-major F, AGGGCACCTTTG CCAGC; Beta-major R, GGCAGCCTCTGCAGCG; Beta-major probe, 6FAM-CGTGATTG TGCTGGGCCACCACCT-TAMRA. Embryonic F, CCTCAAGGAGACCTTTGCTCAT; Embryonic R, CAGGCAGCCTGCACCTCT; Embryonic probe, 6FAM-CAACATGTTGG TGATTGTCCTTTCT-TAMRA. To obtain the relative expression, we first calculated the gene expression levels relative to GAPDH (the fold change to GAPDH), which were then normalized to the level of control non-dox group.
Western Blotting
Day 4.5 or day 9 EB cell lysates were prepared by using 1X RIPA Buffer (ThermoScientific) in combination with Complete Protease Inhibitor Cocktail (Roche) and PhosSTOP (Roche). Protein concentration was measured with Bradford reagent (Sigma), and 30μg samples were prepared using 2X Laemmli Buffer (BioRad). Samples were then denatured on a heat block at 100°C for 10 minutes. After electrophoresis on 8% acrylamide gels, proteins were transferred at 400mA for 2 hours of PVDF membranes (Millipore). Subsequently, the membranes were blocked for 1 hour with 5% BSA in 1X TBS-Tween20. The following primary antibodies were applied at the indicated dilution in Primary Antibody Signal Boost Immunoreaction Enhancer (Calbiochem); 1:1000 dilution of phosphorylated SMAD1/5/8 (Cell Signaling), 1:4000 dilution of SMAD1 (Abcam), 1:3000 cTNI (Abcam), 1:100 SCL. GAPDH (Abcam) was diluted at 1:5000, and ACTIN (Millipore) was diluted at 1:2000 with 5% BSA in 1X TBS-Tween20. All primary antibodies were incubated overnight at 4°C on a shaker. The membranes were washed for 3 × 10 minutes in 1X TBS-Tween20, before secondary antibody application. ECL peroxidase-labeled anti-mouse and anti-rabbit antibodies (GE Biosciences) were diluted at 1:20,000 with 5% BSA in 1X TBS-Tween20. Both secondary antibodies were incubated for 1 hour on a shaker at room temperature. The membranes then washed 3 × 10 minutes in 1X TBS-Tween20. SuperSignal West Pico Chemiluminescent Substrate (ThermoScientific) was utilized to detect the HRP signal. The western blots were quantified by measuring ImageJ (http://imagej.nih.gov/ij/index.html) and data for each antibody were normalized to the value of GAPDH.
Statistical analysis
Differences between non-induced and induced samples were assessed by using the Student’s t test. Differences between multiple groups were assessed by ANOVA.
RESULTS
Endoglin induction leads to enhanced hematopoiesis
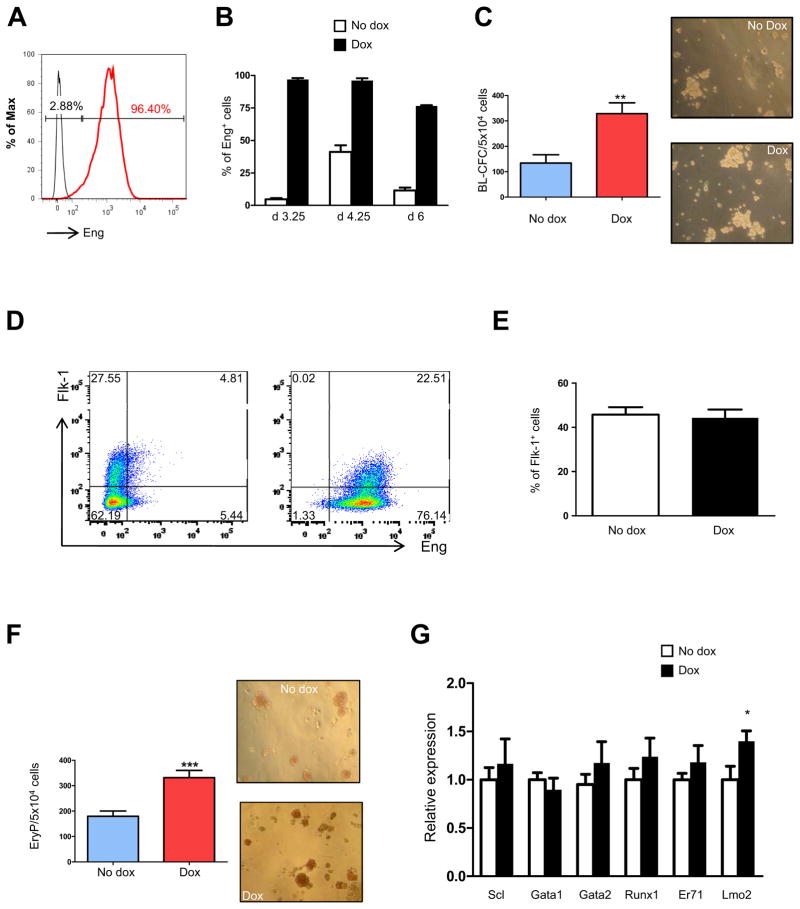
We assessed the effect of endoglin overexpression in blood development by generating an ES cell line with inducible expression of endoglin. Conditional expression of Eng was confirmed by FACS analyses (Fig. 1A). As shown in Fig. 1B, expression of endoglin was dramatically up-regulated in dox-induced cultures when compared to their respective endogenous levels (non-induced controls) at days 3.25, 4.25, and 6 of EB differentiation (Fig. 1B). Induction of endoglin during early EB development, from day 2 to 3.25, generated significantly more Blast Colony-Forming Cells (BL-CFCs) compared to non-induced controls (Fig. 1C). This correlated with an increase in the frequency of Flk-1+Eng+ progenitors (Fig. 1D), although the levels of Flk-1 per se remained unchanged upon dox induction (Fig. 1D, E). On day 4 of EB differentiation the number of hemangioblast progenitors dropped significantly, as expected based on the transient nature of this progenitor (16, 17), nevertheless a stimulatory effect by endoglin could still be observed in the presence in dox-induced cultures (an average of 10 vs. 6 BL-CFCs in dox-induced and controls, respectively). At this point, which coincides with the peak of primitive erythroid (EryP) activity during EB development (16), we found that endoglin induction resulted in increased numbers of EryP colonies (Fig. 1F). Real time PCR analyses of day 3.25 EBs showed significant up-regulation of Lmo2 (Fig. 1G), while changes in the expression levels of other hemangioblast/hematopoietic-associated transcription factors were not statistically significant (Fig. 1G).
Fig. 1. Induction of endoglin stimulates hemangioblast and primitive hematopoietic development.
iEng ES cells were differentiated into EBs. Dox was added to the culture medium from day 2 of EB differentiation. (A) Histogram confirms substantial endoglin induction of day 3.25 iEng EBs following their treatment with dox for 30 hours. (B) Graph shows levels of endoglin expression in control non-induced, reflecting endogenous levels of Eng, and in dox-induced groups during EB development. (C) At the same time point, cells were assessed for hemangioblast activity in BL-MCM, which consists of methylcellulose containing VEGF, SCF, and TPO. Error bars indicate standard errors from 3 independent experiments performed in duplicate. *p<0.05. Right panels, representative morphology of blast colonies obtained from non-induced and induced iEng ES cells. Colonies are shown at the same magnification (100x). (D) A representative FACS profile for endoglin and Flk-1 expression at day 3.25, and (E) respective graphic representation on the frequency of Flk-1+ cells. Error bars indicate standard errors from 3 independent experiments. (F) Primitive erythroid colony activity of non-induced and Eng-induced day 4.25 EBs. Dox was added from day 2 of EB differentiation. Error bars indicate standard errors from 3 independent experiments performed in duplicate. Right panels show representative morphology of EryP colonies showed at the same magnification (100x). (G) Gene expression analysis for Scl, Gata1, Gata2, Runx1, ER71 (Etv2), and Lmo2. Transcripts are normalized to control non-dox group. Error bars indicate standard errors from 3 independent experiments performed in triplicate. *p<0.05.
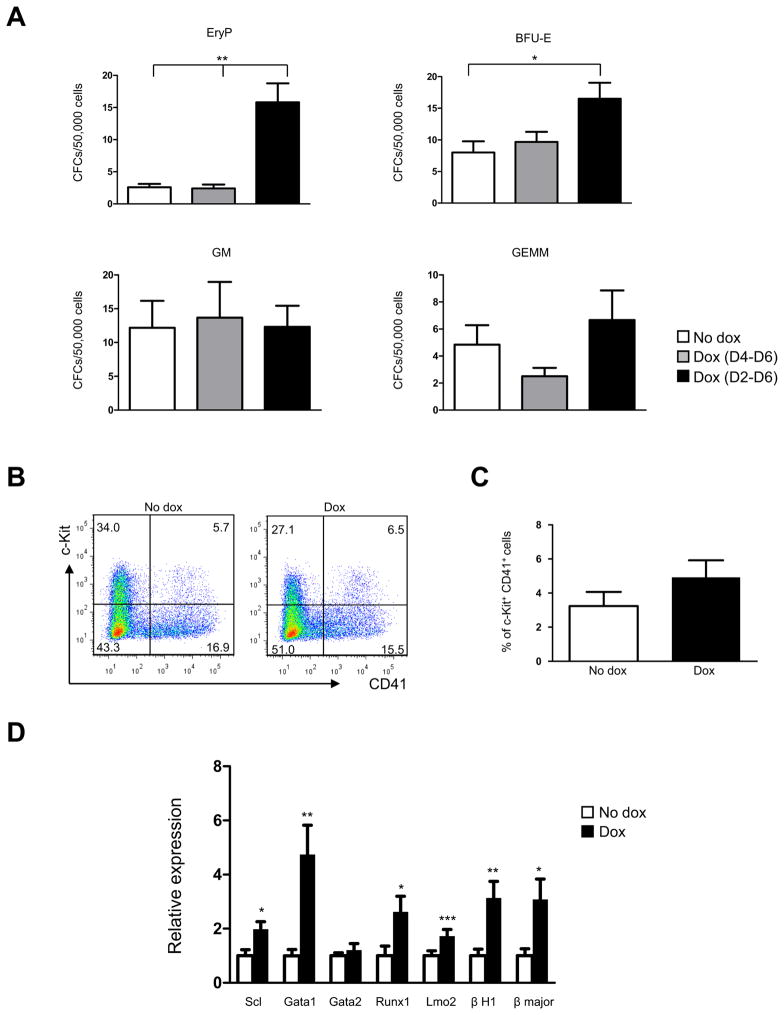
To determine the effect of Eng overexpression on definitive hematopoietic development, we assayed the hematopoietic colony activity of iEng day 6 EBs that had been induced with dox from day 2 or day 4 of EB differentiation. As observed in Fig. 2A, early induction was necessary for endoglin to exert a stimulatory effect on colony activity, as later induction from day 4 to 6 of EB differentiation did not alter the frequency of hematopoietic colonies. When endoglin expression was induced from day 2, we observed increased numbers of EryP colonies, although at EB day 6 the frequency of this primitive progenitor is normally much lower. Increases were also observed in definitive burst forming units-erythroid (BFU-E) (Fig. 2A), while myeloid (GM) and multi-lineage (GEMM) progenitors were unaffected. Although no changes were observed in the frequency of c-Kit+CD41+ hematopoietic progenitors cells (Fig. 2B–C), significant up-regulation of critical hematopoietic regulators was observed at EB day 6, including Scl, Gata1, Runx1, and Lmo2 (Fig. 2D). Consistent with the augmented erythropoiesis, we observed increased levels of globins (Fig. 2D). These findings corroborate the premise that endoglin positively modulates the hematopoietic transcriptional network.
Fig. 2. Effect of endoglin overexpression in definitive hematopoiesis.
(A) iEng ES cells were differentiated as EBs for 6 days and assayed for definitive hematopoietic activity. In these studies, dox was added to the EB medium from either day 2 or 4 of EB differentiation. Error bars indicate standard errors from 3 independent experiments, *p<0.05, **<0.01. (B) A representative FACS profile for c-Kit and CD41 expression in day 6 iEng EBs, and (C) respective graphic representation denoting the percentage of cKit+CD41+ cells in non-induced and induced iEng ES cells. Error bars indicate standard errors from 3 independent experiments. In these experiments dox was added to the cultures from day 2 to day 6 of EB differentiation. (D) Gene expression analysis for Scl, Gata1, Gata2, Runx1, Lmo2, embryonic and adult globins from induced and non-induced iEng day 6 EBs. Dox was added from d2 to d6 of EB differentiation. Transcripts are normalized to control non-dox group. Error bars indicate standard errors from 3 independent experiments performed in triplicate. *p<0.05, **p<0.01, ***p<0.001.
Endoglin overexpression inhibits endothelial and cardiac cell lineages
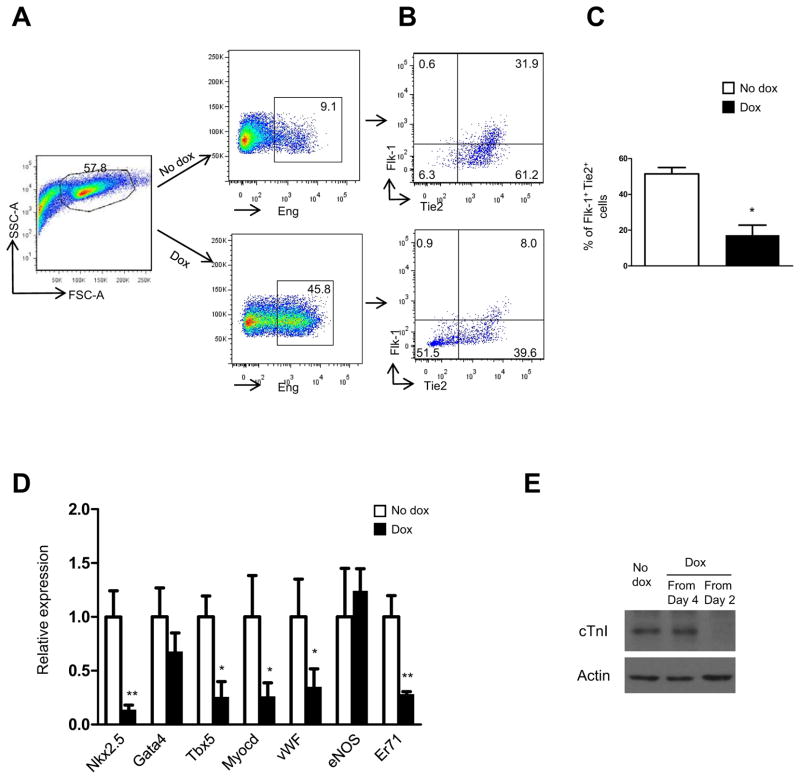
To determine the effect of endoglin induction in other mesodermal lineages, we analyzed the expression levels of Flk-1 and Tie-2 in non-induced (Fig. 3A, upper panel) and dox-induced (Fig. 3A, lower panel) day 6 EBs by flow cytometry. The endothelial lineage was compromised, as evidenced by the reduced levels of Flk-1+Tie-2+ cells (Fig. 3B, C). This was confirmed by gene expression analyses, which revealed down-regulation of the endothelial markers vWF and Er71 (Fig. 3D). Interestingly we also found significant inhibition of genes associated with the cardiac lineage, including Nkx2.5, Tbx5, and Myocardin (Fig. 3D), the latter is also a regulator of the smooth muscle lineage (18). Suppression of cardiogenesis was confirmed by western blot analyses to cardiac Troponin I (cTnI) (Fig. 3E). As observed for induction of the blood lineage (Fig. 2A), inhibition of the cardiac lineage was detected only when endoglin was induced from day 2 to day 6 of EB differentiation (Fig. 3E).
Fig. 3. Increased hematopoiesis happens at the expense of other mesodermal lineages.
(A) FACS plots show endoglin staining of non-induced (upper right) and Eng-induced (lower right) day 6 EBs (exposed to dox from day 2). Eng-expressing cells gated in (A) were then analyzed for the expression of (B) Flk-1 and Tie2, which together mark endothelial progenitors. Representative FACS plots demonstrate that endoglin induction inhibits the endothelial lineage (B). (C) Panel show respective graphic representation with the percentage of Flk-1+Tie2+ in non-induced and induced iEng ES cells. Error bars indicate standard errors from 3 independent experiments. *p<0.05 (E) Gene expression analysis for cardiac and endothelial markers, including Nkx2.5, Gata4, Tbx5, Myocardin (Myocd), vWF, eNOS, and Er71. Transcripts are normalized to control non-dox group. Error bars indicate standard errors from 2 independent experiments performed in triplicate. *p<0.05, **p<0.01. (F) Western blots for cTnI. Dox was added to EB medium from either day 4 or day 2 to day 9 of EB differentiation.
Endoglin overexpression effects require BMP signaling
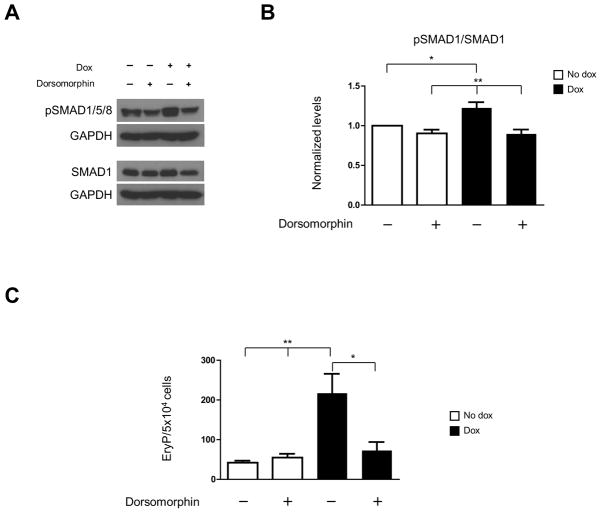
To address whether Eng induction activates the downstream effector of TGFβ superfamily signaling pathway, the Smad1, we verified the phosphorylation level of SMAD1/5/8 (pSMAD1/5/8). Upon dox-induction from EB day 2, levels of pSMAD1/5/8 were significantly increased as compared to non-induced culture (Fig. 4A, B). Data correlated to the drastically increase in the numbers primitive erythroid precursors (Fig. 4C). Since BMP signaling is well known to be crucial for early hematopoietic development (19, 20), we assessed whether addition of dorsomorphin, a BMP signaling inhibitor (21), to dox-induced EB cultures would counteract the effect of endoglin overexpression on erythropoiesis. EB cultures were treated with dorsomorphin at 2μM for 24 hours from day 3.5 to 4.5. This time point was chosen to avoid an early effect of this inhibitor on mesoderm development. While no changes were observed in control (non-induced) cultures in terms of phosphorylated Smad1 (Fig. 4A, B) and primitive erythroid colony activity (Fig. 4C), addition of dorsomorphin for 24 hours (d3.5 –d4.5) to endoglin-induced cultures brought down phosphorylation of Smad1 to control level (Fig. 4A, B). Importantly, these reduced levels of Smad1 phosphorylation in the presence of dorsomorphin in iEng EB cultures were accompanied by a significant reduction in the number of EryP colonies (Fig. 4C). These results suggest that the stimulatory effect of endoglin on hematopoiesis may happen through enhanced BMP signaling.
Fig. 4. Endoglin overexpression stimulates primitive hematopoiesis via BMP signaling pathway.
iEng ES cells were differentiated into EBs for 4.5 days. Dox was added to the culture medium from day 2, whereas Dorsomorphin was added to the culture medium at day 3.5 of EB differentiation. Cells were characterized at day 4.5 as follows: (A) Western blot for the BMP downstream effector, Smad1 and its phosphorylated form Smad1/5/8 (pSmad1/5/8). GAPDH was used as loading control. (B) Quantification of phosphorylated Smad1. After normalization to GAPDH levels, results were plotted as ratio between phosphorylated Smad1/5/8 and total Smad1. Error bars indicate standard errors from 3 independent experiments. (C) Respective iEng EB cultures (± dox; ±dorsomorphin) were assayed for primitive erythroid development. Error bars indicate standard errors from 3 independent experiments. *p<0.05, **p<0.01.
Scl rescues endoglin−/− defective hematopoiesis
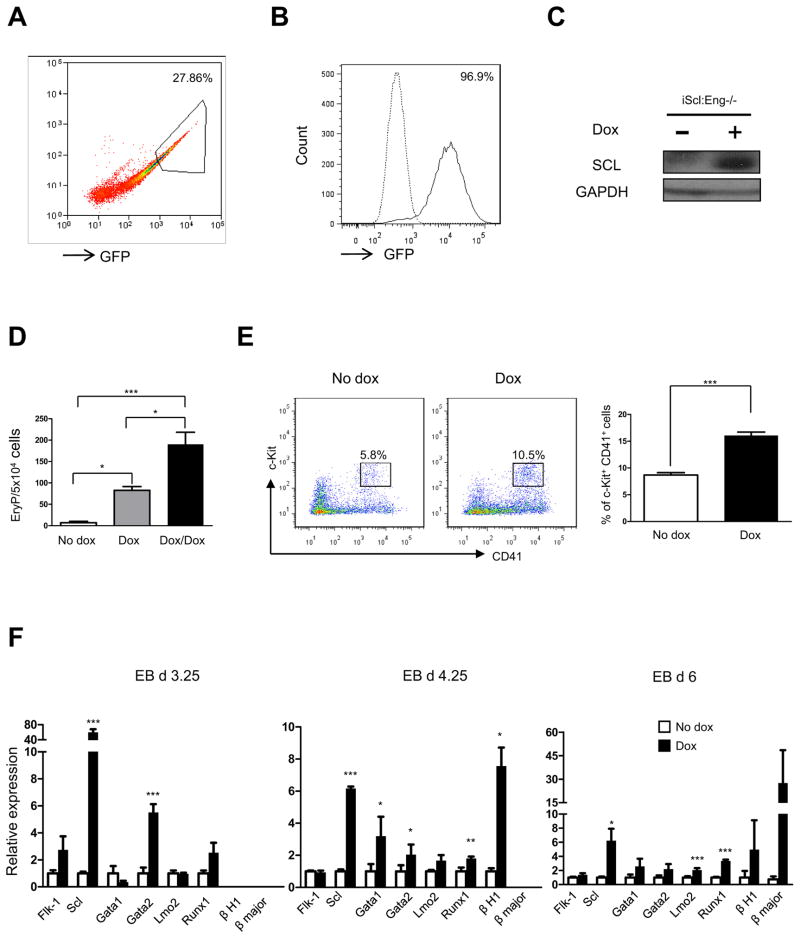
Because lack of endoglin is accompanied by reduced levels of the master hematopoietic regulator Scl (13), whereas induction of endoglin leads to upregulation of this transcription factor, we hypothesized that endoglin acts upstream of Scl. Based on this assumption, we investigated the ability of Scl to rescue the Eng−/− defective hematopoietic phenotype by inserting an inducible lentiviral Scl transgene into Eng−/− ES cells (iScl:Eng−/−). Expression of the transgene was detected by an ires-GFP reporter downstream of the Scl gene (Fig. 5A, B). Western blot analyses confirmed Scl induction in these cells (Fig. 5C).
Fig. 5. Scl rescues defective primitive erythroid development in Eng−/− ES cells.
(A) Eng−/− ES cells infected with an inducible lentiviral vector encoding Scl-iresGFP were selected following three rounds of sorting for GFP. Representative FACS plots show (A) sorting gate for GFP (high expressers) at the second round purification of iScl:Eng−/− ES cells, and (B) induction of Scl expression, as indicated by GFP expression, in day 3.25 iScl:Eng−/−EBs following dox induction from day 2 of EB differentiation. Solid line represents levels of GFP in Scl-induced EBs, whereas dashed line denotes non-induced control (no dox). (C) Western blot confirms induction of Scl in dox-induced iScl:Eng−/− day 3.25 cultures. GAPDH was used as loading control. (D) At day 4.25 of EB differentiation, iScl:Eng−/− ES cells were assayed for primitive erythroid activity. Dox was added to the EB cultures from day 2 of EB differentiation. In one experimental arm, dox was also added to the hematopoietic medium (black bar). Error bars indicate standard errors from 3 independent experiments. *p<0.05, **p<0.01. (E) A representative FACS profile for c-Kit and CD41 expression in non-induced and SCL-induced Eng−/− day 6 EBs, and respective graphic representation on the frequency of c-Kit+CD41+ cells. Error bars indicate standard errors from 4 independent experiments. ***p<0.001. Dox was added from day 2 to day 6 of EB differentiation. (F) Relative levels of Flk-1, Scl, Lmo2, Runx1, Gata1, Gata2, embryonic and adult globins in iScl:Eng−/− EBs at days 3.25, 4.25, and 6 of differentiation. Transcripts are normalized to control non-dox group. Error bars indicate standard errors from 2 independent experiments performed in triplicate.*p<0.05,** p<0.01, *** p<0.001.
We then determined the ability of Scl-induced Eng−/− ES cells to generate BL-CFCs and EryPs, which were found previously to be significantly reduced in the absence of endoglin (13). Whereas induction of Scl did not alter the reduced number of BL-CFCs found in the absence of endoglin (data not shown), this was not the case for primitive erythropoiesis. As observed in Fig. 5D, Scl induction during EB differentiation (D2–D6) rescued the Eng−/− phenotype, as evidenced by the approximately 10-fold increase in the number of EryPs. This number doubled when dox was maintained also in the hematopoietic culture medium (3434) (Fig. 5D). This dramatic effect on hematopoietic development correlated with increased percentage of c-Kit+CD41+ cells (Fig. 5E) as well as higher expression of the hematopoietic genes Gata1, Gata2, Lmo2, Runx1, globins, and as expected, Scl (Fig. 5F). These data indicate that Scl acts downstream of endoglin and can compensate for the lack of endoglin during primitive erythropoiesis.
DISCUSSION
Our findings reveal endoglin as a critical regulator of the hematopoietic program. Endoglin has previously been shown to be co-expressed with CD45 in hematopoietic cells obtained from ES/OP9 co-cultures (22). In adult bone marrow, this receptor enriches for the long-term repopulating hematopoietic stem cells (HSCs) (23), and more recently, this was also observed in the AGM, the first site of HSCs (24). We have previously shown that lack of endoglin results in suppressed primitive hematopoiesis, as shown by decreased primitive erythroid colony activity and expression levels of key hematopoietic transcription factors, including Scl, Gata1, and Gata2 (12, 13). Here we show that increasing the expression level of this receptor during EB differentiation has exactly the opposite effect, leading to enhanced hematopoiesis, as evidenced by the increased numbers of primitive and definitive erythroid CFCs (Figs. 1F and 2A) and up-regulation of Scl, Gata1, Lmo2 and Runx1 (Figs. 1G and 2D), all critical regulators of the hematopoietic program (25–31). Taken together, both the loss-of-function and overexpression data indicate that endoglin regulates Scl expression, as well as that of its binding partners, Lmo2 and Gata1. This assumption is corroborated by the fact that overexpression of SCL rescues the defective erythroid phenotype of Eng-deficient embryoid bodies (Fig. 5).
Another important finding is that activation of the hematopoietic program by continuous expression of endoglin happens at the expense of the endothelial and cardiac lineages, as progenitors for both these lineages are significantly reduced in the presence of endoglin induction (Fig. 3). These results suggest that the levels of endoglin are critical to fine-tune early lineage decisions during embryonic development.
In terms of mechanist insight, it is known that endoglin interacts with TGFβ1 and TGFβ3, but also interacts with other members of the TGF-β superfamily, including activin-A, BMP-7, and BMP-2 in association with their respective ligand binding receptor kinases (1). BMPs are of particular interest, as these ligands are crucial for proper mesoderm patterning (32) and blood specification (20). BMPs phosphorylate SMAD1/5/8 upon forming a heteromeric complex with BMPRII or ActRII, and one of their type I receptors (ALK-2, ALK-3, or ALK-6) (33, 34). Knockouts for bmp4 (20), bmp2 (35), and their common receptors alk3 (36) and bmprII (37) are embryonic lethal with reduced mesoderm (32). In particular, BMP4 is necessary for hematopoietic commitment of mesoderm, promoting expression of Scl, Gata1, Gata2, and Lmo2 (31, 38–42). Addition of the BMP signaling inhibitor dorsomorphin to endoglin-induced EB cultures, at a time point in which mesoderm had already developed (day 3.5), abolished the stimulatory effects of endoglin on erythropoiesis, suggesting that this effect is mediated by BMP signaling. Which specific BMP is not determined, however given the known role of BMP4 in hematopoietic specification (43), it seems likely that endoglin interacts with BMP4.
Therefore we provide evidence that endoglin modulates the hematopoietic transcriptional network, most likely through regulation of BMP4 signaling. However a direct interaction between endoglin and BMP4 has not yet been reported, and thus requires further investigation.
Acknowledgments
This project was supported by NIH grants R01 HL085840-01 and U01 HL100407.
Footnotes
AUTHORSHIP CONTRIBUTIONS
J.B. designed and performed research, analyzed data, and wrote the manuscript; L.B., A.M. and T.T. performed research and analyzed data; and R.C.R.P. designed research, analyzed, and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barbara NP, Wrana JL, Letarte M. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-beta superfamily. J Biol Chem. 1999;274:584–594. doi: 10.1074/jbc.274.2.584. [DOI] [PubMed] [Google Scholar]
- 2.Yamashita H, Ichijo H, Grimsby S, Moren A, ten Dijke P, Miyazono K. Endoglin forms a heteromeric complex with the signaling receptors for transforming growth factor-beta. J Biol Chem. 1994;269:1995–2001. [PubMed] [Google Scholar]
- 3.Cheifetz S, Bellon T, Cales C, et al. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J Biol Chem. 1992;267:19027–19030. [PubMed] [Google Scholar]
- 4.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebrin F, Goumans MJ, Jonker L, et al. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004;23:4018–4028. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McAllister KA, Grogg KM, Johnson DW, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DW, Berg JN, Baldwin MA, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho RLC, Jonker L, Goumans MJ, et al. Defective paracrine signalling by TGF-beta in yolk sac vasculature of endoglin mutant mice: a paradigm for hereditary haemorrhagic telangiectasia. Development. 2004;131:6237–6247. doi: 10.1242/dev.01529. [DOI] [PubMed] [Google Scholar]
- 9.Lebrin F, Deckers M, Bertolino P, ten Dijke P. TGF-beta receptor function in the endothelium. Cardiovasc Res. 2005;65:599–608. doi: 10.1016/j.cardiores.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Bourdeau A, Dumont DJ, Letarte M. A murine model of hereditary hemorrhagic telangiectasia. J Clin Invest. 1999;104:1343–1351. doi: 10.1172/JCI8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur HM, Ure J, Smith AJ, et al. Endoglin, an ancillary TGFbeta receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev Biol. 2000;217:42–53. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Magli A, Catanese J, Xu Z, Kyba M, Perlingeiro RC. Modulation of TGF-{beta} signaling by endoglin in murine hemangioblast development and primitive hematopoiesis. Blood. 2011;118:88–97. doi: 10.1182/blood-2010-12-325019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlingeiro RC. Endoglin is required for hemangioblast and early hematopoietic development. Development. 2007;134:3041–3048. doi: 10.1242/dev.002907. [DOI] [PubMed] [Google Scholar]
- 14.Iacovino M, Hernandez C, Xu Z, Bajwa G, Prather M, Kyba M. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 2009;18:783–792. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlingeiro RCR, Kyba M, Bodie S, Daley GQ. A role for thrombopoietin in hemangioblast development. Stem Cells. 2003;21:272–280. doi: 10.1634/stemcells.21-3-272. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy M, Firpo M, Choi K, et al. A common precurson for primitive erythropoiesis and definitive haematopoiesis. Nature. 1997;386:488–493. doi: 10.1038/386488a0. [DOI] [PubMed] [Google Scholar]
- 17.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Chang PS, Wang Z, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 19.Sadlon TJ, Lewis ID, D’Andrea RJ. BMP4: its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells. 2004;22:457–474. doi: 10.1634/stemcells.22-4-457. [DOI] [PubMed] [Google Scholar]
- 20.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 21.Yu PB, Hong CC, Sachidanandan C, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SK, Bourdeau A, Letarte M, Zuniga-Pflucker JC. Expression and function of CD105 during the onset of hematopoiesis from Flk1(+) precursors. Blood. 2001;98:3635–3642. doi: 10.1182/blood.v98.13.3635. [DOI] [PubMed] [Google Scholar]
- 23.Chen CZ, Li M, de Graaf D, et al. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc Natl Acad Sci U S A. 2002;99:15468–15473. doi: 10.1073/pnas.202614899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roques M, Durand C, Gautier R, et al. Endoglin expression level discriminates long-term hematopoietic from short-term clonogenic progenitor cells in the aorta. Haematologica. doi: 10.3324/haematol.2011.046235. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Stacy T, Binder M, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 27.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 28.Robb L, Lyons I, Li R, et al. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci U S A. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 30.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pimanda JE, Ottersbach K, Knezevic K, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 33.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 34.Aoki H, Fujii M, Imamura T, et al. Synergistic effects of different bone morphogenetic protein type I receptors on alkaline phosphatase induction. J Cell Sci. 2001;114:1483–1489. doi: 10.1242/jcs.114.8.1483. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- 36.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 37.Beppu H, Kawabata M, Hamamoto T, et al. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 38.Maeno M, Mead PE, Kelley C, et al. The role of BMP-4 and GATA-2 in the induction and differentiation of hematopoietic mesoderm in Xenopus laevis. Blood. 1996;88:1965–1972. [PubMed] [Google Scholar]
- 39.Zhang C, Evans T. BMP-like signals are required after the midblastula transition for blood cell development. Dev Genet. 1996;18:267–278. doi: 10.1002/(SICI)1520-6408(1996)18:3<267::AID-DVG7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 40.Huber TL, Zhou Y, Mead PE, Zon LI. Cooperative effects of growth factors involved in the induction of hematopoietic mesoderm. Blood. 1998;92:4128–4137. [PubMed] [Google Scholar]
- 41.Sanchez M, Gottgens B, Sinclair AM, et al. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 42.Gottgens B, Nastos A, Kinston S, et al. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung YS, Zhang WJ, Arentson E, Kingsley PD, Palis J, Choi K. Lineage analysis of the hemangioblast as defined by FLK1 and SCL expression. Development. 2002;129:5511–5520. doi: 10.1242/dev.00149. [DOI] [PubMed] [Google Scholar]