Abstract
There is a need for a noninvasive technique to monitor living pluripotent stem cell condition without any labeling. We present an optical imaging technique that is able to capture information about optical path difference through the cell and cell adhesion properties simultaneously using a combination of quantitative phase microscopy (QPM) and interference reflection microscopy (IRM) techniques. As a novel application of QPM and IRM, this multimodal imaging technique demonstrated its ability to distinguish the undifferentiated status of human induced pluripotent stem (hiPS) cells quantitatively based on the variation of optical path difference between the nucleus and cytoplasm as well as hiPS cell-specific cell adhesion properties.
OCIS codes: (180.3170) Interference microscopy, (110.4190) Multiple imaging
1. Introduction
In current stem cell biology, the greatest challenge is to maintain the undifferentiated status of stem cells. This can be addressed by careful monitoring and characterization of cells. The process of stem cell in undifferentiated status is at present monitored by biological assays, namely, immunocytochemistry. However, this process is time consuming and requires biomarkers or labels. There is a clear need for a truly noninvasive technique which can monitor the degree of undifferentiated condition rapidly. Such a technique will most likely involve a form of optical imaging or spectroscopy but must not involve the addition of any kind of biomarker. Biomarkers are used to sort embryonic stem cells, in conjunction with fluorescent or magnetic labels.
There are issues with the use of fluorescent and magnetic markers. Fluorescent biomarkers have been employed in cell sorting and characterization, but fluorescent techniques have a number of drawbacks, that is, photobleaching prohibits long-term studies, production of free radical singlet oxygen species will damage live cells, finally, the use of biomarkers causes modification to cells’ surface chemistry. Magnetic beads cannot easily be visualized in microscopy; they must all be removed from the cells, because a large mass could cause large mechanical stresses to the cells, which can affect the cells’ behavior. There is thus a requirement from the stem cell community for a rapid, easy, sensitive, nondestructive, noninvasive, label-free technique which can be applied on the single cell level as well as monitor or sort large populations of cells. This review will concentrate on label-free optical measurement techniques, which are noninvasive and have sufficiently high resolution that can be applied at the single cell level.
The first optical technique suitable for noninvasive characterization of cells is quantitative phase imaging. Compared to other traditional optical techniques such as phase contrast microscopy or differential interference contrast microscopy, quantitative phase microscopy (QPM) has been developed to visualize and quantitatively analyze the distribution of phase shift of transmitted light through a specimen with nanometer resolution [1–3]. Since the amount of phase shift indicates the optical path difference (which contains the information of both the thickness and refractive index of the specimen), the QPM technique has been used to discern diverse cellular information under biophysical conditions such as the structural fluctuation of erythrocyte [4,5], cell growth depending on the cell cycle [6] and the measurement of refractive indices of intracellular materials [7,8]. In recent years, numerous novel techniques using QPM have been developed to enable a stable and quantitative measurement for long-term cellular dynamics using low-coherent illumination [7,8] and diffraction phase microscopy [9].
The second optical technique suitable for the characterization of cells is interference reflection imaging which enables the achievement of cell adhesion status without any contrast agents. Interference reflection microscopy (IRM) [10] and reflection interference contrast microscopy (RICM) [11] have been used widely as appropriate tools to study the distribution and dynamics of focal adhesion protein [12,13], cell spreading and migration [14,15], secretion [16], cell mitosis [17], and cytotoxicity [18]. These methods give the image contrast depending on cell-to-substrate distance by the interference generated from a slight variation of optical path difference between the reflection beam from the cellular membrane and from the interface of substrate and culture medium. The reflection contrast provides a semi-quantitative analysis about 3-D information of the adherent surface of living cells [19,20].
We devised the microscope which can perform QPM imaging and IRM imaging simultaneously with nanometer phase resolution. The multimodal QPM-IRM imaging system can be a new tool for label-free acquisition of whole cell topographic information about cell adhesion as well as cell optical thickness, which is the multiplication of cell thickness and cell refractive index. This makes it possible to gain complex characteristics for judging the state of living cells noninvasively, especially suitable for monitoring the undifferentiated condition of pluripotent stem cells.
In general, a human pluripotent stem cell forms a colony which is packed with many cells. Once the cells are dissociated into a single cell, most of the cells never survive without some additives such as ROCK inhibitor. Some of the small cluster of cells survives; however, the condition of colony is not good. The difficulty of stem cell culture is maintaining an undifferentiated state, which represents tightly packed colonies. If researchers can obtain the information as to which cells are beginning differentiation very early, they can adjust the culture conditions to recover the cells.
In this report, the authors showed data relating to the conditions of pluripotency by using the QPM and IRM technique. Good pluripotent stem cells showed relatively long optical path difference in cytoplasm and small contact area to culture dish. Pluripotent stem cells in good status showed small volume as well. Because pluripotent stem cells grow artificially, small cells have a relatively big surface area to obtain nutrients from the culture media. On the other hand, to maximize surface area, the contact point of cells to dish should be a small point which has a weak force. As a result, cells are making colonies to obtain enough force to attach together.
2. Materials and methods
2.1. Cell culture
HeLa cells (from American Type Culture Collection, US) and CHO cells (from European Collection of Cell Cultures, UK) were cultured, respectively, in Dulbecco’s modified Eagle’s medium (GIBCO) and Ham’s F12 medium (GIBCO) with 10% fetal bovine serum and 100 units/ml penicillin plus 100 μg/ml streptomycin at 37°C with 5% CO2. The osteoclast-like (OC) cells were prepared according to the original method developed by Akatsu et al. [21]. Briefly, to form OC cells, mouse primary osteoblastic cells isolated from the calvariae of 1-day-old newborn mice and bone marrow cells collected from the tibiae of 6- to 9-week-old male ddY mice were co-cultured in modified alpha-MEM (GIBCO) with 10% fetal bovine serum for a week. The cells were re-suspended and cultured on 35mm culture dishes for 4 days for microscopic studies. To identify the OC cells, cells were fixed and stained with TRAP staining kit (AK04F, Primary Cell, Japan) after the microscopic studies. Human induced pluripotent stem (hiPS) cells (established by ReproCELL, Japan) were cultured in ReproFF2 medium (ReproCELL) at 37°C with 5% CO2. For judgments of iPS cell conditions, after obtaining the phase contrast image with a microscope (TS100, Nikon, Japan), the cell condition was judged based on researcher’s experiences.
2.2. Optical setup
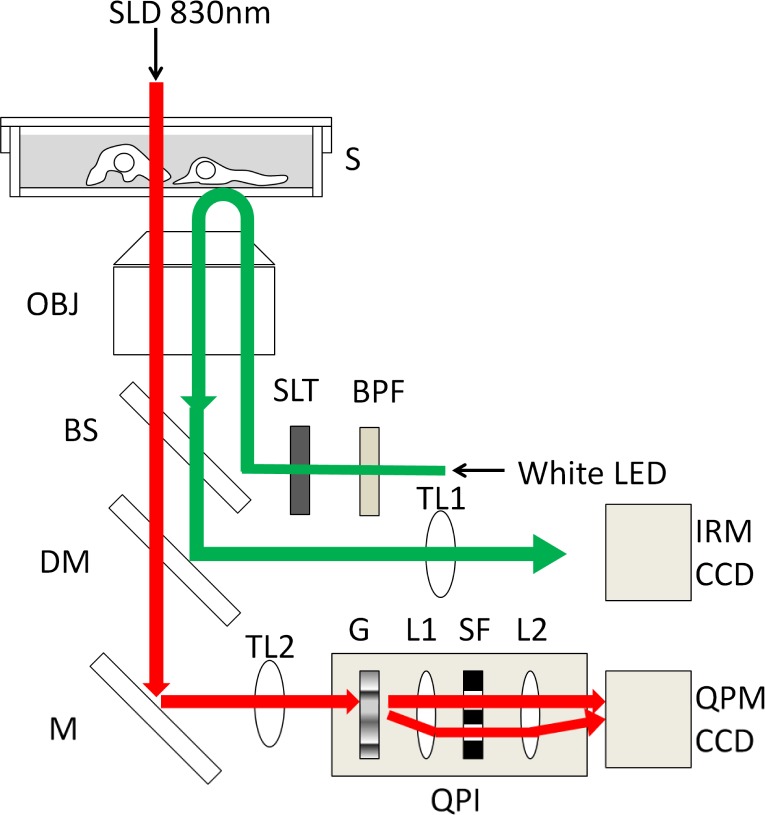
The experimental inverted microscope setup (shown in Fig. 1 ) enabled researchers to acquire QPM images and IRM images of living cells simultaneously. The cells cultured on the 35mm dish were placed on the microscope stage, controlled at 37°C in a humidified atmosphere with 5% CO2. For QPM imaging, the cells were illuminated with a super luminescent diode of 830nm (ASLD83-050-B-FA, Amonics, Hong Kong). The transmitted light through cells was focused by the objective lens (UAPO20XW340/0.7, Olympus, Japan) and led to the quantitative phase imaging unit based on the diffraction phase microscopy described in reference [9]. The diffraction phase microscopy has been developed to realize a single-shot acquisition of interferogram with stable common-path geometry. The phase grating (G), which diffracts the light into many diffraction orders, was placed at the imaging plane after the tube-lens (TL2). The diffracted light passed through a 4-f imaging system consists of a couple of relay lenses (L1 and L2). Only the first and zeroth orders of diffracted lights are allowed to pass the spatial filter placed at the Fourier plane between L1 and L2 and the zeroth order light was also spatially filtered by a pinhole as shown in Fig. 2(a) . Then, the zeroth and first orders of the light were imaged onto the CCD camera (C8484-05G01, 1344x1024 pixels, 12bit, Hamamatsu Photonics, Japan). A spatially modulated full-field interferogram from these two images was generated on the image plane of a CCD camera at an exposure time of 100ms. The interferogram was led to a quantitative phase image using fringe pattern analysis. The background phase image in no specimen was removed. The amount of phase difference of each pixel indicated the optical path difference (OPD) through the cell. OPD(x,y) of each pixel equals λ × Δφ(x,y)/2π, where λ is the wavelength of illumination light and Δφ(x,y) is a measured phase difference between the cell and the outer medium. OPD(x,y) is also equal to (n1 – n2) × d(x,y), where n1 and n2 are the refractive indices of the cell and outer medium, respectively, and d(x,y) is cell thickness. Thus, the OPD is the product of cell thickness and the difference of refractive indices between the cell and culture medium. We tested the quantitative characteristics of our QPM setup by using a specimen whose shape is known. We prepared the glass slides in which five kinds of different depth of grooves from 450 to 10,000 Å were fabricated and filled with an adhesive polymer. The phase difference of each groove showed a good linearity between 10 to 80 nm in OPD (shown in Fig. 2(b)). As a contrast with the living cells, polystyrene beads (10 μm in diameter, product name; Polybead microspheres, manufacturer; Polysciences, US) were also observed with the QPM. The outer medium was replaced by immersion oil instead of culture medium to reduce the difference of refractive indices between the object and the outer medium.
Fig. 1.
Schematic diagram of QPM-IRM imaging setup. Illumination light of SLD 830nm (red line) for QPM imaging was transmitted through a specimen (S). Transmitted light image was formed by an objective lens (OBJ) and a tube lens (TL2). The image was led to the quantitative phase imaging (QPI) unit. Optical setup of the QPI unit was described in Fig. 2(a) in detail. The QPI unit consisted of a grating apparatus (G), a spatial filter (SF) to be used as a sample and reference image and a couple of relay lenses (L1 and L2). Interference image was formed onto CCD camera (QPM CCD). White LED illumination light (green line) for IRM imaging was selected with a wide-bandwidth band-pass filter (BPF) and irradiated to the bottom side of specimen through an aperture slit (SLT). Reflection interference image form the specimen was formed onto another CCD camera (IRM CCD) by a tube lens (TL1) after being reflected by a long-pass dichroic mirror (DM) through a beam splitter (BS).
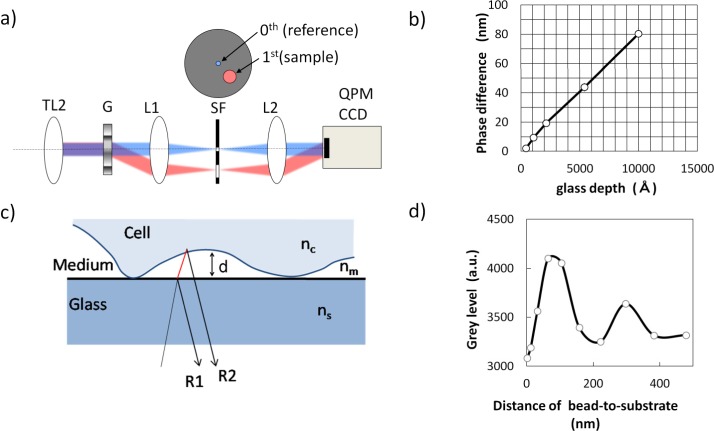
Fig. 2.
The principle of QPM-IRM imaging and quantitative measurement results by using grooved glass slides and polystyrene beads whose physical shapes are known. a) Optical diagram of the quantitative phase imaging unit. TL2: Tube lens, G: Phase grating, SF: Spatial filter, L1 and L2: A couple of relay lenses. b) Linearity of QPM measurement by using grooved glass slides; the measured phase difference is plotted as a function of known physical thickness. c) Schematic diagram of IRM. R1: Reflection light from glass-to-medium interface, R2: Reflection light from medium-to-cell interface. Symbols nc, nm and ns indicate the refractive indices of cell, medium and substrate, respectively. d) IRM intensity plotted as a function of bead-to-substrate distance measured by using a polystyrene bead (10 μm in diameter) attached onto a glass-bottom culture dish.
For IRM imaging, the cells were illuminated by a white light LED (LEDC17, Thorlabs, US) with wide band-pass filter (450 to 750nm, Semrock, US) through the objective lens. Interference of the reflection light from the adherent surface was generated depending on the cell-to-substrate distance. The optical basis of interference occurring around the top surface of the glass substrate is shown in Fig. 2(c). A part of the incident beam was reflected at the interface of glass substrate and the culture medium to give a reflection beam (R1). A part of transmitted beam was reflected at the boundary between the cell and the culture medium and generated another reflection beam (R2). Each reflection beam interfered and generated the image contrast depending on the distance between the glass surface and the cell surface. The intensity of the interfered light is described as a cosine function of the distance as written in reference [19]. When the cell-to-substrate distance is zero, the resultant interfered light intensity is the darkest because the phase of reflection beam R2 is shifted by π to R1. In accordance with the increase of the cell-to-substrate distance from zero, the intensity of the reflected interference light becomes brighter. The reflected intensity becomes the brightest when the cell-to-substrate distance is λ/(4 × nm) = 112nm, where λ is the wavelength of illumination light and nm is the refractive index of the outer medium. Because the coherence length of the illumination light was less than 1 μm, the interference of IRM was observed only within the region of cell-to-substrate interface. The use of low-coherent light for the illumination was advantageous for avoiding the interference between the top and the bottom surface of the glass substrate, whose thickness was about 100 μm. To reduce a background of directly reflected light from the bottom surface of the substrate or the objective lens, the aperture slit was placed at the illumination optics. The full field of the IRM image was separated from QPM image by a long-pass dichroic mirror and formed on another CCD camera (C8484-05G01) at an exposure time of 50ms. Any spatial unevenness of light intensity in the image field was corrected. The offset of background grey level in a no-cell region was normalized at 3,000 counts. IRM images of 10 μm polystyrene beads indicated the interference contrast based on the bead-to-substrate distance within a few hundreds of nanometers (shown in Fig. 2(d)). The reflection interference intensity was the darkest at the position where the bead contacts the substrate. According as the bead-to-substrate distance increases, the IRM intensity increased until it became the brightest at the position where the distance was about 100 nm. Strictly speaking, the pattern of the refection contrast (frequency and amplitude) in case of cell adhesion are not equal to the case of polystyrene bead because of the difference of the refractive index, the angle of reflection surface, and so on [19]. However, it is acceptable to conjecture that the close contact region of cell adhesion can be seen with a certain darker region as well. For this reason, we extracted a darker region below the specified grey level as a close contact region of cells in the following image analysis.
As well, bright field (BF) image was acquired for the morphological information of each cell by transmitted illumination using a 660nm LED light source (LEDC45, Thorlab). Image registration between QPM and IRM was aligned pixel by pixel within a single pixel error. We succeeded in observing HeLa cells on this microscope overnight continuously without any damages (data were not shown).
2.3. Image analysis
From the QPM images, the average OPD of the nucleus region and cytoplasm region were measured with image analysis software (AQUACOSMOS, Hamamatsu Photonics). Each region was determined by manually tracing the shape of BF images in the same field of view. The ratio of OPD (cytoplasm/nucleus) was calculated for each cell. In the IRM images, we found that the hiPS cells showed many small contact regions to the substrate. In order to evaluate this adhesion characteristics, the maximal size of the dark spots, where the cells contact the substrate, was calculated from the IRM images by image analysis software (HC-image Analysis, Hamamatsu Photonics), as follows. First, we manually traced the boundary of each cell in the BF image to determine the region of individual cells. The software automatically superimposed the region of each cell on the IRM image. In order to reduce the random optical noise in the image, a spatial averaging by 3 by 3 pixels window was processed on the IRM image. Then the area where the IRM image intensity was lower than a threshold was extracted as close-contact spots. The spots whose sizes were smaller than four pixels (0.42 μm2) were excluded because such small spots could be artifacts. After the image processing above, many close-contact spots in each cell were extracted and the size of the largest close-contact spot in each cell was used as the parameter of adhesion. The cell populations under testing were quantitatively assessed by means of this parameter.
2.4. Statistics
For comparison between hiPS cells and the other cells, the average OPD of the subcellular region and the maximal size of close contact spots were measured in 15 to 20 cells. In the study of “good” and “no good” status of hiPS colonies, subcellular OPD and cell adhesion were measured from 619 “good” cells and 433 “no good” cells in individual (5 to 6) colonies. Student’s t-test was employed as a statistical hypothesis test to compare “unpaired” two samples under null hypothesis. To define statistical significance, the p-value, which is the probability that random sampling would lead a difference between samples, was calculated. When the p-value was less than the significance level which was often 0.01, the null hypothesis was rejected and the result was expressed to be statistically significant.
3. Results and discussion
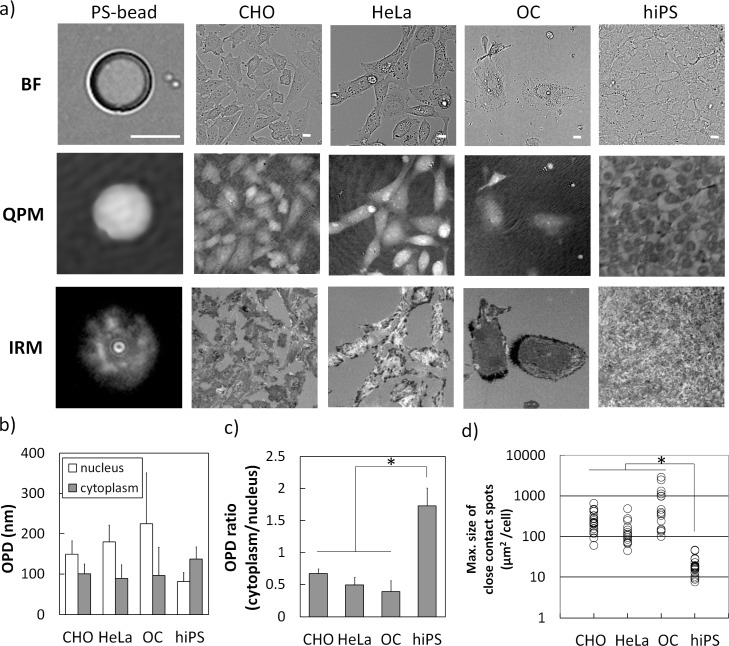
In this paper, we report the possibilities of noninvasive optical technique using QPM-IRM imaging to distinguish the condition of living cells. The representative images of living stem cells and the other differentiated cells as well as polystyrene beads are shown in Fig. 3(a) . The images of beads were helpful to infer the state of cells. Bright-field (BF) images showed the morphological information of the samples. QPM imaging of beads showed a sphere shape along the bead’s thickness. IRM image of beads showed the interference fringes based on the bead-to-substrate distance only in the close region within a few hundreds of nanometers.
Fig. 3.
QPM-IRM image characteristics of hiPS cells and other types of cells: a) Bright-field (BF) and QPM-IRM images of living cells and 10μm polystyrene bead (PS-bead) were shown. Full grey scale of QPM image indicated OPD of 1 μm. The gray scale of IRM indicated intensity level from 2850 to 3150. The following data were measured from 15 to 20 individual cells (b to d). Scale bar is 10μm. b) The average OPD of nucleus region (white bar) and cytoplasm region (grey bar). Each data was expressed as the mean ± S.D. c) The ratio of cytoplasm-to-nucleus OPD of each cell was expressed as the mean ± S.D. *, P < 0.001. d) The maximal size of close contact spots of each adherent cell was expressed in dot plotted chart (white circles). *, P < 0.001. P means the p-value of the statistical significance test. Refer to subsection 2.4.
We found a clear difference in the subcellular distribution of optical path difference (OPD) between hiPS cells and the other differentiated cells in QPM images. CHO cells, HeLa cells and OC cells showed a hill-like distribution of OPD. In the other words, the average OPD of cytoplasm region was lower than that of nucleus region. In contrast, hiPS cells signified the opposite results. Each hiPS cell in the colonies showed a concave distribution in the QPM image, i.e., OPD of cytoplasm region was greater than that of nucleus region (shown in Fig. 3(b)). The ratio of OPD (cytoplasm/nucleus) could be a simple index in order to distinguish the hiPS cells and the other types of differentiated cells, that indicated clear differences between hiPS cells (1.72 ± 0.27) and the other cells (0.38 ± 0.17 to 0.66 ± 0.07) (shown in Fig. 3(c)).
In the point of cell adhesion shown in IRM images, we also found that each hiPS cell showed a number of small adhesion spots and looked like tiptoe-standing on the substrate, although the other adherent cells had rather larger adhesion areas. For analyzing cell adhesion quantitatively, the maximal size of adhesion spots extracted from each cell was compared (shown in Fig. 3(d)). The maximal size of adhesion spots of the hiPS cells was 19.8 ± 10.2 μm2, on the other hand, those of other adherent cells were from 147.7 to 808.8 μm2 as mean value. These results were statistically convincing that the adhesion spot size of hiPS cells was obviously smaller than the other adherent cells.
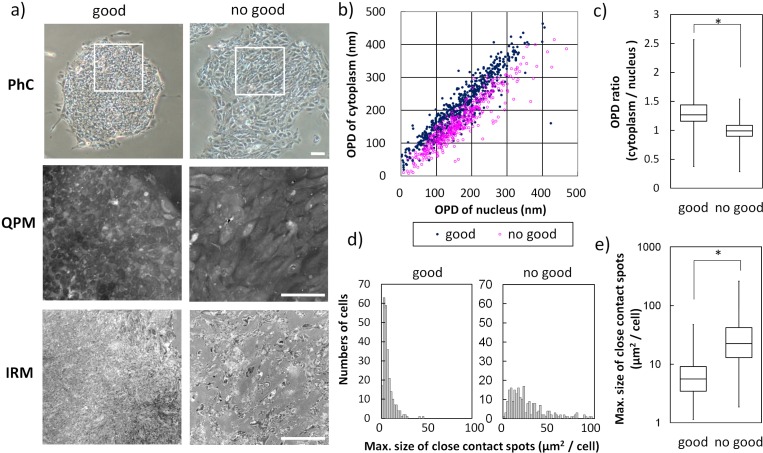
Finally we report the differences between two conditions of hiPS cells which were judged to “good” or “no good” colonies based on researcher’s experiences. In the QPM images of the hiPS cells, OPD distribution of each hiPS cell in “good” colonies was shown as a concave pattern, however, OPD of “no good” hiPS cells changed to a hill-like distribution (shown in Fig. 4(a) middle). Each OPD of cytoplasm of “no good” hiPS cells became lower than that of “good” hiPS cells and statistical analysis of OPD ratio (cytoplasm/nuclear) drew that “good” hiPS colonies were more than 1 (1.32 ± 0.27) and “no good” hiPS colonies were as much as 1 (1.00 ± 0.18) as mean value (shown in Figs. 4(b) and 4(c)). In the IRM images, “good” hiPS cells showed a small adhesion of tiptoe-standing. In contrast, “no good” hiPS cells showed a larger size of adhesion spots (shown in Fig. 4(a) lower). Quantitative analysis of the maximal size of close contact spots indicated that “good” hiPS cells had small and uniform adhesion spot size and “no good” hiPS cells did not keep such adherent characteristics (shown in Fig. 4(d)). The maximal size of the close contact area of “no good” hiPS cells was about 5 times as large as that of “good” hiPS cells (as mean value, 38.4 and 7.4 μm2, respectively) (shown in Fig. 4(e)). Thus, “no good” hiPS cells lost the characteristics expressed in the “good” state. This result suggests that the evaluation parameter indicating higher OPD ratio (cytoplasm/nuclear) and smaller adhesion area could become one of the indexes to judge the appropriate culture condition of hiPS cells.
Fig. 4.
The quantitative differences between hiPS cells judged to “good” and “no good” status: a) The representative images of “good” (left) and “no good” (right) colonies of hiPS cells. Phase contrast (PhC) microscopy images and QPM-IRM images of white-lined region of PhC. Full gray scale of QPM image indicated OPD of 1 μm. Gray scale of IRM image indicated intensity level from 2850 to 3150. Scale bar is 100 μm. The following data were measured from 619 “good” cells and 433 “no good” cells (b to d). b) The average OPD of nucleus region and of cytoplasm region were expressed as two axis scattergram (“good” in dark blue and “no good” in magenta). c) The ratio of OPD (cytoplasm/nucleus) of each cell was expressed as a box plot. *; P < 0.001. d) Maximal size of close contact adhesion spots of each cell in two groups were expressed as a histogram of number of cells. e) Statistical analysis of maximal size of close contact spots of two groups were shown in a box plot. *, P < 0.001. P means the p-value of statistical significance test. Refer to subsection 2.4.
4. Conclusion
As a newly devised microscopy technique, this combination of full-field quantitative phase microcopy and interference reflection microscopy has demonstrated applications in the evaluation of cellular structure, height of cell, and adherent surface. This optical technique presented topographic information about intact cells in state of cultured conditions, which were not obtained by conventional phase contrast microscopy or fluorescence microscopy or flowcytometer performed on fixed or dissociated stem cells. There were many differences between hiPS cells and other types of cells. Human iPS cells in an undifferentiated state showed longer optical path differences in the cytoplasm area rather than the nucleus area. In contrast, other types of cells were completely opposite, with a shorter optical path difference in the cytoplasm area rather than the nucleus area. Once iPS cells were judged “no good”, the optical path difference of the peripheral was decreased and cells started spreading. These results suggest the possibility that undifferentiated hiPS cells in colonies are physically thicker in cytoplasm or contain higher refractive index materials packed in cytoplasm than in nucleus area. Our future work includes independent measurement of the physical thickness and the optical refractive index of the same sample. The candidates of label-free optical technique to determine the refractive index of hiPS cells would be the low-coherence quantitative phase microscope [8], the tomographic phase microscope [22] and the full-field optical coherence microscopy [23]. On another side, adhesion of “good” hiPS cells was pin-point and like a tip-toe standing. This could be supporting the fact that hiPS cells cannot grow after dissociation because of weak contact force, in other words, they could need to form a colony to enhance the adherent force to the dish surface (like one pencil cannot stand alone but bundled pencils can stay one position stably). From a growing vigor point, it is convincing that stem cells require small volume and rather big surface area from top to tiptoe for active mass transport via plasma membrane. The cell's ability to either get substances from the outside or eliminate waste is related to the surface area. How much food and other materials from the outside and how much waste the cell has to get rid of are both related to the volume.
These observations showed the possibility that this optical method might be used for characterizing other cell types, such as embryonic stem cells used for drug screening or stem cell therapy. There is a good agreement with the higher cytoplasm-to-nucleus ratio of optical path difference, smaller cell adhesion spot size and also small cell volume in embryonic stem cells. Simplifying image analysis and introducing a pattern recognition algorithm to distinguish cells would lead to on-line monitoring of cell culture condition or cell separation. To classify stem cell states more accurately, several kinds of undifferentiation markers should be used in future studies. Since this optical technique based on phase measurement has its own benefits and drawbacks, it is more suited to characterization of stem cells in different ways and on different “platforms.”
References and links
- 1.Popescu G., Deflores L. P., Vaughan J. C., Badizadegan K., Iwai H., Dasari R. R., Feld M. S., “Fourier phase microscopy for investigation of biological structures and dynamics,” Opt. Lett. 29(21), 2503–2505 (2004). 10.1364/OL.29.002503 [DOI] [PubMed] [Google Scholar]
- 2.Ikeda T., Popescu G., Dasari R. R., Feld M. S., “Hilbert phase microscopy for investigating fast dynamics in transparent systems,” Opt. Lett. 30(10), 1165–1167 (2005). 10.1364/OL.30.001165 [DOI] [PubMed] [Google Scholar]
- 3.Li X., Yamauchi T., Iwai H., Yamashita Y., Zhang H., Hiruma T., “Full-field quantitative phase imaging by white-light interferometry with active phase stabilization and its application to biological samples,” Opt. Lett. 31(12), 1830–1832 (2006). 10.1364/OL.31.001830 [DOI] [PubMed] [Google Scholar]
- 4.Popescu G., Ikeda T., Best C. A., Badizadegan K., Dasari R. R., Feld M. S., “Erythrocyte structure and dynamics quantified by Hilbert phase microscopy,” J. Biomed. Opt. 10(6), 060503 (2005). 10.1117/1.2149847 [DOI] [PubMed] [Google Scholar]
- 5.Park Y. K., Diez-Silva M., Popescu G., Lykotrafitis G., Choi W., Feld M. S., Suresh S., “Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum,” Proc. Natl. Acad. Sci. U.S.A. 105(37), 13730–13735 (2008). 10.1073/pnas.0806100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mir M., Wang Z., Shen Z., Bednarz M., Bashir R., Golding I., Prasanth S. G., Popescu G., “Optical measurement of cycle-dependent cell growth,” Proc. Natl. Acad. Sci. U.S.A. 108(32), 13124–13129 (2011). 10.1073/pnas.1100506108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Millet L. J., Mir M., Ding H., Unarunotai S., Rogers J. A., Gillette M. U., Popescu G., “Spatial light interference microscopy (SLIM),” Opt. Express 19(2), 1016–1026 (2011). 10.1364/OE.19.001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi T., Iwai H., Miwa M., Yamashita Y., “Low-coherent quantitative phase microscope for nanometer-scale measurement of living cells morphology,” Opt. Express 16(16), 12227–12238 (2008). 10.1364/OE.16.012227 [DOI] [PubMed] [Google Scholar]
- 9.Popescu G., Ikeda T., Dasari R. R., Feld M. S., “Diffraction phase microscopy for quantifying cell structure and dynamics,” Opt. Lett. 31(6), 775–777 (2006). 10.1364/OL.31.000775 [DOI] [PubMed] [Google Scholar]
- 10.Verschueren H., “Interference reflection microscopy in cell biology: methodology and applications,” J. Cell Sci. 75, 279–301 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Bereiter-Hahn J., Fox C. H., Thorell B., “Quantitative reflection contrast microscopy of living cells,” J. Cell Biol. 82(3), 767–779 (1979). 10.1083/jcb.82.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders R. M., Holt M. R., Jennings L., Sutton D. H., Barsukov I. L., Bobkov A., Liddington R. C., Adamson E. A., Dunn G. A., Critchley D. R., “Role of vinculin in regulating focal adhesion turnover,” Eur. J. Cell Biol. 85(6), 487–500 (2006). 10.1016/j.ejcb.2006.01.014 [DOI] [PubMed] [Google Scholar]
- 13.Holt M. R., Calle Y., Sutton D. H., Critchley D. R., Jones G. E., Dunn G. A., “Quantifying cell-matrix adhesion dynamics in living cells using interference reflection microscopy,” J. Microsc. 232(1), 73–81 (2008). 10.1111/j.1365-2818.2008.02069.x [DOI] [PubMed] [Google Scholar]
- 14.Sengupta K., Aranda-Espinoza H., Smith L., Janmey P., Hammer D., “Spreading of neutrophils: from activation to migration,” Biophys. J. 91(12), 4638–4648 (2006). 10.1529/biophysj.105.080382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassiman J. J., van der Schueren B., van Leuven F., van den Berghe H., “Qualitative and quantitative differences in spreading of human fibroblasts on various protein coats. Modulation by treatment of the cells with amines,” J. Cell Sci. 54, 79–95 (1982). [DOI] [PubMed] [Google Scholar]
- 16.Llobet A., Wu M., Lagnado L., “The mouth of a dense-core vesicle opens and closes in a concerted action regulated by calcium and amphiphysin,” J. Cell Biol. 182(5), 1017–1028 (2008). 10.1083/jcb.200807034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh G., Yumura S., “A novel mitosis-specific dynamic actin structure in Dictyostelium cells,” J. Cell Sci. 120(24), 4302–4309 (2007). 10.1242/jcs.015875 [DOI] [PubMed] [Google Scholar]
- 18.Choi C. K., Margraves C. H., English A. E., Kihm K. D., “Multicontrast microscopy technique to dynamically fingerprint live-cell focal contacts during exposure and replacement of a cytotoxic medium,” J. Biomed. Opt. 13(5), 054069 (2008). 10.1117/1.2993143 [DOI] [PubMed] [Google Scholar]
- 19.Limozin L., Sengupta K., “Quantitative reflection interference contrast microscopy (RICM) in soft matter and cell adhesion,” ChemPhysChem 10(16), 2752–2768 (2009). 10.1002/cphc.200900601 [DOI] [PubMed] [Google Scholar]
- 20.Weber I., “Reflection interference contrast microscopy,” Methods Enzymol. 361, 34–47 (2003). 10.1016/S0076-6879(03)61004-9 [DOI] [PubMed] [Google Scholar]
- 21.Akatsu T., Tamura T., Takahashi N., Udagawa N., Tanaka S., Sasaki T., Yamaguchi A., Nagata N., Suda T., “Preparation and characterization of a mouse osteoclast-like multinucleated cell population,” J. Bone Miner. Res. 7(11), 1297–1306 (1992). 10.1002/jbmr.5650071109 [DOI] [PubMed] [Google Scholar]
- 22.Choi W., Fang-Yen C., Badizadegan K., Oh S., Lue N., Dasari R. R., Feld M. S., “Tomographic phase microscopy,” Nat. Methods 4(9), 717–719 (2007). 10.1038/nmeth1078 [DOI] [PubMed] [Google Scholar]
- 23.Choi W. J., Jeon I., Ahn S. G., Yoon J. H., Kim S., Lee B. H., “Full-field optical coherence microscopy for identifying live cancer cells by quantitative measurement of refractive index distribution,” Opt. Express 18(22), 23285–23295 (2010). 10.1364/OE.18.023285 [DOI] [PubMed] [Google Scholar]