Abstract
When growth regulatory genes are damaged in a cell, it may become cancerous. Current technological advances in the last decade have allowed the characterization of the whole genome of these cells by directly or indirectly measuring DNA changes. Complementary analyses were developed to make sense of the massive amounts of data generated. A large majority of these analyses were developed to construct interaction networks between genes from, primarily, expression array data. We review the current technologies and analyses that have developed in the last decade. We further argue that as cancer genomics evolves from single gene validations to gene network inferences, new analyses must be developed for the different technological platforms.
Keywords: gene network, inference, microarrays, rna-seq, cancer
Introduction
Cancer is uncontrolled accelerated cellular growth and is responsible for approximately 13 percent of deaths worldwide [1]. Over the last half century, our understanding of cancer development has evolved from environmental to genetic causes [2,3]. While it is likely a combination of the two [4,5], much research in the last three decades have focused on genetic predispositions with the prevailing common disease/common variant hypothesis [6,7]. Namely, the disease (cancer) is driven by a common set of alleles, possibly spanning across multiple loci, in the population. While predisposition from certain alleles confer susceptibility to cancer [8], random somatic mutations throughout the lifetime of the organism without predisposition may also lead to similar results. In either case, genomic instability, a signature feature of cancer [6], leads to somatic mutations including single nucleotide polymorphisms (SNPs), insertions and deletions of large or small segments of DNA, chromosomal translocations, inversions, and other structural rearrangements due to broken and rejoined DNA, epigenetic modifications (usually chromatin modifications), and DNA acquisition from other sources such as infections from HPV [9], which all may lead to aberrant expression profiles or altered protein function due to amino acid substitution.
Most acquired mutations throughout the lifetime of an individual are likely benign; however, when a mutation alters a gene or the expression of a gene that confers growth, malignant neoplasms arise. The cells within the growing tumor are progeny of the original cell, whose driver or set of driver genes initiated expansive growth. A malignant tumor mirrors the selective process originally described by Darwin through a selective process, rewarding those that grow and expand unchecked. In the last decade, an attempt to understand how these mutations lead to cancer was initiated through the development of many “micro”-techniques. These include the use of the microscope, biochemical and cell biological techniques, as well as advanced genomic-based tools, with significant focus in the last decade on the latter [10].
Since genes regulate the growth of cells, a natural approach to understanding cancer is to identify “what genes have gone awry?” At the time this question was posed, the technologies to answer these questions were not well developed. Thus, genomic-based tools grew out of this need. Southern blotting, which identifies DNA sequences using oligonucleotide probe hybridization, along with the development of the microarray chip, a chip with thousands of embedded probes corresponding to a specific gene, led to a massive increase in the number of genes one could simultaneously investigate. In the last decade, two types of array technologies played a significant role in our understanding of how cancer cells differ from normal cells — one measuring gene expression and the other gene copy number. Expression analysis of cancer cells focus on gene regulation by measuring under/overexpressed mRNA in a sample tumor, while the second focuses on genomic structural changes via copied and deleted regions of DNA. Both technologies have their respective successes and difficulties. For example, array comparative genomic hybridization (aCGH) data, or copy number data, explicitly informs which regions of the genome have been altered. However, it is still uncertain which specific genes within the altered region are aberrantly expressed. Further, expression analysis can specify the genes that are under/overexpressed, but do not inform about the causal molecular mechanisms underlying the gene expression change.
In 2005, building upon the concepts of Sanger sequencing, rather than microarray hybridization, sequencing DNA of interest became the next-generation in genomic discovery technologies [11-15]. Next-generation sequencing (NGS) is the ability to sequence DNA samples using a reference library in a massive parallel capacity, revolutionizing sample resolution and the time necessary to sequence those samples [12]. While cost initially prohibited this technology for widespread use, now NGS is moderately more expensive than microarrays and in the next 5 months will likely be similarly priced [16]. NGS technologies are an improvement over microarrays for multiple reasons [17]. First, microarray technology requires a priori knowledge of the genes of interest, introducing probe bias, whereas NGS technologies do not. Second, microarray segments may cross-hybridize to incorrect probes introducing noise in the signal, while NGS technologies rely on sequencing, which counts single nucleotides. As a consequence of this single base-pair resolution, NGS technologies can identify point mutations in cancers [18,19]. Finally, nanograms of material is needed for NGS, while microarrays rely on orders of magnitude more, increasing the reliance on PCR, and thus, PCR biases have a larger impact on results. Most microarray technologies now have appropriate analogues to NGS, e.g., expression arrays to RNA-seq [20] and aCGH to CNV-seq [21] are among a few. It should be noted, however, that while NGS will likely replace array-based technologies, the amount of samples currently available is still insufficient for many types of investigations. Thus, until sufficient NGS samples are collected, microarrays will still be needed.
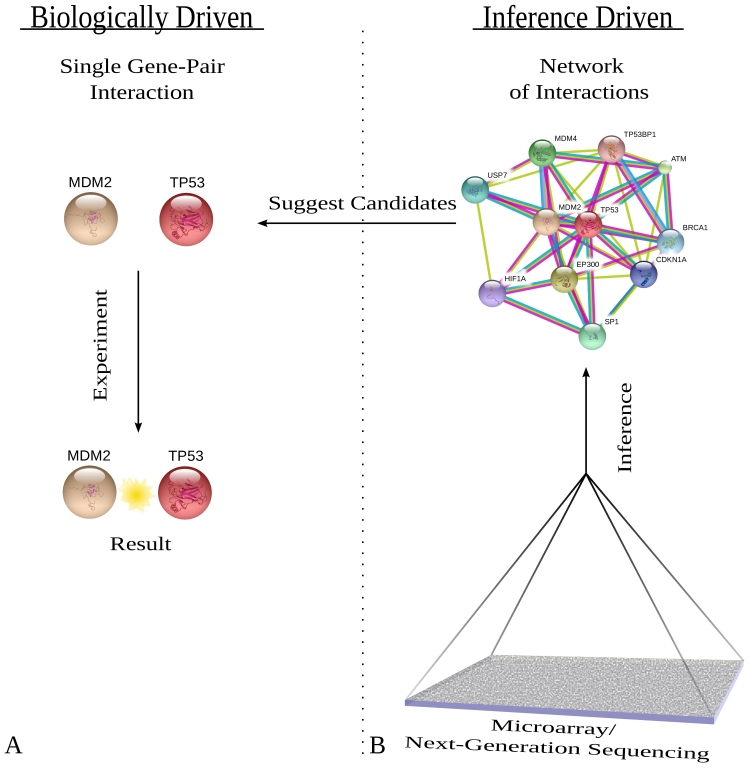
Before microarrays or NGS technologies, researchers focused on single gene hypotheses (Figure 1A). While this is a thorough systematic scientific approach to cancer biology, it is time consuming since few genes are investigated at a time, it is biased, and the cancer is often simulated with a biological model. Other steps have been made to improve this type of interrogation through the use of RNA interference that “knocks-down” mRNA transcripts [22]. While these scans allow for many genes to be interrogated at once, it is still necessary to define the set of genes to investigate and in what type of tissue and under what conditions.
Figure 1.
A shows the typical process of investigating hypothetical gene interactions. A hypothesis is made, experiments are performed, and then a result is obtained. B shows a typical process in which a microarray informs about hypothetical networks. After many whole genome-wide arrays have been created, the data is collected together in some algorithmic way (discussed in text) and (a) network(s) of interactions are inferred. Those networks can then be suggested for biological validation. Networks of gene relations were generated from a curated protein database with p53 as the center of the network [43].
From one perspective, it seems that these approaches could be supplemented by suggesting multiple gene candidates for validation by starting with genomic cancer data (Figure 1B), i.e., let the pattern of expression or copy number tell us what genes are interacting. Indeed, current technologies inform on the order of tens-of-thousands of genes, and thus, it is necessary to amplify the genes that convey abnormal expression or copy number resulting from causative mutations. However, even NGS technology has variability due to random fluctuations in the cell, and thus, large sample sizes must be used to investigate these genes. Since collective samples are used, probabilistic methods must be employed to identify genes of interest. However, simply identifying genes of interest does not convey a pathway or necessarily generate an informative cancer model. It is the major goal of cancer biology to map DNA alterations to the causative function of the genes altered. One way to understand causation is to look at networks of interacting genes, i.e., genes whose expression affects other genes. How to discover collectively active genes in an unbiased way, however, is not obvious. Thus, we must also, in a systematic, well-defined, mathematical way, amplify networks of interest using appropriate models. Luckily, probabilistic network models are optimized for specifically these tasks. The derived networks from modeling and genomic data may then improve understanding of gene interactions in cancer progression and help link causative mutations to disease.
Topics
The Statistical Tools: Are All Network Identification Tools Created Equal?
Armed with a massive amount of probes on a single array or a complete genomic library from NGS technologies, the whole genome can now be investigated in one experiment. While creating a revolution in cancer, genomic technologies still suffer from difficulties in data analysis [23-25]. Core issues include noise and testing too many hypotheses. Since most genes are not aberrantly expressed in a cell, gene expression fluctuates about its healthy homeostatic mean. Thus, each gene has a variable range of expression values that may be any random value. If we are not careful, we can mistakenly associate a large deviation from expression as significant, even though the expression was just a fluctuation in the cancer sample. Biologists attempt to mitigate this difficulty by increasing the number of technical replicates, limiting technical errors, as well as increasing biological replicates reducing the impact of “passenger” genes — genes that are altered, but non-drivers in the cancer.
Despite these efforts, it is still difficult to distinguish between a significant change and a normal statistical fluctuation. For example, suppose a gene is suspected to be upregulated. The expression mean is found from biological replicates of our cancer and healthy replicates. These means can be compared using a t-test, which makes the assumption that the t-statistic follows the t-distribution. If our measurement is significantly differentially expressed, then the t-statistic will be in the far tails of the t-distribution, returning a small p-value — a measure of how extreme an observation is [26]. This problem is further compounded when we test tens-of-thousands of genes where there is a greater chance of seeing a large statistical fluctuation. We need to be even stricter in what we call a significant expression change as opposed to a normal statistical fluctuation. Typically, the p-value is corrected using, for example, a Bonferonni correction. This then boils down to filtering out what gene is important, what gene is not, and what genes your analysis suggests are important but really are not. The last of these three are called “false positives.” These investigations can be further improved upon through the use of the false discovery rate that determines how likely the “positive” finding is a true positive (a real result) [27,28].
Often, identification of gene candidates in cancer samples is insufficient to build a cancer model, thus sometimes we must attempt to characterize the samples in some general way based on the genomic alterations measured. One way to accomplish this task is through cluster analyses, such as hierarchical clustering, employed by grouping genes with similar expression [29], which often leads to discovery of tumor subtypes. These types of investigations define “distances” representing similarity between gene expression profiles and grouping those similar ones together. The question here is, what patterns of gene expression emerge and are they consistent across samples? These types of clustering analyses, referred to as unsupervised learning, often do not inform why genes cluster; however, it can offer inferences into why. Other times, making predictions with expression data is used; supervised learning, for example, utilizes large sets of data to “train” a model, such as Random Forest models [30], artificial neural networks [31], and support vector machines [32], and then make predictions.
While these analyses help identify interesting genes, aggregate information, and make predictions, they do not construct networks of interacting genes. In this context, “interacting” encompasses chemical, syntenic, and indirect regulation of one gene on another via proteins and other factors such as non-coding RNAs. Underlying these analyses sits the hope that one could start from genome-wide experiments and let the aggregated results inform which genes are interacting in a network, not just correlated in a network. The important distinction here is interacting versus correlations. Correlations describe only a statistical relationship between genes, returning a statistic that only informs from sampling the model distribution, whereas interactions return the model distribution itself (Note: we explicitly define interactions in the next section). Probabilistic models have thus been designed to build gene networks using, e.g., Bayesian Networks [33], information-theoretic models [34], deterministic models [35], and sparse-network methods [36,37]. All of these models are, in some way, attempting to construct or help construct a graph, i.e., an abstract construct that connects vertices (often genes) with edges (an interaction between those genes). Each method has its own set of advantages and disadvantages in terms of accuracy and computational time, and each method invokes its own set of assumptions about the nature of the interactions [38]. For example, deterministic models commonly referred to as ODEs (ordinary differential equations) explicitly model the interactions in terms of relatively simple equations, without noise, but must be fit to a large number of biological parameters that are often unknown. Information-theoretic models, while successful in identifying transcription factors in cancer [39], cannot handle loops in networks and suffer from noise from indirect interactions, which effectively removes information from the system. Bayesian networks while successful are typically computationally expensive.
A Physics Approach … Going Backward
Physicists traditionally make sense of natural phenomena by concocting a mathematical model while ensuring experimental agreement. For example, magnetic forces may be represented by a mathematical model constructed between particles and can generally be written in terms of energies or interactions between those particles. When dealing with statistical quantities, those interactions are put into the framework of the probabilistic model, which defines the probability of being in a particular state. For example, the probability of being in the state with particle-1 up and particle-2 down, is a function of the interactions between the two particles. Recently, physicists have taken an interest in the inverse-problem, i.e., using experimental data to reverse-engineer the interactions typically a priori defined. Thus, back to the geneticist, rather than defining interactions and examining the resultant statistical dynamics of altered gene states, we use the experimental probabilities of being in a particular expression or copy number state, for example, gene-1 deleted and gene-2 amplified, to determine the interactions between the genes.
As shown by Lezon et al. [40], one can take this approach and determine the explicit relationship between gene interactions and statistically measured quantities, such as a Pearson correlation. Fortuitously, the probabilistic model parallels the common Spin-Glass or Ising system physicists have investigated for decades. With the analogy that genes are interacting particles, Lezon et al. successfully showed that the gene interactions are not equal to the expression covariance matrix as typically calculated from expression technologies, but equal to the inverse of the covariance matrix. At first glance, this may be surprising, but these results exemplify the fact that algorithms that supplement statistical correlations for gene interactions are incomplete, otherwise, the gene interactions would be equal to the covariance matrix.
Fortunately, this approach does not suffer from indirect correlations as some of the other approaches do, delivers the explicit forces one would typically a priori define between genes from measured data, may infer large interactions between genes even when those genes have low correlations [41], and it can be vastly improved upon through the use of dimension reduction algorithms such as the James-Stein shrinkage estimator and graphical lasso [36,37]. One difficulty, however, from this approach is that directed graphs are not generated (who causes who is unknown), as modified Bayesian networks can, such as BANJO [33]. Ultimately, however, any underlying network discovered with strong interactions can only be validated through further experimentation (Figure 1), use of protein-protein interaction databases, pathway interrogations, and utilizing other genomic based technologies.
Conclusions
Building gene networks from existing data is a bottom-up approach attempting to fill in the gaps and understand gene relations. While many network algorithms calculate statistical correlations between genes, they often do not describe direct causal gene interactions, which are the explicit biological model we hope to capture. Improving these computational methods is likely to be the future of reverse-engineering gene networks, and here we have highlighted some promising approaches that have borrowed concepts from statistical physics. In addition, since fundamentally different biochemical genomic technologies represent different observables, for example, ChIP-seq [42], combining them with other measurables will likely lead to dimension reduction, improving both gene candidate false positives as well as reduce noise in gene interaction networks. Finally, other non-genomic-based technologies, such as protein-protein interaction [43] and pathway [44] databases, have been curated, which may be used to supplement these investigations as well as validate discovered networks. It is the hope of cancer biology that this data may be integrated into a complete model defining the cancer genome.
Abbreviations
- SNPs
single nucleotide polymorphisms
- aCGH
array comparative genomic hybridization
- NGS
next-generation sequencing
- ODEs
ordinary differential equations
References
- American Cancer Society. Global Cancer Facts & Figures 2nd Edition. Atlanta: American Cancer Society; 2011. [Google Scholar]
- Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66(6):1191–1308. [PubMed] [Google Scholar]
- Peto J. Cancer epidemiology in the last century and the next decade. Nature. 2011;411(6835):390–395. doi: 10.1038/35077256. [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33(Suppl):238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- Johnson GC, Todd JA. Strategies in complex disease mapping. Curr Opin Genet Dev. 2000;10(3):330–334. doi: 10.1016/s0959-437x(00)00075-7. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH. et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Talbot SJ, Crawford DH. Viruses and tumours―an update. Eur J Cancer. 2004;40(13):1998–2005. doi: 10.1016/j.ejca.2003.11.039. [DOI] [PubMed] [Google Scholar]
- Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26(4):5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- von Bubnoff A. Next-generation sequencing: the race is on. Cell. 2008;132(5):721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Guo J, Yu L, Turro NJ, Ju J. An integrated system for DNA sequencing by synthesis using novel nucleotide analogues. Acc Chem Res. 2010;43(4):551–563. doi: 10.1021/ar900255c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies ― the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program. Genome.com [Internet] Available from: www.genome.gov/sequencingcosts .
- Hurd PJ, Nelson CJ. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct Genomic Proteomic. 2009;8(3):174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1055. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9(7):554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- Upton GJ, Sanchez-Graillet O, Rowsell J, Arteaga-Salas JM, Graham NS, Stalteri MA. et al. On the causes of outliers in Affymetrix GeneChip data. Brief Funct Genomic Proteomic. 2009;8(3):199–212. doi: 10.1093/bfgp/elp027. [DOI] [PubMed] [Google Scholar]
- Shaffer JP. Multiple hypothesis testing. Annu Rev Psychol. 1995;46(1):561–584. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64(3):479–498. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- Marchevsky AM, Patel S, Wiley KJ, Stephenson MA, Gondo M, Brown RW. et al. Artificial neural networks and logistic regression as tools for prediction of survival in patients with Stages I and II non-small cell lung cancer. Mod Pathol. 1998;11(7):618. [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20(18):3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9(1):67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- Cherepinsky V, Feng J, Rejali M, Mishra B. Shrinkage-based similarity metric for cluster analysis of microarray data. Proc Natl Acad Sci USA. 2003;100(17):9668–9673. doi: 10.1073/pnas.1633770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadeiras M, von Bayern M, Sinha A, Shahzad K, Latif F, Lim WK. et al. Drawing networks of rejection―a systems biological approach to the identification of candidate genes in heart transplantation. J Cell Mol Med. 2011;15(4):949–956. doi: 10.1111/j.1582-4934.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezon TR, Banavar JR, Cieplak M, Maritan A, Fedoroff NV. Using the principle of entropy maximization to infer genetic interaction networks from gene expression patterns. Proc Natl Acad Sci USA. 2006;103(50):19033–19038. doi: 10.1073/pnas.0609152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman E, Berry MJ 2nd, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440(7087):1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J. et al. STRING 8―a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Suppl 1):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]