Abstract
Raman spectroscopy is used to gather information on the mineral and organic components of bone tissue to analyze their composition. By measuring the Raman signal of bone through spatially offset Raman spectroscopy the health of the bone can be determined. We’ve customized a system with 8 collection channels that consist of individual fibers, which are coupled to separate spectrometers and cooled CCDs. This parallel detection system was used to scan gelatin phantoms with Teflon inclusions of two sizes. Raman signals were decoupled from the autofluorescence background using channel specific polynomial fitting. Images with high contrast to background ratios of Raman yield and accurate spatial resolution were recovered using a model-based diffuse tomography approach.
OCIS codes: (110.6960) Tomography, (170.5660) Raman spectroscopy
1. Introduction
Current medical standards for bone imaging use x-rays, in the form of either standard x-ray, computed tomography (CT), or dual energy x-ray absorptiometry (DEXA), to determine the health of bone by measuring the mineral component. However, research has shown that the organic component of the bone matrix, specifically the collagen complex, is rich with information pertaining to the bone health for both cases of disease and healing after fracture [1,2]. The ability to measure these organic components has the potential to significantly improve diagnostic bone imaging, and in this study the technology to tomographically image optical biochemical signatures of bone from thick tissues is examined using a maximally sensitive detector geometry.
It is possible to measure organic components using spectroscopic techniques that take advantage of Raman scattering, an inelastic scattering phenomena used for determining the vibrational and rotational modes of molecules and compounds [3]. This optical technique can be used for monitoring bone health as it has the ability to provide information on both the organic and mineral states of the material without the addition of chemical or fluorescent markers [4–8]. Bone’s composite structure lends itself to the generation of strong Raman bands, which can be associated with the quantities of phosphate, carbonate and collagen amide components, as well as less intense collagen bands associated with collaged amino acids, notably proline, hydroxyproline and phenylalanine. These individual components can be used to determine the composition of the carbonated apatite and octacalcium phosphate within the bone matrix, which are used as bone health markers [9]. Previous research using samples from both animal models and ex vivo human patients has shown that it is possible to determine disease states and bone maturation from Raman scattering data [10–12]. The ability to successfully translate in vivo Raman scattering data to accurate diagnosis of bone health could provide a powerful tool for the clinician.
Previous work on these measurement systems showed that transmission mode sampling was able to reconstruct a Raman signal approximately 100-fold greater than those made in reflection mode [13]. This difference was attributed to the ability of the transmission measurements to collect Raman signal that was generated deeper within the tissue and that the tissue itself attenuates the excitation signal as a function of depth, thereby reducing background, this method is referred to as spatially offset Raman spectroscopy (SORS) [8,14,15]. However, the collection of deeper signals also results in increased levels of auto-fluorescence and increased elastic scattering events, which lead to reduced spatial resolution upon reconstruction of the Raman signal [16].
This study focuses on a system that combines both acquisition modes with the potential to use MR images to generate spatial distributions. The unique feature of this system design is that each fiber bundle is connected to an individual spectrometer and temperature-controlled CCD, thereby allowing for higher light throughput and hence increased sensitivity per unit time. The design is presented and tested using Teflon phantoms to assess sensitivity and ability to tomographically recover different sized regions.
2. Materials and methods
2.1. Experimental system
An MRI-coupled multispectrometer optical imaging platform described elsewhere was modified to facilitate optical tomography using Raman signals [17]. One of the unique features of this system is that each of the 8 detection fibers is coupled to its own scientific grade spectrometer with a cooled CCD detector. Since the entire CCD chip can be used to acquire Raman spectra, this arrangement provides superior light sensitivity and dynamic range compared to systems which employ multichannel detection on one CCD chip, a critical consideration when measuring the relatively low intensities of Raman peaks. Parallel detection also facilitates rapid tomographic acquisition times. For Raman imaging, the slit widths were set to 75 µm and the 1200 lp/mm gratings were used, centered at 900 nm. The light source was a 200 mW 830 nm Raman laser (Invictus, Kaiser Optical Systems, Ann Arbor, MI) that was multiplexed into each of the eight fibers sequentially, while the remaining seven fibers were used to measure the light remitted from the tissue. Thus, a total of 56 optical projections were measured and for each projection, the Raman spectrum and the excitation intensity were measured. Light was coupled from the laser to the fiber bundles through an automated rotary stage programmed through LabView software with 50 mW of light delivered to the surface. Each fiber bundle contains a central illumination fiber surrounded by seven detection fibers of 400-micron diameter and NA of 0.37. The fiber bundles were arranged at equal angular increments around the phantom surface. In Raman imaging mode, 850 nm long pass filters were inserted in the light path at the entrance to each spectrometer. These filters were removed when measuring the excitation intensity and a neutral density filter with optical density = 5 was inserted in the source path. The acquisition time for each source detector pair was determined with an automated algorithm to ensure that saturation of the CCD did not occur. Raman measurements had a maximum acquisition time of 50 seconds, while excitation light was measured for 5 seconds. Three replicate data sets were taken at each source and detector location, for a total experimentation time around 22 minutes.
2.2. Phantoms
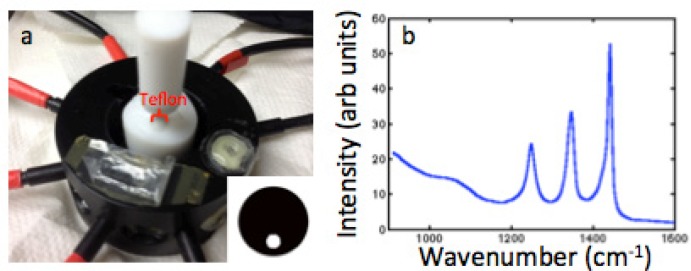
Gelatin phantoms were constructed using agar, 1% Intralipid, 0.01% India ink (for targeted transport scattering and absorption coefficients of μs′ = 1.0mm−1 and μa = 0.01mm−1), and water and had an outer diameter of 27mm to mimic the size of rat legs and the characteristics of tissue [18]. To simulate the Raman signal from bone, Teflon rods with diameters of 5 and 12.5mm were used as inclusions within the gelatin phantoms. A phantom with the 5mm Teflon inclusion is shown in Fig. 1(a) placed inside of the fiber interface with 8 fibers placed around the surface. The Raman spectrum of pure Teflon measured with one channel of the system is shown in Fig. 1(b). These three characteristic peaks arise between 1200 and 1400 wavenumbers, which is similar to the region containing the components of bone.
Fig. 1.
(a) Gelatin phantom with Teflon inclusion inside fiber holder with inset image showing the location of the inclusion. (b) Measured Teflon spectrum without background subtraction.
2.3. Data processing
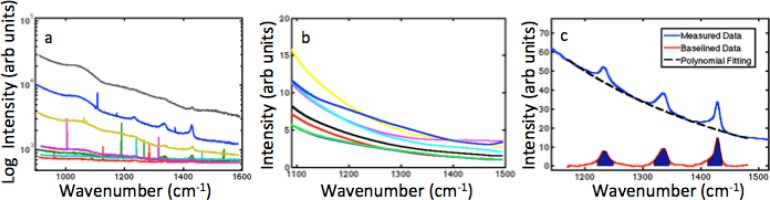
For each Raman measurement, three sequential acquisitions were recorded and the median of these three spectra is computed to reduce the appearance of spectral spikes caused by cosmic rays [19]. These spikes appear very narrow yet with high amplitude compared to the Raman spectrum. An example of a single 7-channel acquisition is presented in Fig. 2(a) and shows these spikes in the measured spectra. The median filtering process largely removes this noise. The resulting spectra are processed with a 16-point Hamming window to remove high frequency noise components present in the pixel-to-pixel variations in the data over the entire measured wavenumber region of 555cm−1 to 1295cm−1. Inter-detection wavelength calibration is then completed based on a Neon emission calibration standard.
Fig. 2.
(a) Measured spectra from 7 parallel detection channels, showing narrow spikes present before median filtering. (b) Polynomial fits for background signal when measuring a homogeneous gelatin phantom. (c) Truncated measured signal with Teflon Raman peaks; the polynomial fit to the background and the difference between them, highlighting the portion of the spectra that is integrated in order to construct the Raman data.
One of the major challenges of measuring Raman spectra from deep tissue is extracting the relatively weak Raman signals from an often dominant background autofluroescence. Background signal can arise from fluorescence in the fibers and filters as well as from non-Raman interactions within the sample and dark noise of the system. The exact origins of these signals are unknown but visible in the spectra since the quantum efficiency of Raman scattering is on the scale of 10-7. Therefore even minor impurities will have enough fluorescence to generate a background signal. It is common practice in the Raman community to use a polynomial fit in order to separate these components from the Raman signal [20,21]. Most Raman measurements are taken by stacking the signals one upon another along the y-axis of a CCD; so choosing a single polynomial fit takes into account any characteristics of that detector. In this system each channel is linked to its own spectrometer and CCD, therefore each channel may have a different response and a different polynomial that best fits the data.
To determine the order of the polynomial to be used in each channel for this system, spectra were acquired using a homogeneous gelatin phantom. The acquired signal, truncated to the region of interest, 1100 to 1500 cm−1, was then fit with polynomials from 3rd order to 5th order and the cumulative error was calculated between the background signal and the polynomial. The order of the polynomial with the lowest error for each collection channel was stored for calculation once the Teflon Raman signal was present. All fittings were fit with 4th or 5th order polynomials, but the coefficients were calculated for each measurement due to variations in background when the Teflon inclusion is added. Figure 2(b) shows the variation in the polynomial fits for the homogeneous gelatin phantom for the 8 different channels. When a Raman signal is present in the spectrum, the data points containing the peaks are removed from consideration before the fitting of the polynomial to ensure that the fit is not biased.
The result of the polynomial fitting is then subtracted from the process spectra. The result should represent the pure Teflon Raman signal. This is illustrated in Fig. 2(c), which shows the original spectrum, the polynomial fit to the background signal, and the resulting Teflon signal. By integrating the region underneath the Raman peaks that was excluded during the polynomial fitting it is possible to generate a single value for each of the 56 source detector pairs, which represents the Raman signal intensity for that optical projection.
In addition to integrating the Raman signals, the excitation intensities are integrated and corrected for filtering intensity. Optical data are then calibrated to the image reconstruction model using the procedure described in Davis et al. [17]. Briefly, this process involves dividing the Raman intensity data by the excitation intensity, which is known as the born ratio, and then multiplying by the modeled excitation intensity calculated by assuming the optical properties are known. Once completed, data are ready for image reconstruction.
2.4. Image Reconstruction
All images are reconstructed using the open source NIRFAST software package [22]. Prior to image reconstruction, a finite element mesh, which models the tissue geometry is created in NIRFAST. This is can be accomplished by segmenting DICOM images from another imaging modality and generating a mesh in NIRFAST or, for simple shapes like the circular geometries used herein, by specifying the shape geometry mesh resolution and allowing the software to generate a mesh.
The software produces images by iteratively matching the measured data to a diffusion model of light propagation in tissue. In this case, the fluorescence reconstruction algorithms are used since the imaging approach is identical so far as the modeling is concerned. This produces images of fluorescence yield, which is the product of the quantum yield of the fluorophore and the absorption coefficient of the fluorescent compound at the excitation wavelength. For consistency, we will call this parameter Raman yield herein. Since the optical properties are not recovered explicitly, they must be estimated either through another modality or literature values.
Raman yield reconstructions were completed with two different techniques for comparison. The mathematical difference of these methods has been explained previously [23]. The first method used the diffusion equation and the location of surface measurements to reconstruct interior values of Raman yield. The second method incorporated a priori knowledge of the interior region boundaries with the surface measurements and restricted the Raman yield results to homogeneous values.
After reconstruction, the contrast to background ratio was calculated by determining the mean value of the reconstruction results for the nodes of the mesh within the region of expected Teflon signal and dividing by the mean value of the reconstruction for the nodes representing the gelatin background of the phantom.
3. Results
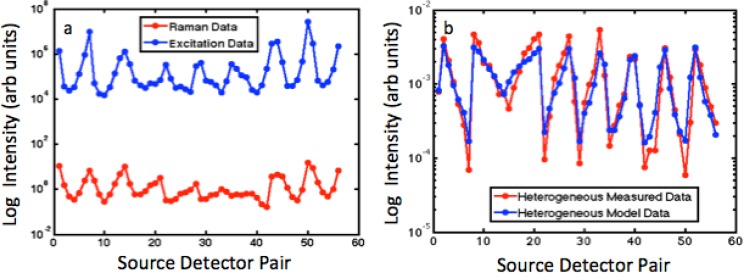
Figure 3(a) shows the integrated intensities of the Teflon Raman signal and excitation signal for each of the 56 optical projections through the phantom. As expected, larger perturbations in the Raman data are observed as compared to the excitation data since the Raman measurements describe an asymmetrical phantom with high contrast.
Fig. 3.
(a) Plot of the log intensity of Raman and Excitation for each source and detector pair for 8 sources and 7 detection channels. (b) Born ratio of the measured data along with the born ratio calculated for a heterogeneous diffusion model.
Similar trends in the data are seen in Fig. 3(b), which plots the Born ratio at each source detector pair for the measured data and for the calculated data from a heterogeneous forward diffusion model. The measured data has a slightly greater change in magnitude than the modeled data.
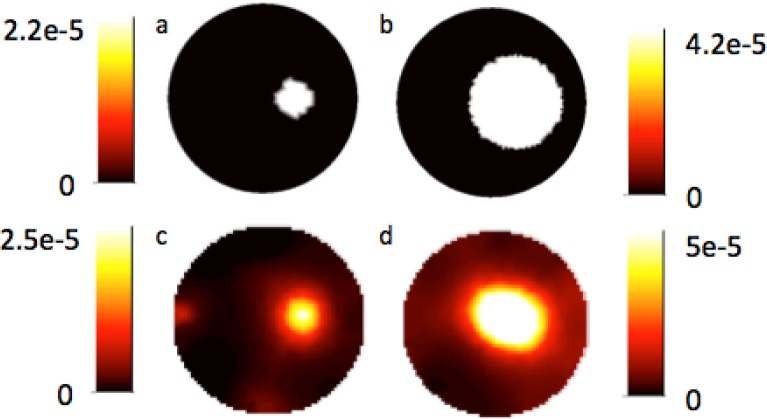
Images of Teflon Raman yield recovered from both gelatin phantoms when no interior prior information is included are shown in Figs. 4(c) and 4(d). The addition of prior information results in the Teflon Raman yield shown in Figs. 4(a) and 4(b), these images also represent the position and size of the inclusions in the phantoms.
Fig. 4.
(a) and (b) Experimental reconstructed Raman yield for gelatin-based phantoms with Teflon inclusions using spatial prior information to restrict the recovered values to be homogeneous in the two regions. (c) and (d) Experimental reconstructed Raman yield for phantoms when no prior spatial information is included in the iterative algorithm.
The Raman yield was recovered with high spatial correlation for both phantoms without the inclusion of prior information. Raman yield was reconstructed with a contrast to background ratio of 8.1 and 9.8 for the 5mm and 12.5mm sized Teflon inclusions, respectively. The size of the reconstructed Teflon inclusion was smaller than the true size, with diameters from line profiles measured as 4.3mm and 9.3mm. When prior information was included with the phantom the contrast to background was raised to 31.7 and 57.3 while the overall maximum reconstructed value decreased.
For the reconstructions completed with no prior information, the 12.5mm Teflon inclusion has the greatest Raman signal, but also the highest background value. In the 5mm Teflon inclusion phantoms the background is very low except for areas within the mesh where edge artifacts occur at the surface aligned with the source and detector placement.
4. Discussion
The recovered images of Raman yield in Figs. 4(c) and (d) show excellent agreement with the true spatial position of the Teflon inclusion and have relatively high values of contrast to background, in agreement with earlier fluorescent experiments comparing various reconstruction methods [23]. When spatial prior information is included when reconstructing Raman yield the recovered contrast is increased by at least 3-fold. In this case, geometrical information was used to determine the interior region boundaries, but DICOM images could be used in the future for segmentation of regions.
As expected the Raman yield was greater for the 12.5mm Teflon inclusion than for the 5mm inclusion. This increase in signal can be explained by the presence of more material to generate the Raman scattered photons. For imaging bone in vivo, the Raman active material will not consist of such a large portion of the cross section as the 12.5 inclusion and in some cases might consist of two smaller regions of interest.
One advantage of the multisystem parallel detection scheme used here is the ability to measure a large dynamic range around the circumference of the tissue volume since individual detection channels can be controlled by adjusting integration times, or adding neutral density filters. The excitation data in Fig. 3(a) range over three to four orders of magnitude. A system that arranges the collection fibers in a line at the entrance to the spectrograph with a single CCD is unable to provide adequate dynamic range performance to measure these signals accurately. Additionally, single spectrometer systems, which are multiplexed sequentially, would require significantly longer acquisition times. For example, the total scan time for the phantoms in this study was 22 minutes. Acquiring equivalent source-detector data with a single-spectrometer sequentially multiplexed system would require at least 154 minutes.
The low variation in Raman signal for source detector pairs 28 to 42 can be attributed to the fact that the source is placed far from the Raman generating inclusion and therefore most excitation light will not pass through the inclusion before being detected. As the source completes its rotation and comes close to the Teflon inclusion again the parabolic nature of the data is restored.
The phantoms imaged in this study were relatively simple with homogeneous background and homogeneous inclusions. In vivo imaging will be challenged by the heterogeneity of both the bone and surrounding tissue. Some of these effects should be reduced by the incorporation of the Born ratio when processing the data [24]. By reconstructing the integrated Raman peaks rather than simple peak intensities we are able to increase the amount of signal and decrease the effects of noise on the data. Traditional methods of Raman scattering analysis use ratio techniques. These techniques could still be applied with this method of Raman yield reconstruction. An additional challenge when imaging Raman scattering of low intensities is reducing the background signal collected. To address this challenge, work will be conducted to include long pass filtering at the collection end of the fibers. Gelatin phantoms with bone inclusions will allow for testing of new developments while keeping the background homogeneous.
5. Conclusion
Raman tomography was demonstrated in tissue-simulating phantoms using a multichannel optical tomography system with parallel spectroscopic detection for the first time. Images of Raman yield with high contrast to background ratios and accurate spatial resolution were recovered with and without the use of prior interior information of the mesh. Parallel collection channels reduced the number and complexity of post-processing steps necessary while increasing the dynamic range of data obtained.
Acknowledgments
This work was supported through NIH grant R01AR04796.
References and links
- 1.Bailey A. J., Wotton S. F., Sims T. J., Thompson P. W., “Post-translational modifications in the collagen of human osteoporotic femoral head,” Biochem. Biophys. Res. Commun. 185(3), 801–805 (1992). 10.1016/0006-291X(92)91697-O [DOI] [PubMed] [Google Scholar]
- 2.Paschalis E. P., Shane E., Lyritis G., Skarantavos G., Mendelsohn R., Boskey A. L., “Bone fragility and collagen cross-links,” J. Bone Miner. Res. 19(12), 2000–2004 (2004). 10.1359/jbmr.040820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahadevan-Jansen A., Mitchell M. F., Ramanujamf N., Malpica A., Thomsen S., Utzinger U., Richards-Kortum R., “Near-infrared Raman spectroscopy for in vitro detection of cervical precancers,” Photochem. Photobiol. 68(1), 123–132 (1998). 10.1111/j.1751-1097.1998.tb03262.x [DOI] [PubMed] [Google Scholar]
- 4.Krafft C., Steiner G., Beleites C., Salzer R., “Disease recognition by infrared and Raman spectroscopy,” J Biophotonics 2(1-2), 13–28 (2009). 10.1002/jbio.200810024 [DOI] [PubMed] [Google Scholar]
- 5.Crane N. J., Popescu V., Morris M. D., Steenhuis P., Ignelzi M. A., Jr, “Raman spectroscopic evidence for octacalcium phosphate and other transient mineral species deposited during intramembranous mineralization,” Bone 39(3), 434–442 (2006). 10.1016/j.bone.2006.02.059 [DOI] [PubMed] [Google Scholar]
- 6.Boivin G., Meunier P. J., “The mineralization of bone tissue: a forgotten dimension in osteoporosis research,” Osteoporos. Int. 14(Suppl 3), S19–S24 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Matousek P., Stone N., “Emerging concepts in deep Raman spectroscopy of biological tissue,” Analyst (Lond.) 134(6), 1058–1066 (2009). 10.1039/b821100k [DOI] [PubMed] [Google Scholar]
- 8.Stone N., Baker R., Rogers K., Parker A. W., Matousek P., “Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): future possibilities for the diagnosis of breast cancer,” Analyst (Lond.) 132(9), 899–905 (2007). 10.1039/b705029a [DOI] [PubMed] [Google Scholar]
- 9.Morris M. D., Finney W. F., Rajachar R. M., Kohn D. H., “Bone tissue ultrastructural response to elastic deformation probed by Raman spectroscopy,” Faraday Discuss. 126, 159–168, discussion 169–183 (2004). 10.1039/b304905a [DOI] [PubMed] [Google Scholar]
- 10.McCreadie B. R., Morris M. D., Chen T. C., Sudhaker Rao D., Finney W. F., Widjaja E., Goldstein S. A., “Bone tissue compositional differences in women with and without osteoporotic fracture,” Bone 39(6), 1190–1195 (2006). 10.1016/j.bone.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 11.Draper E. R., Morris M. D., Camacho N. P., Matousek P., Towrie M., Parker A. W., Goodship A. E., “Novel assessment of bone using time-resolved transcutaneous Raman spectroscopy,” J. Bone Miner. Res. 20(11), 1968–1972 (2005). 10.1359/JBMR.050710 [DOI] [PubMed] [Google Scholar]
- 12.Morris M. D., Mandair G. S., “Raman assessment of bone quality,” Clin. Orthop. Relat. Res. 469(8), 2160–2169 (2011). 10.1007/s11999-010-1692-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan S., Schulmerich M., Cole J. H., Dooley K. A., Kreider J. M., Pogue B. W., Morris M. D., Goldstein S. A., “Image-guided Raman spectroscopic recovery of canine cortical bone contrast in situ,” Opt. Express 16(16), 12190–12200 (2008). 10.1364/OE.16.012190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller M. D., Majumder S. K., Mahadevan-Jansen A., “Spatially offset Raman spectroscopy of layered soft tissues,” Opt. Lett. 34(7), 926–928 (2009). 10.1364/OL.34.000926 [DOI] [PubMed] [Google Scholar]
- 15.Matousek P., Draper E. R., Goodship A. E., Clark I. P., Ronayne K. L., Parker A. W., “Noninvasive Raman spectroscopy of human tissue in vivo,” Appl. Spectrosc. 60(7), 758–763 (2006). 10.1366/000370206777886955 [DOI] [PubMed] [Google Scholar]
- 16.Kepshire D. S., Davis S. C., Dehghani H., Paulsen K. D., Pogue B. W., “Subsurface diffuse optical tomography can localize absorber and fluorescent objects but recovered image sensitivity is nonlinear with depth,” Appl. Opt. 46(10), 1669–1678 (2007). 10.1364/AO.46.001669 [DOI] [PubMed] [Google Scholar]
- 17.Davis S. C., Pogue B. W., Springett R., Leussler C., Mazurkewitz P., Tuttle S. B., Gibbs-Strauss S. L., Jiang S. S., Dehghani H., Paulsen K. D., “Magnetic resonance-coupled fluorescence tomography scanner for molecular imaging of tissue,” Rev. Sci. Instrum. 79(6), 064302 (2008). 10.1063/1.2919131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen S. J., Patterson M. S., Wilson B. C., “The use of India ink as an optical absorber in tissue-simulating phantoms,” Phys. Med. Biol. 37(4), 985–993 (1992). 10.1088/0031-9155/37/4/012 [DOI] [PubMed] [Google Scholar]
- 19.J. R. Janesick, Scientific Charge-Coupled Devices (SPIE Press, Bellingham, Wash., 2001), p. xvi. [Google Scholar]
- 20.Esmonde-White F. W. L., Esmonde-White K. A., Morris M. D., “Minor distortions with major consequences: correcting distortions in imaging spectrographs,” Appl. Spectrosc. 65(1), 85–98 (2011). 10.1366/10-06040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieber C. A., Mahadevan-Jansen A., “Automated method for subtraction of fluorescence from biological Raman spectra,” Appl. Spectrosc. 57(11), 1363–1367 (2003). 10.1366/000370203322554518 [DOI] [PubMed] [Google Scholar]
- 22.Dehghani H., Eames M. E., Yalavarthy P. K., Davis S. C., Srinivasan S., Carpenter C. M., Pogue B. W., Paulsen K. D., “Near infrared optical tomography using NIRFAST: Algorithm for numerical model and image reconstruction,” Commun. Numer. Methods Eng. 25(6), 711–732 (2009). 10.1002/cnm.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis S. C., Dehghani H., Wang J., Jiang S., Pogue B. W., Paulsen K. D., “Image-guided diffuse optical fluorescence tomography implemented with Laplacian-type regularization,” Opt. Express 15(7), 4066–4082 (2007). 10.1364/OE.15.004066 [DOI] [PubMed] [Google Scholar]
- 24.Soubret A., Ripoll J., Ntziachristos V., “Accuracy of fluorescent tomography in the presence of heterogeneities: study of the normalized Born ratio,” IEEE Trans. Med. Imaging 24(10), 1377–1386 (2005). 10.1109/TMI.2005.857213 [DOI] [PubMed] [Google Scholar]