Abstract
Background
Sexual dimorphism in various bone phenotypes, including bone mineral density (BMD), is widely observed; however the extent to which genes explain these sex differences is unclear. To identify variants with different effects by sex, we examined gene-by-sex autosomal interactions genome-wide, and performed eQTL analysis and bioinformatics network analysis.
Methods
We conducted an autosomal genome-wide meta-analysis of gene-by-sex interaction on lumbar spine (LS-) and femoral neck (FN-) BMD, in 25,353 individuals from eight cohorts. In a second stage, we followed up the 12 top SNPs (P<1×10−5) in an additional set of 24,763 individuals. Gene-by-sex interaction and sex-specific effects were examined in these 12 SNPs.
Results
We detected one novel genome-wide significant interaction associated with LS-BMD at the Chr3p26.1-p25.1 locus, near the GRM7 gene (male effect = 0.02 & p-value = 3.0×10−5; female effect = −0.007 & p-value=3.3×10−2) and eleven suggestive loci associated with either FN- or LS-BMD in discovery cohorts. However, there was no evidence for genome-wide significant (P<5×10−8) gene-by-sex interaction in the joint analysis of discovery and replication cohorts.
Conclusion
Despite the large collaborative effort, no genome-wide significant evidence for gene-by-sex interaction was found influencing BMD variation in this screen of autosomal markers. If they exist, gene-by-sex interactions for BMD probably have weak effects, accounting for less than 0.08% of the variation in these traits per implicated SNP.
Keywords: gene-by-sex, interaction, BMD, association, aging
Introduction
Osteoporosis is a common condition that affects at least 30% of women and 12% of men at some point in life (1). Females show a greater incidence of both stress fractures early in life (2) and fragility fractures later in life (3). Areal bone mineral density (BMD), evaluated by dual-energy x-ray absorptiometry (DXA), is to date the most widely used assessment of bone strength and a reliable clinical predictor of osteoporotic fracture (4) in both men and women. Twin studies have demonstrated that BMD is a highly heritable trait (h2 ~ 75%) in both women and men up to the age of 70 (5).
Strong sexual dimorphism in bone phenotypes, including BMD, has been reported, possibly explaining the observed differences in fracture risk between the sexes (6). One explanation for this sex-specific predisposition to osteoporosis and fracture risk is the possibility that the observed differences between men and women are driven by genetic effects determining bone fragility. There are known sex differences in bone traits in mice (7-10). Similarly, some genome-wide linkage analyses in humans have reported sex-specific results. In a whole-genome linkage analysis stratified by sex, sex-specific QTLs were found in the Framingham sample (11), as well as by other groups (6). Furthermore, in a meta-analysis by Ioannidis et al (12) that included data from the 9 whole-genome linkage scans for BMD, several sex-specific QTLs were observed. Notably, some sex-specific QTLs did not appear in any of the individual studies of BMD linkage, possibly because the individual studies had limited power.
Previous published GWAS of bone phenotypes, such as BMD (13-18), did not test for SNP-by-sex interactions, nor did they mainly investigate sex-specific results. As the sample sizes of GWAS expanded with the growth of consortia, it has become more feasible to study sex-specificity of BMD (19). However, direct comprehensive assessment of SNP-by-sex interactions for BMD has not been previously reported. The availability of large-scale GWAS collaborations allows for massive testing of SNP-by-sex interaction effects (20) and for rigorous replication of proposed discovered interactions that would allay the risk of false positives (21). To study potential genetic contributions to the sexual dimorphism in BMD, we first performed a comprehensive study of genome-wide gene-by-sex interactions in cohorts with both men and women who are part of the Genetic Factors for Osteoporosis (“GEFOS”) consortium. In addition, we tested SNPs previously reported to be associated with BMD in a recent meta-analysis of GWAS studies for sex by SNP interactions.
Methods
Subjects and Bone mineral density measurements
The discovery samples included 25,353 subjects of European ancestry (9,056 men and 16,297 women) from 8 cohorts (AMISH, CHS, DECODE, ERF, FHS, HABC, RSI, RSII, see Supplementary Table 1) who are members of the GEFOS consortium. For replication, we recruited an additional 24,763 individuals (6,814 men and 17,949 women) from 19 independent studies. Two (HKO: Asian and SAFOS: Mexican American) of replication cohorts consist of samples of non-European ancestry. All cohorts measured BMD (g/cm2) at the femoral neck (FN) and lumbar spine (LS) using dual-energy X-ray absorptiometry (DXA). Sample size varies (340 ~ 7,605) across different cohorts and average age for each cohort ranged from 19 to 80. Supplementary Table S1 displays the sample characteristics for each cohort.
Genotyping and genotype imputation
Discovery GWAS cohorts were genotyped with various platforms, including Illumina 370CNV (CHS and DECODE), Illumina Infinium HumanHap550 Beadchip (RSI and RSII), the Affymetrix Nsp 250K chip, the Sty 250K chip and the 50K gene-centered MIP chip (FHS), Illumina Human 1M-Duo BeadChip (HABC), Affymetrix 500k or 6.0 (AMISH), and both Illumina and Affymetrix (ERF). Imputation for non-genotyped SNPs was used to perform meta-analysis of results from the same set of SNPs across all individual cohorts. Imputation quality was assessed using the ratio of empirical observed variance of the allele dosage to the expected binomial variance. The imputations were conducted using Hidden-Markov Model implemented in MACH (22,23), IMPUTE (24,25) or BIMBAM (26). The imputation reference panel was the HapMap CEU Phase II (release 22, build 36). Details of the imputation procedures in each discovery cohort and the filtering criteria for each SNP can be found in Supplementary Table S2.
Five of the 19 cohorts for follow-up used various genotyping platforms and applied their cohort-specific filtering criteria. Samples from the other 14 replication studies, all members of the GENOMOS consortium, were de novo genotyped by K-Biosciences (http://www.kbioscience.co.uk/) using a competitive allele specific PCR (KASPar) assay. Most of the de-novo genotyped studies opted for amplified DNA. We evaluated the genotype concordance between genomic DNA genotypes and amplified DNA genotypes in an independent panel of 82 SNPs within 96 samples (19). The genotype accuracy between before and after amplification was 99.97%. A Y-chromosome specific assay was evaluated in all samples to confirm the sex of the individual sample. Sample mismatches between the sex specific assay and the reported sex in the questionnaire were removed from analysis. Aspects of reproducibility (I.e. assessment of duplicate sample, positive control and negative control genotyping) were considered during the stage of assay design for all markers. For the QC of cohort specific genotyping sex concordance checks were assessed using a Y-chromosome assay to identify potential plating errors for which if unresolved samples were excluded. Genotyping concordance using a different platform was also assessed in the EDOS study using Illumina OmniExpress technology for a distinct project on a subset of the EDOS cohort which was originally genotyped at Kbiosciences. The concordance of the shared markers with the microarray and the SNPs genotyped by Kbiosciences was 99.7%. Genotyping comparisons between discovery and follow-up samples were not compared. The following inclusion thresholds were applied for the cohorts typed by K-Biosciences: sample call rate > 80%, SNP call rate > 90%, Hardy Weinberg Equilibrium P-value > 1×10−6. Detailed information on all replication cohorts can be found in Supplementary Table S2.
Statistical Methods for phenotype-genotype association analysis and Meta-analysis
A two-stage approach was applied to test gene-by-sex interaction. In stage 1, using a fixed effects inverse variance approach, we performed a meta-analysis of summary statistics for the gene-by-sex interaction effects in genome-wide analyses of FN- and LS-BMD obtained from 8 discovery cohorts. We used a p-value threshold of P≤5×10−8 for genome-wide significance and P≤1×10−5 for suggestive signals. In the second stage, we then evaluated SNPs with interaction p-values < 1.0×10−5 obtained from stage 1 in replication cohorts. In addition, we evaluated the sex-specific effects for these top SNPs from our GWAS analysis. We also conducted candidate gene analysis to investigate the gene-by-sex interaction for previously reported BMD associated loci from the largest GWAS meta-analysis to date (19).
Stage 1: GWAS Discovery Analysis
Only cohorts with both sexes were included in our discovery stage. Each discovery cohort conducted cohort-specific genome-wide association analyses using linear regression with a main effect for each SNP and SNP-by-sex interaction terms using an additive model for each SNP. We adjusted for age, height, weight, study site (for multisite cohort studies), sex, and also principal components to control for population stratification. The “dosage” information for imputed genotype was used to account for uncertainty of imputation. In addition, we conducted sex-specific association analyses for the top SNPs (P≤1×10−5) to examine their effect on BMD within each sex. We excluded SNPs with poor imputation quality (Observed variance/Expected variance < 0.3 for MACH and BIMBAM imputed datasets and < 0.4 for IMPUTE imputed datasets) and low minor allele frequency (MAF <0.03 for sex-specific analysis and MAF < 0.05 for gene-by-sex interaction analysis).
Stage 2: Replication Analysis
Twelve SNPs from the first stage were followed for replication. For each cohort we performed sex-specific association analysis and for the cohorts with both sexes, SNP-by-sex interaction was examined directly as with the discovery cohorts. For cohorts with both sexes, we meta-analyzed their association results. For cohorts with only one sex, we meta-analyzed the sex-specific association results for the main effects of a SNP and then estimated the gene-by-sex interaction effects by with its corresponding standard error , where and are sex-specific SNP effects from sex-stratified analysis. Utilizing all samples, the final result was obtained through meta-analyzing, with inverse variance as weight, the direct gene-by-sex interactions test result from two-sex sample cohorts and the interaction results estimated from contrasting the one-sex sample cohorts.
Power Analysis for Interaction
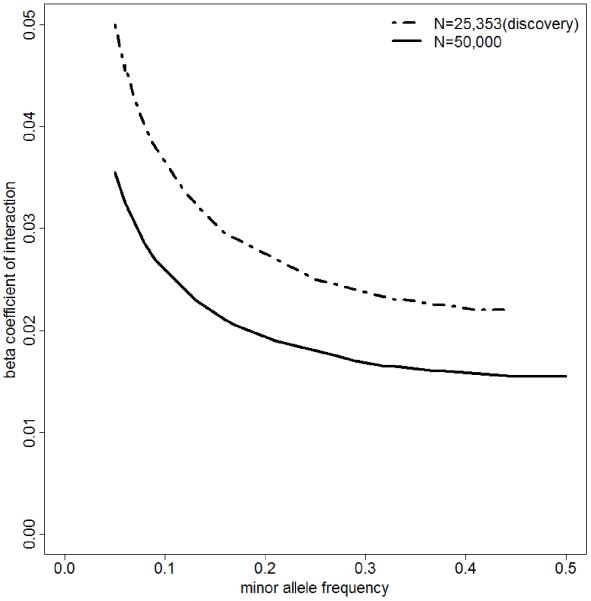
Before beginning second-stage analyses, we estimated that we would have samples of size 50,000 in total. So we conducted a power analysis on the detection of gene-by-sex interaction based on the sample size of our discovery cohorts (25,353) and the estimated sample size of discovery + follow-up cohorts (50,000). Using the effect sizes of top SNPs we observed in our discovery cohort, we explored the relationship between detectable (with 80% power at genome-wide significant level of 5×10−8) interaction effect sizes and allele frequency. We explored all different scenarios corresponding to the range of observed effect sizes of our top SNPs, but their power analysis results are similar so we only present one scenario in Figure 1, corresponding to the parameter estimates of rs1405534 in LSBMD (effect size of −0.0048 for SNP and −0.0219 for SEX).
Figure 1. Power Analysis.
This plot presents the relationship between detectable gene-by-sex interaction effect size with 80% power and minor allele frequency. Specifically, we used the parameter estimates of rs1405534 in LSBMD (SD = 0.187 at genome-wide significant level of 5×10−8) along its relevant parameter estimates (effect size of −0.0048 for SNP and of −0.0219 for SEX), as the setting for our power analysis. The sample size used is 25,353 for discovery cohort and 50,000 (estimated) for discovery + replication cohort. With the sample of size 50,000, we would have 80% power to detect gene-by-sex interaction effect of 0.018 or larger in LSBMD for the SNP with minor allele frequency of 0.25. This would explain 0.08% variation of LSBMD. With 25,353 individuals in the Discovery stage, we did not have adequate power to identify interaction reaching genome-wide significant level (p < 5×10−8) at any allele frequency.
Gene Expression Quantitative Trait Loci (eQTL) Analysis
In parallel to our second stage analysis, we also investigated our top findings from the first stage for the possibility of a meaningful biologic process underlying the genes that these SNPs belonged to. We conducted cis-expression quantitative trait locus (eQTL) analysis within a 500kb flanking region of each of the top SNP to evaluate whether the SNP-by-sex interactions also influence transcript levels of genes in human primary osteoblasts and lymphocytes. In each locus, we selected either the gene in which the interacting SNPs were located or its closest nearby gene. Expression experiments in primary osteoblasts and lymphocytes were conducted in different study samples. For un-genotyped SNPs, surrogate SNPs with LD r2 ≥ 0.5 and within 100kb of the targeted SNPs were used in primary osteoblast samples.
Lymphocytes
A gene expression profile with 24,385 RefSeq annotated genes (Illumina Sentrix Human-6 BeadChips) and genome-wide genotyping of ~2.5 million SNPs were available from Hapmap samples (lymphoblastoid cell lines from 128 women and 142 men). Blood sample collection, RNA and DNA isolation, expression profiling, and DNA genotyping have been described in detail (27). A mixed-effect regression model implemented in R was used to test SNP-sex interactions, adjusting for sex, ethnicity and age.
Primary Osteoblasts
A gene expression profile with 18,144 known genes (Illumina Human Ref8v2 BeadChips) and genome-wide genotyping of 561,303 SNPs (Illumina 550k Duo chips) were available (GSE15678) in 95 human Caucasian primary osteoblast samples (42 women and 53 men). Human trabecular bone came from the shaft of proximal femora obtained from donors undergoing total hip replacement. Primary osteoblasts were derived from bone tissue. Tissue collection, RNA and DNA isolation, expression profiling, and DNA genotyping have been described in detail (28). A linear regression model implemented in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) was used to test SNP-sex interactions, adjusting for sex and year of birth.
Gene-Set Enrichment Tests of Functional Similarity
To explore functional similarity of the genes from our top SNP-sex interactions, we performed a gene-set enrichment test to examine the probability of selected candidate genes clustering in particular biological/functional pathways as defined by the Gene Ontology (GO) project (29).The GO Consortium provides controlled vocabularies, which model “Biological Process”, “Molecular Function” and “Cellular Component” that are structured into directed acyclic graphs based on published literature and databases. Gene products may be annotated to one or more GO nodes. To determine whether any GO terms annotate a specified list of genes at a frequency greater than that would be expected by chance, a p-value was calculated using the hypergeometric distribution (30). To correct for multiple testing, the false discovery rate (FDR) was estimated (31).
Results
GWAS SNP-by-Sex Interaction for Bone Mineral Density
We observed one significant locus associated with SNP-by-sex interaction for LS-BMD at 3p26 with the most significant marker being rs10510373 (closest gene GRM7, P = P≤3.41×10−8) and 11 SNPs with suggestive signals for interactions for either LS-BMD or FN-BMD. We carried forward these 12 SNPs from stage 1 GWAS for replication in 24,763 replication samples. The genome-wide association plots are shown in Supplementary Figure S1 and the quantile-quantile plots are displayed in Supplementary Figure S2, which shows that there was no systematic inflation of test statistics (with genomic inflation factor 1.010 for FNBMD and 1.003 for LSBMD). The results for Stage 1, Stage 2, Stage1 + Stage2 analysis for the 12 SNPs are displayed in Table 1. Of the 12 SNPs carried forward for replication, no SNP reached genome-wide significance for the combined Stage 1 + Stage 2 analyses.
Table 1.
Stage 1 and replication results for meta-analysis of sex-by-gene interaction. Positive Beta implies larger effect of coded allele in men.
| Stage 1 (N =25353) | Stage 2 (N=24763) | Stage 1 + Stage 2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chr | BP (b36) | Genes In or Nearby |
Alleles* | Allele Frequency |
Beta | SE | P-value | Beta | SE | p-value | Beta | SE | p-value |
| FNBMD | ||||||||||||||
| rs17284960 | 5 | 163556080 | MAT2B | t/c | 0.378 | −0.011 | 0.002 | 7.50×10−6 | 0.0012 | 0.003 | 6.84×10−1 | −0.0061 | 0.0019 | 1.05×10−3 |
| rs10756362 | 9 | 12334883 | TYRP1 | t/g | 0.427 | 0.011 | 0.002 | 5.76×10−6 | 0.0031 | 0.0031 | 3.05×10−1 | 0.0079 | 0.0019 | 2.16×10−5 |
| rs11788458 | 9 | 113657842 | UGCG | a/g | 0.855 | 0.017 | 0.003 | 5.42×10−7 | −0.0045 | 0.0037 | 2.20×10−1 | 0.0072 | 0.0025 | 3.50×10−3 |
| rs3748317 | 14 | 93923432 | SERPINA1 | a/c | 0.150 | −0.016 | 0.004 | 7.39×10−6 | 0.0024 | 0.0035 | 4.97×10−1 | −0.0067 | 0.0025 | 7.22×10−3 |
| LSBMD | ||||||||||||||
| rs2295294 | 1 | 10113471 | UBE4B | a/t | 0.623 | 0.019 | 0.004 | 8.60×10−6 | −0.0057 | 0.0035 | 9.84×10−2 | 0.0043 | 0.0026 | 1.07×10−1 |
| rs7417366 | 1 | 166730211 | XCL2 | a/g | 0.648 | 0.021 | 0.004 | 7.42×10−8 | 0.0027 | 0.0039 | 4.92×10−1 | 0.0118 | 0.0028 | 2.18×10−5 |
| rs10510373 | 3 | 7886027 | GRM7 | c/g | 0.112 | 0.034 | 0.006 | 3.41×10−8 | −0.0138 | 0.0052 | 7.59×10−3 | 0.006 | 0.0039 | 1.29×10−1 |
| rs4832734 | 4 | 37379393 | RELL1 | t/c | 0.834 | 0.023 | 0.005 | 1.89×10−6 | −0.0046 | 0.0051 | 3.71×10−1 | 0.01 | 0.0035 | 4.35×10−3 |
| rs6830890 | 4 | 112738575 | C4orf32 | a/g | 0.127 | −0.028 | 0.006 | 7.98×10−7 | 0.0074 | 0.0048 | 1.19×10−1 | −0.0073 | 0.0036 | 4.22×10−2 |
| rs1405534 | 8 | 25066043 | DOCK51 | c/g | 0.491 | 0.015 | 0.003 | 8.99×10−6 | 0.0004 | 0.0033 | 8.92×10−1 | 0.0076 | 0.0024 | 1.29×10−3 |
| rs12900333 | 15 | 76465331 | CRABP1 | t/c | 0.119 | −0.028 | 0.006 | 6.20×10−6 | 0.0096 | 0.0051 | 5.81×10−2 | −0.0056 | 0.0039 | 1.48×10−1 |
| rs2717096 | 18 | 73058030 | GALR1 | a/g | 0.442 | −0.019 | 0.004 | 1.50×10−6 | 0.0022 | 0.0033 | 5.08×10−1 | −0.0065 | 0.0025 | 8.97×10−3 |
Alleles = Coded Allele/non-Coded Allele
We also conducted sex-specific analysis for the 12 SNPs with at least suggestive interaction signals in Stage 1 to examine their sex-specific signal with BMD (Table 2). None of these 12 SNPs showed a genome-wide significant sex-specific signal.
Table 2.
Gender specific results for the top SNPs from the discovery cohorts
| Male Sample | Female Sample | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stage 1 | Stage 2 | Stage 1 + Stage 2 | Stage 1 | Stage 2 | Stage 1 + Stage 2 | |||||||||||||||
| SNP ID | Genes In or Nearby |
Alleles* | BETA | SE | P-value | BETA | SE | P-value | BETA | SE | P-value | BETA | SE | P-value | BETA | SE | P-value | BETA | SE | P-value |
| FNBMD | ||||||||||||||||||||
| rs17284960 | MAT2B | t/c | −0.008 | 0.002 | 1.69×10−5 | −0.003 | 0.005 | 5.76×10−1 | −0.005 | 0.002 | 2.21×10−3 | 0.001 | 0.001 | 3.00×10−1 | −0.001 | 0.002 | 4.89×10−1 | 0.001 | 0.001 | 3.95×10−1 |
| rs10756362 | TYRP1 | t/g | 0.006 | 0.002 | 3.08×10−3 | 0.000 | 0.005 | 9.38×10−1 | 0.005 | 0.002 | 3.67×10−3 | −0.003 | 0.001 | 1.22×10−2 | −0.002 | 0.002 | 3.52×10−1 | −0.002 | 0.001 | |
| rs11788458 | UGCG | a/g | 0.007 | 0.003 | 1.18×10−2 | −0.004 | 0.005 | 3.93×10−1 | 0.003 | 0.002 | 1.98×10−1 | −0.007 | 0.002 | 5.40×10−5 | 0.001 | 0.002 | 8.11×10−1 | −0.004 | 0.001 | |
| rs3748317 | SERPINA1 | a/c | −0.009 | 0.003 | 1.09×10−3 | 0.001 | 0.005 | 7.71×10−1 | −0.004 | 0.002 | 9.24×10−2 | 0.005 | 0.002 | 4.60×10−3 | 0.001 | 0.002 | 7.37×10−1 | 0.002 | 0.001 | 6.66×10−2 |
| LSBMD | ||||||||||||||||||||
| rs2295294 | UBE4B | a/t | 0.011 | 0.003 | 3.77×10−4 | −0.003 | 0.004 | 4.30×10−1 | 0.003 | 0.002 | 2.11×10−1 | −0.006 | 0.002 | 9.84×10−3 | 0.000 | 0.002 | 8.46×10−1 | −0.001 | 0.001 | 5.54×10−1 |
| rs7417366 | XCL2 | a/g | 0.011 | 0.003 | 5.75×10−4 | −0.006 | 0.006 | 2.88×10−1 | 0.007 | 0.002 | 1.91×10−3 | −0.008 | 0.002 | 1.22×10−4 | −0.004 | 0.003 | 9.41×10−2 | −0.004 | 0.001 | 9.02×10−4 |
| rs10510373 | GRM7 | c/g | 0.020 | 0.005 | 3.03×10−5 | 0.002 | 0.007 | 7.05×10−1 | 0.009 | 0.003 | 6.03×10−3 | −0.007 | 0.003 | 3.32×10−2 | −0.001 | 0.004 | 8.98×10−1 | −0.001 | 0.002 | 6.66×10−1 |
| rs4832734 | RELL1 | t/c | 0.018 | 0.004 | 1.99×10−6 | −0.009 | 0.007 | 2.19×10−1 | 0.010 | 0.003 | 7.10×10−4 | −0.003 | 0.003 | 2.14×10−1 | 0.008 | 0.003 | 6.35×10−3 | 0.001 | 0.002 | 7.55×10−1 |
| rs6830890 | C4orf32 | a/g | −0.021 | 0.005 | 5.64×10−6 | 0.007 | 0.006 | 2.18×10−1 | −0.007 | 0.003 | 1.56×10−2 | 0.004 | 0.003 | 1.98×10−1 | −0.004 | 0.003 | 2.36×10−1 | −0.001 | 0.002 | 7.29×10−1 |
| rs1405534 | DOCK5 | c/g | 0.009 | 0.003 | 1.35×10−3 | −0.005 | 0.004 | 2.37×10−1 | 0.001 | 0.002 | 5.57×10−1 | −0.004 | 0.002 | 2.40×10−2 | −0.007 | 0.002 | 1.71×10−3 | −0.005 | 0.001 | 1.17×10−5 |
| rs12900333 | CRABP1 | t/c | −0.019 | 0.005 | 7.56×10−5 | 0.001 | 0.007 | 8.56×10−1 | −0.008 | 0.003 | 1.36×10−2 | 0.000 | 0.003 | 9.20×10−1 | −0.002 | 0.003 | 4.60×10−1 | −0.002 | 0.002 | 1.99×10−1 |
| rs2717096 | GALR1 | a/g | −0.012 | 0.003 | 2.74×10−4 | −0.001 | 0.004 | 8.67×10−1 | −0.005 | 0.002 | 2.85×10−2 | 0.002 | 0.002 | 4.00×10−1 | −0.002 | 0.002 | 2.89×10−1 | 0.000 | 0.001 | 7.49×10−1 |
Alleles = Coded Allele/non-Coded Allele
Differential Expression and eQTL
As listed in Table 3, except for RELL1, gene expression levels of 11 top associated genes were obtained in human lymphocytes. Gene expression levels in lymphocytes of these 11 genes were higher in men than the expression levels in women. However, none of them were considered statistically significant after correcting for multiple comparisons using Bonferroni correction (p-value cutoff= 0.05/11 = 0.0046). A significant eQTL was found for SNP rs3748371, as a polymorphic allele T in rs3748371 was associated with higher SERPINA1 expression (data not showen). However, no significant SNP-sex interactions on gene expression were shown in human lymphocyte samples.
Table 3.
Expression analysis (eQTL and differential expression by sex) for top SNPs identified from the Discovery stage
| Human Lymphocytes eQTL |
Human Primary Osteoblast eQTL |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Targeted SNPs |
Differential* Expression by sex P-values |
SNPxSex Interaction P-values |
Differential* Expression by sex P-values |
Surrogate SNPs |
LD (r2 or D’) |
Distance (kb) |
MAF | alleles | SNPxSex Interaction P-values |
| FN BMD | ||||||||||
| MAT2B | rs17284960 | 4.06E-04 | 0.682 | 0.043 | rs17284960 | 1.00 | 0.0 | 0.42 | C/T | 0.47 |
| TYRP1 | rs10756362 | 0.117 | 0.398 | 0.711 | rs7860071 | 0.67 | 98.7 | 0.30 | T/C | 0.68 |
| UGCG | rs11788458 | 0.048 | 0.867 | 0.723 | rs11788458 | 1.00 | 0.0 | 0.14 | C/T | 0.62 |
| SERPINA1 | rs3748317 | 0.677 | 0.418 | 0.558 | rs709932 | 1.00 | 4.5 | 0.14 | T/C | 0.47 |
| LS BMD | ||||||||||
| UBE4B | rs2295294 | 0.251 | 0.166 | 0.960 | rs6696978 | 0.95 | 18.7 | 0.30 | G/A | 0.36 |
| XCL2 | rs7417366 | 0.490 | 0.615 | 0.509 | rs7417366 | 1.00 | 0.0 | 0.35 | C/T | 0.66 |
| GRM7 | rs10510373 | 0.097 | 0.014 | 0.948 | rs10510378 | 0.95 | 55.9 | 0.13 | C/T | 0.08 |
| RELL1 | rs4832734 | N.A | N.A | N.A | N.A | N.A | N.A | N.A | N.A | N.A |
| C4orf32 | rs6830890 | 0.250 | 0.974 | 0.620 | N.A | N.A | N.A | N.A | N.A | N.A |
| DOCK5 | rs1405534 | 0.038 | 0.910 | 0.857 | rs1358268 | 1.00 | 4.0 | 0.46 | T/C | 0.40 |
| CRABP1 | rs12900333 | 0.879 | 0.737 | 0.309 | rs2037348 | 0.84 | 24.7 | 0.22 | C/T | 0.74 |
| GALR1 | rs2717096 | 0.463 | 0.565 | 0.988 | rs2717096 | 1.00 | 0.0 | 0.45 | A/G | 0.76 |
gene expression is considered significantly higher in men than the expression level in women for p < 4.55×10−3 (0.05/11)
Except for RELL1, gene expression levels of 11 top associated genes were obtained in human primary osteoblasts. Gene expression levels in these 11 genes were not significantly different between men and women. For targeted SNPs not genotyped in human primary osteoblast samples, surrogate SNPs with LD r2 ≥ 0.5 and within 100kb of the targeted SNPs were selected and estimate their eQTL (ie, SNP rs6696978 as a surrogate SNP of targeted SNP, rs2295294 in UBE4B loci). No surrogate SNP (genotyped) can be found for targeted SNP rs6830890 in C4orf32 loci; therefore, the eQTL in this region was not estimated. No significant SNP-sex interactions on gene expression (eQTL) were found in human primary osteoblast samples.
Gene-Set Enrichment Tests
To test the probability of our candidate genes clustering into a particular biological pathway, we performed a gene set enrichment test on 12 genes listed in Table 3. Due to lack of biological or functional annotation, RELL1 and C4orf32 were excluded from analyses. The most significant clustering (Fisher exact test p=6.03×10−4; Benjamini-Hochberg multiple testing correction p=0.045) of genes involved nervous system development and function, including UGCG, GRM7, TYRP1 and UBE4B genes. However, by looking at more specific biological function ontology terms under nervous system development and function, no significantly enriched functions were found.
Previously, we have identified 55 genome-wide significant loci associated with BMD with the most significant p-values < 5×10−8 (19), and we found an additional 10 loci that were considered as suggestive signals with the most significant p-values < 5×10−6. We performed a gene-set enrichment analysis on these 65 loci and found functional enrichments on ossification (p=1.8×10−7), bone formation (p=2.8×10−6), mineralization of trabecular bone (6.1×10−6), cell-cell connection among osteoclast and osteoblast and chondrocyte (p=1.2×10−6), degenerative mitral valve disease (p=1.5×10−6) and Wnt/β-catenin Signaling (p=3.4×10−5). To estimate whether additional enriched functional pathways could be identified by adding gene-sex interaction loci to the 65 BMD associated loci, we performed a gene-set enrichment analysis on 75 loci (65 BMD associated loci with 10 gene-sex interaction loci with functional information). These ten gene-sex interaction loci were not significantly clustered together with any of the 65 BMD associated loci in any known functional and biological pathways or gene sets. No additional enriched functional and biological pathways were found.
Discussion
This is the first genome-wide association analysis focusing on SNP-by-sex interaction of BMD phenotypes. We identified 12 SNP-sex interaction loci with suggestive genome-wide significance (p < 10−5) in 25,353 adult men and women which did not replicate in an additional 24,763 adult men and women. Also we observed no replicated SNP by sex interaction for any of the top SNPs found to be significantly associated with BMD in the largest GWAS meta-analysis performed to date (Detail described in Supplementary text and Supplementary Table 3). To account for the varying linkage disequilibrium structure across different populations, we also analyzed the data from the sample of European ancestry (data not shown). Although we observed improvement of the p-values (smaller p-values in European ancestry samples only), but no SNP by sex interaction was replicated for any of the top SNPs in this sample. Hence, the conclusion remained the same.
Recent studies (32) suggest that sex-specific genetic architecture influences many human phenotypes, including reproductive, physiological, and complex disease traits. Some of the underlying mechanisms might be attributed to differential gene regulation in males and females, particularly in sex steroid responsive genes (different hormonal milieu). In the field of skeletal genetics, in particular, sex-specific findings were reported in linkage studies (both in humans and animal models) and candidate gene association studies (reviewed in (6)).
Similar to the studies in mice, whole-genome linkage studies in humans for BMD also provided evidence of sex-specific QTLs with max LOD of 3.29 (12). More recently, this was confirmed by Peacock et al. (33), who found male-specific QTLs on chromosomes 7q34, 14q32, and 21q21 to be linked with aBMD. Several groups studying candidate genes also found evidence of sex-specific associations. For example, a SNP in the glucocorticoid receptor (GR) gene was associated with extreme BMD in Chinese men but not women(34); SNPs in the VKORC1 gene were associated with decreased BMD in Mexican-American and Black men but not women from NHANES III(35). Finally, sex-specific associations of the Pirin (PIR) gene with lumbar spine BMD was shown in a large Chinese cohort(36). None of these findings overlap with our top hits.
Analyzing sex-specific associations between SNPs and BMD tests a different hypothesis than showing that there are interactions between genetic variants and gender on BMD. The former hypothesis tests whether the association between SNPs and BMD differs from zero either within males or females, whereas interaction tests whether the magnitude of association between SNPs and BMD differs significantly between sexes. Given that most genetic variants have very subtle effects, it is expected that the lists of discovered genes and variants associated with BMD in analyses limited to men and in analyses limited to women may differ simply because of power considerations, even if the effects of these variants are not genuinely different in the two sexes. In addition, the observation that men have higher BMD than women does not necessarily imply that genetic variation contributes to the differential distribution of BMD between men and women, despite the fact that sex is by itself genetically-determined.
There are obvious gender-specific hormonal milieus contributing to the sexual dimorphism on bone health. Notably, differences in response to estrogen and testosterone have been shown for male and female chondrocytes, osteoblasts, myoblasts, and other cells (37,38). In an earlier study, sex differences in response to progesterone have also been reported in cells derived from rat lumbar vertebrae (39). Most recently, using human peripheral blood mononuclear precursor cells from adult males and females that were differentiated into osteoclasts, Wang & Stern (40) demonstrated sex-specific actions of estrogen and androgen. Thus, 17b-estradiol and testosterone largely affected expression of different genes from a custom designed array containing 94 genes related to bone and hormone metabolism. If such sex-specific expression of genes related to bone metabolism does in fact occur, this was not reflected by our analyses.
Attempts to evaluate whether there are functionally significant sex-specific differences arising from the interaction results, differences in gene expression between males and females and eQTL on SNP-by-sex interactions were analyzed in human samples from two tissue types: primary osteoblast and lymphocytes. We failed to find significant differential gene expression between men and women among the 12 transcripts prioritized by the SNP-by-sex interaction analyses despite the presence of significant eQTL (eSNP). However, it is important to note that a lack of evidence from expression experiments does not necessarily exclude gene interaction with sex influencing BMD variation, given that (i) experimental models such as osteoclastogenesis, osteoblastogenesis or early skeletal development, do not represent all relevant processes related to the skeleton at the organism level; (ii) variation in a gene leading to disease may affect protein function but not expression; and (iii) absence of detectable SNP-by sex interactions may be due to modest effect sizes or due to different environmental conditions.
There are several potential limitations of this study. First, this analysis was restricted to autosomal SNPs, although we do not expect that most of BMD associated SNPs will be assembled at the sex chromosomes, as there is not much of X-linked heritability of BMD. Second, areal BMD, despite being a sexually-dimorphic phenotype, is not necessarily the most optimal skeletal phenotype because it does not adequately represent bone size. To avoid identifying genes responsible for bone size, we adjusted height in the model to account for this concern. We also repeated our analysis without height adjustment and the results were very similar. However given the limitation of 2-D aBMD, volumetric density measures would be also worthwhile assessing in the future for genetics of sex differences. Third, our analyses were corrected for body weight and height which are important determinants of the skeletal differences observed between men and women. From this perspective gene-sex interactions effects influencing BMD variation through weight and height (size) parameters would have been missed. We evaluated here (mostly) direct skeletal effects. Finally, despite our large sample size, we did not identify genome-wide significant SNP-by-sex interactions from using 25,353 individuals for the discovery stage followed by 24,763 individuals for replication. As shown in Figure 1, with the sample of size 50,000, we would have 80% power to detect gene-by-sex interaction effect of 0.018 or larger in LSBMD (SD = 0.187) for the SNP (which explain around 0.08% variation of a quantitative trait) with minor allele frequency of 0.25. With 25,353 individuals in the Discovery stage, we did not have adequate power to identify interaction reaching genome-wide significant level (p < 5×10−8) at any allele frequency. The power analyses were conducted under the assumption that the interaction effects are similar to what we observed for the main effects, in our first stage analysis. We are aware that the real effect size may be relatively smaller (a “winner’s curse” effect) and acknowledge that despite the large size of the populations studied, we were still underpowered to detect these magnitudes of interaction effect sizes. This result also implies that the gene-by-sex effect, if exists, is smaller in magnitude than the previously observed SNP effects. Although we did not replicate the top SNP-by-sex interaction loci in additional samples, possibly due to insufficient statistical power and/or potential heterogeneity across discovery studies and replication studies, we cannot completely rule out the possibility that these top loci from the discovery stage are in fact involved in sex-specific regulation of BMD.
Using bioinformatics approaches, we performed biological functional interaction network analysis with a couple of pre-specified pathways; namely sex steroids and Wnt signaling (detailed methods and results are described in the supplementary materials). Using this approach, we were able to indirectly link some of our top SNP-by-sex loci to β-estradiol (supplementary Figure S3). For example, UBE4B has been found to negatively regulate the level of p53 and to inhibit p53-dependent transactivation and apoptosis(41). Based on chromatin immunoprecipitation assays, studies found p53 as being recruited to the ER promoter along with other transcription factors and that this complex was formed in a p53-dependent manner, which suggests that p53 regulates ER expression through transcriptional control of the ER promoter (42). This type of evidence from biological experiments may suggest that if there are true SNP-by-sex interactions on BMD, they may be indirect, acting through more complex networks of genes. Thus, UGCG and SERPINA1 (two of the most significant findings from the discovery stage) were found to have a direct functional interaction with Wnt signaling pathways (supplementary Figure S4). SERPINA1 not only interacts with CTNNB1 protein, it also decreases activation of the NFkB complex (43), which also supports its involvement in bone metabolism.
In conclusion, our results suggest that an SNP-by-sex interaction effect if present may be too small and heterogeneous to be detected with the current sample size, and/or with the limitations of the areal BMD phenotypes. Therefore, future investigations of true differences in genetic associations between the sexes will require even larger samples or more sexually-dimorphic skeletal phenotypes such as volumetric skeletal measurements.
Supplementary Material
Acknowledgement
Amish: The Old Order Amish Study was supported by NIH research grants, R01 AG18728, R01HL088119, R01AR046838, U01 HL084756. Partial funding was also provided by the Nutrition and Obesity Research Center of Maryland (P30DK072488). LMY-A was supported by F32AR059469 from NIAMS/NIH.
CABRIO: Supported by grants from Instituto de Salud Carlos III-FIS (Spanish Health Ministry) PI 06/0034 and PI08/0183.
The Canadian Multicentre Osteoporosis Study (CaMos) was supported by a grant from the Canadian Institutes of Health Research (CIHR).
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133 and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center of Advancing Translational Technologies CTSI grant UL1TR000124 and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Erasmus Rucphen Family Study (ERF): The genotyping for the ERF study was supported by EUROSPAN (European Special Populations Research Network) and the European Commission FP6 STRP grant (018947; LSHG-CT-2006-01947). The ERF study was further supported by grants from the Netherlands Organisation for Scientific Research, Erasmus MC, the Centre for Medical Systems Biology (CMSB) and the Netherlands Brain Foundation (HersenStichting Nederland). We are grateful to all patients and their relatives, general practitioners and neurologists for their contributions and to P. Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection. We would also like to acknowledge Internationale Stichting Alzheimer Onderzoek (ISAO) and Hersenstichting Netherlands.
Framingham Osteoporosis Study (FOS):The study was funded by grants from the US National Institute for Arthritis, Musculoskeletal and Skin Diseases and National Institute on Aging (DPK: R01 AR/AG41398; DK: R01 AR050066; YHH: R21 AR056405). The Framingham Heart Study of the National Heart, Lung, and Blood Institute of the National Institutes of Health and Boston University School of Medicine were supported by the National Heart, Lung, and Blood Institute’s Framingham Heart Study (N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (N02-HL-6-4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center
GEOS: The Genetics of Osteoporosis Study was funded in part by the Canadian Institutes of Health Research from Institute of Aging (# 165446), Institute of Genetics (#179433) and Institute of Musculoskeletal health (#221765).
Health ABC: This research was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106 and in part by the Intramural Research Program of the NIH, National Institute on Aging. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C.
HKOS: This project is supported by Hong Kong Research Grant Council (HKU 768610M); The Bone Health Fund of HKU Foundation; The KC Wong Education Foundation; Small Project Funding (201007176237); Matching Grant, CRCG Grant and Osteoporosis and Endocrine Research Fund, and the Genomics Strategic Research Theme of The University of Hong Kong. The authors are very grateful to the participants and clinical staff at the Queen Mary Hospital of The University of Hong Kong.
RSI, RSII, RSIII: The GWA study was funded by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are very grateful to the participants and staff from the Rotterdam Study, the participating general practioners and the pharmacists. We would like to thank Dr. Tobias A. Knoch, Luc V. de Zeeuw, Anis Abuseiris, and Rob de Graaf as well as their institutions the Erasmus Computing Grid, Rotterdam, The Netherlands, and especially the national German MediGRID and Services@MediGRID part of the German D-Grid, both funded by the German Bundesministerium fuer Forschung und Technology under grants #01 AK 803 A-H and # 01 IG 07015 G for access to their grid resources.
San Antonio Family Osteoporosis Study: The SAFOS was supported by NIH research grants R01 AR43351 and P01-HL45522.
Footnotes
Disclosure Page: K.S, U.T, U.S and G.T are employed by deCODE Genetics. BMP serves on a DSMB for a clinical trial of a device (Zoll Lifecor), and on a Steering Committee for the Yale Open Data Access Project funded by Medtronic. These are unrelated to the paper and not likely to be conflicts, but he wanted to disclose them. All other authors state that they have no conflicts of interest.
Author Contributions:
Overseeing: Susana Balcells, John Blangero, Maria Luisa Brandi, L. Adrienne Cupples, Morten Frost, David Goltzman, Jesús González-Macías, Tamara B. Harris, Albert Hofman, John P.A Ioannidis, Magnus Karlsson, Douglas P. Kiel, Panagoula Kollia, Annie WaiChee Kung, Bente Lomholt Langdahl, Östen Ljunggren, Mattias Lorentzon, Janja Marc, Laura Masi, Dan Mellström, Braxton D. Mitchell, Claes Ohlsson, José M. Olmos, Ben A. Oostra, Janez Prezelj, Richard L. Prince, José A. Riancho, J Brent Richards, Fernando Rivadeneira, Bruce M. Psaty, François Rousseau, Pak Chung Sham, Timothy Spector, Kari Stefansson, Elizabeth A. Streeten, Unnur Styrkarsdottir, Unnur Thorsteinsdottir, Frances A. Tylavsky, Andre G. Uitterlinden, Roser Urreizti, Cornelia M. vanDuijn, Wim Van Hul, María T. Zarrabeitia
Genotyping: Melanie A. Carless, L. Adrienne Cupples, Karol Estrada, Sylvie Giroux, Lizbeth Herrera, Rita I. Khusainova, Elza K. Khusnutdinova, Joshua R. Lewis, Yongmei Liu, Mattias Lorentzon, Janja Marc, Carolina Medina-Gómez, Lizbeth Herrera, Simona Mencej-Bedrac, Ling Oei, Claes Ohlsson, Ben A. Oostra, Fernando Rivadeneira, Jerome I Rotter, Timothy Spector, Unnur Styrkarsdottir, Andre G. Uitterlinden, Joyce B.J. vanMeurs, Liesbeth Vandenput
Phenotyping: Nerea Alonso, Maria Luisa Brandi, Martha Castano-Betancourt, Zoe Dailiana, Morten Frost, Natalia García-Giralt, Sylvie Giroux, Albert Hofman, Lise Bjerre Husted, Magnus Karlsson, Douglas P. Kiel, Theodora Koromila, Annie WaiChee Kung, Yongmei Liu, Östen Ljunggren, Mattias Lorentzon, Janja Marc, Laura Masi. Dan Mellström, Xavier Nogues, Claes Ohlsson, José M. Olmos, Janez Prezelj, Richard L. Prince, José A. Riancho, J Brent Richards, Fernando Rivadeneira, François Rousseau, Gunnar Sigurdsson, Elizabeth A. Streeten, Unnur Styrkarsdottir, Andre G. Uitterlinden, Wim Van Hul, Joyce B.J. vanMeurs, Su-Mei Xiao, Kun Zhu, M.Carola Zillikens
Data analysis: Najaf Amin, Serkalem Demissie, Joel Eriksson, Karol Estrada, Elin Grundberg, Yi-Hsiang Hsu, Candace M. Kammerer, Douglas P. Kiel, Theodora Koromila, Annie WaiChee Kung, Tony Kwan, Guo Li, Ching-Ti Liu, Gudmar Thorleifsson, Janja Marc, Simona Mencej-Bedrac, Ryan L. Minster, Jeffrey R. O’Connell, Ling Oei, Tomi Pastinen, Millan S. Patel, Janez Prezelj, Fernando Rivadeneira, Pak Chung Sham, Unnur Styrkarsdottir, Su-Mei Xiao, Laura M. Yerges-Armstrong, Hou-Feng Zheng, Yanhua Zhou
Meta-analysis: Karol Estrada, Evangelos Evangelou, John P.A Ioannidis, Ching-Ti Liu, Yanhua Zhou
Writing group: L. Adrienne Cupples, Karol Estrada, Yi-Hsiang Hsu, David Karasik, Douglas P. Kiel, Ching-Ti Liu, Richard L. Prince, Fernando Rivadeneira, Laura M. Yerges-Armstrong
Reference
- 1.Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes & Development. 2006;20(18):2492–2506. doi: 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- 2.Beck TJ, Ruff CB, Shaffer RA, Betsinger K, Trone DW, Brodine SK. Stress fracture in military recruits: Gender differences in muscle and bone susceptibility factors. Bone. 2000;27(3):437–444. doi: 10.1016/s8756-3282(00)00342-2. [DOI] [PubMed] [Google Scholar]
- 3.Cummings SR, Melton LJ. Epidemiology and outcomes of osteoporotic fractures. Lancet. 2002;359(9319):1761–1767. doi: 10.1016/S0140-6736(02)08657-9. [DOI] [PubMed] [Google Scholar]
- 4.Kanis JA, Stevenson M, McCloskey EV, Davis S, Lloyd-Jones M. Glucocorticoid-induced osteoporosis: a systematic review and cost-utility analysis. Health Technology Assessment. 2007;11(7):1. doi: 10.3310/hta11070. [DOI] [PubMed] [Google Scholar]
- 5.Flicker L, Hopper JL, Rodgers L, Kaymakci B, Green RM, Wark JD. Bone-Density Determinants in Elderly Women - a Twin Study. Journal of Bone and Mineral Research. 1995;10(11):1607–1613. doi: 10.1002/jbmr.5650101102. [DOI] [PubMed] [Google Scholar]
- 6.Karasik D, Ferrari SL. Contribution of gender-specific genetic factors to osteoporosis risk. Annals of Human Genetics. 2008;72:696–714. doi: 10.1111/j.1469-1809.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 7.Beamer WG, Shultz KL, Ackert-Bicknell CL, Horton LG, Delahunty KM, Coombs HF, Donahue LR, Canalis E, Rosen CJ. Genetic dissection of mouse distal chromosome 1 reveals three linked BMD QTLs with sex-dependent regulation of bone phenotypes. Journal of Bone and Mineral Research. 2007;22(8):1187–1196. doi: 10.1359/jbmr.070419. [DOI] [PubMed] [Google Scholar]
- 8.Ishimori N, Stylianou IM, Korstanje R, Marion MA, Li RH, Donahue LR, Rosen CJ, Beamer WG, Paigen B, Churchill GA. Quantitative trait loci for BMD in an SM/J by NZB/BlNJ intercross population and identification of Trps1 as a probable candidate gene. Journal of Bone and Mineral Research. 2008;23(9):1529–1537. doi: 10.1359/JBMR.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan S, Edderkaoui B, Baylink DJ, Beamer WG, Shultz KL, Wergedal JE. Genetic regulation of femoral bone mineral density: Complexity of sex effect in chromosome 1 revealed by congenic sublines of mice. Bone. 2007;41(3):340–345. doi: 10.1016/j.bone.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Orwoll ES, Belknap JK, Klein RF. Gender specificity in the genetic determinants of peak bone mass. Journal of Bone and Mineral Research. 2001;16(11):1962–1971. doi: 10.1359/jbmr.2001.16.11.1962. [DOI] [PubMed] [Google Scholar]
- 11.Karasik D, Cupples LA, Hannan MT, Kiel DP. Age, gender, and body mass effects on quantitative trait loci for bone mineral density: the Framingham Study. Bone. 2003;33(3):308–316. doi: 10.1016/s8756-3282(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 12.Ioannidis JPA, Ralston SH, Ng MY, Sham PC, Zintzaras E, Lewis CM, Deng HW, Econs MJ, Karasik D, Devoto M, Kammerer CM, Spector T, Andrew T, Cupples LA, Duncan EL, Foroud T, Kiel DP, Koller D, Langdahl B, Mitchell BD, Peacock M, Recker R, Shen H, Sol-Church K, Spotila LD, Uitterlinden AG, Wilson SG, Kung AWC. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. Journal of Bone and Mineral Research. 2007;22(2):173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. Bmc Medical Genetics. 2007;8 doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HAP, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FMK, Wilson SG, Zhou YH, Ralston SH, van Duijn CM, Spector T, Kiel DP, Stefansson K, Ioannidis JPA, Uitterlinden AG. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nature Genetics. 2009;41(11):1199–U58. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang TL, Guo Y, Liu YJ, Shen H, Liu YZ, Lei SF, Li J, Tian Q, HW D. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int. 2011 doi: 10.1007/s00198-011-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nature Genetics. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 17.Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang LM, Richards JB, Estrada K, Zhou Y, van Nas A, Moffatt MF, Zhai G, Hofman A, van Meurs JB, Pols HAP, Price RI, Nilsson O, Pastinen T, Cupples LA, Lusis AJ, Schadt EE, Ferrari S, Uitterlinden AG, Rivadeneira F, Spector TD, Karasik D, Kiel DP. An Integration of Genome-Wide Association Study and Gene Expression Profiling to Prioritize the Discovery of Novel Susceptibility Loci for Osteoporosis-Related Traits. Plos Genetics. 2010;6(6) doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Styrkarsdottir U HB, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. New England Journal of Medcine. 2008;358(22):2344–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 19.Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y, Duncan EL, Ntzani EE, Oei L, Albagha OME, Amin N, Kemp JP, Koller DL, Li G, Liu C, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao S, Yerges-Armstrong LM, Zheng H, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson JF, Wilson SG, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, García-giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim G, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee S, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi M, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, González-Macías J, Kähönen M, Karlsson M, Khusnutdinova E, Koh J, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren Ö, Lorenc RS, Marc J, Mellström D, Obermayer-Pietsch B, Olmos JM, Pettersson U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NLS, Urreizti R, van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gómez C, Palsson S.Th., Reppe S, Rotter JI, Sigurdsson G, van Meurs JBJ, Verlaan D, Williams FMK, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw K, Lehtimäki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HAP, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AWC, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens M, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JPA, Kiel DP, Rivadeneira F. New genomic loci for bone mineral density, osteoporosis and risk of fracture. Nature Genetics. 2012;55(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu LY, Schaub MA, Sirota M, AJ B. Sex differences in disease risk from reported genome-wide association study findings. Human Genetics. 2011 doi: 10.1007/s00439-011-1081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patsopoulos NA, Tatsioni A, Ioannidis JPA. Claims of sex differences - An empirical assessment in genetic associations. Jama-Journal of the American Medical Association. 2007;298(8):880–893. doi: 10.1001/jama.298.8.880. [DOI] [PubMed] [Google Scholar]
- 22.Abecasis GR, Li Y, Willer CJ, Ding J, Scheet P. MaCH: Using Sequence and Genotype Data to Estimate Haplotypes and Unobserved Genotypes. Genetic Epidemiology. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Willer C, Sanna S, Abecasis G. Genotype Imputation. Annual Review of Genomics and Human Genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. Plos Genetics. 2009;5(6) doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature Reviews Genetics. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 26.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. American Journal of Human Genetics. 2006;78(4):629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurles ME, Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastinen T, Grundberg E, Kwan T, Ge B, Lam KCL, Koka V, Kindmark A, Mallmin H, Dias J, Verlaan DJ, Ouimet M, Sinnett D, Rivadeneira F, Estrada K, Hofman A, van Meurs JM, Uitterlinden A, Beaulieu P, Graziani A, Harmsen E, Ljunggren O, Ohlsson C, Mellstrom D, Karlsson MK, Nilsson O. Population genomics in a disease targeted primary cell model. Genome Research. 2009;19(11):1942–1952. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botstein D, Ashburner M, Ball CA, Blake JA, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Consortium GO. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherlock G, Boyle EI, Weng SA, Gollub J, Jin H, Botstein D, Cherry JM. GO:TermFinder - open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. [Google Scholar]
- 32.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nature Reviews Genetics. 2008;9(12):911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peacock M, Koller DL, Lai DB, Hui S, Foroud T, Econs MJ. Bone mineral density variation in men is influenced by sex-specific and non sex-specific quantitative trait loci. Bone. 2009;45(3):443–448. doi: 10.1016/j.bone.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng HW, Peng YM, Lei SF, Guo Y, Xiong DH, Yan H, Wang L, Guo YF. Sex-specific association of the glucocorticoid receptor gene with extreme BMD. Journal of Bone and Mineral Research. 2008;23(2):247–252. doi: 10.1359/JBMR.071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford DC, Brown-Gentry K, Rieder MJ. VKORC1 Common Variation and Bone Mineral Density in the Third National Health and Nutrition Examination Survey. Plos One. 2010;5(12) doi: 10.1371/journal.pone.0015088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang NLS, Di Liao C, Ching JKL, Suen EWC, Chan IHS, Orwoll E, Ho SC, Chan FWK, Kwok AWL, Kwok T, Woo J, Leung PC. Sex-specific effect of Pirin gene on bone mineral density in a cohort of 4000 Chinese. Bone. 2010;46(2):543–550. doi: 10.1016/j.bone.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Corsi KA, Pollett JB, Phillippi JA, Usas A, Li G, Huard J. Osteogenic Potential of Postnatal Skeletal Muscle-Derived Stem Cells Is Influenced by Donor Sex. Journal of Bone and Mineral Research. 2007;22(10):1592–1602. doi: 10.1359/jbmr.070702. [DOI] [PubMed] [Google Scholar]
- 38.Tosi LL, Boyan BD, Boskey AL. Does sex matter in musculoskeletal health? - The influence of sex and gender on musculoskeletal health. Journal of Bone and Joint Surgery-American Volume. 2005;87A(7):1631–1647. doi: 10.2106/JBJS.E.00218. [DOI] [PubMed] [Google Scholar]
- 39.Ishida Y, Heersche JNM. Progesterone stimulates proliferation and differentiation of osteoprogenitor cells in bone cell populations derived from adult female but not from adult male rats. Bone. 1997;20(1):17–25. doi: 10.1016/s8756-3282(96)00315-8. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Stern PH. Sex-Specific Effects of Estrogen and Androgen on Gene Expression in Human Monocyte-Derived Osteoclasts. Journal of Cellular Biiochemistry. 2011;112:3714–3721. doi: 10.1002/jcb.23297. [DOI] [PubMed] [Google Scholar]
- 41.Leng RP, Wu H. UBE4B, a ubiquitin chain assembly factor, is required for MDM2-mediated p53 polyubiquitination and degradation. Cell Cycle. 2011;10(12):1912–1915. doi: 10.4161/cc.10.12.15882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs-Young R, Shirley SH, Rundhaug JE, Tian J, Cullinan-Ammann N, Lambertz I, Conti CJ. Transcriptional Regulation of Estrogen Receptor-alpha by p53 in Human Breast Cancer Cells. Cancer Research. 2009;69(8):3405–3414. doi: 10.1158/0008-5472.CAN-08-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapiro L, Pott GB, Ralston AH. Alpha-1-antitrypsin inhibits human immunodeficiency virus type 1. Faseb Journal. 2001;15(1):115–122. doi: 10.1096/fj.00-0311com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.