Abstract
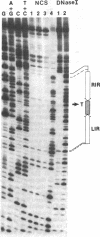
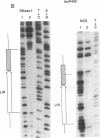
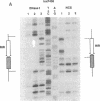
The interaction between the P1 recombinase protein Cre and the DNA site at which it acts, loxP, has been studied by using nuclease protection techniques. The region of DNA protected by Cre against nuclease attack by DNase I or neocarzinostatin is a 34-base-pair (bp) region containing two 13-bp inverted repeats separated by an 8-bp spacer region. These protected sequences have previously been shown to be required for efficient Cre-mediated recombination at loxP. The results of the above protection experiments suggest (i) that no more than 34 bp may be required for loxP recombination and (ii) that the asymmetry of loxP recombination is due to the 8-bp spacer sequence. With neocarzinostatin, a specific nucleotide within the 8-bp spacer region is not protected. This nucleotide is located in a 2-bp sequence shown to be involved in a loxP crossover event, suggesting that this region remains exposed after Cre binding. Protection experiments have also been done with loxP sites that have either the left or right inverted repeat deleted. The nuclease protection pattern of these sites reveals that each loxP site consists of two binding domains for Cre, each being composed of one 13-bp inverted repeat and the contiguous 4 bp of the 8-bp spacer region.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984 Feb 10;259(3):1509–1514. [PubMed] [Google Scholar]
- Abremski K., Hoess R., Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983 Apr;32(4):1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R., Vapnek D. Integration of the plasmid prophages P1 and P7 into the chromosome of Escherichia coli. J Mol Biol. 1979 May 15;130(2):161–173. doi: 10.1016/0022-2836(79)90424-8. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Ziese M., Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Poon R., Beerman T. A., Goldberg I. H. Characterization of DNA strand breakage in vitro by the antitumor protein neocarzinostatin. Biochemistry. 1977 Feb 8;16(3):486–493. doi: 10.1021/bi00622a023. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A. Bacteriophage lambda int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981 Aug 25;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D., Hoess R. Bacteriophage P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol. 1981 Aug 25;150(4):487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]