Abstract
Motivated by the limitations of liposomal drug delivery systems, we designed a novel histidine-based AB2-miktoarm polymer (mPEG-b-(polyHis)2) equipped with a phospholipid-mimic structure, low cytotoxicity, and pH-sensitivity. Using “core-first” click chemistry and ring-opening polymerization, mPEG2kDa-b-(polyHis29kDa)2 was successfully synthesized with a narrow molecular weight distribution (1.14). In borate buffer (pH 9), the miktoarm polymer self-assembled to form a nano-sized polymersome with a hydrodynamic radius of 70.2 nm and a very narrow size polydispersity (0.05). At 4.2 µmol/mg polymer, mPEG2kDa-b-(polyHis29kDa)2 strongly buffered against acidification in the endolysosomal pH range and exhibited low cytotoxicity on a 5 d exposure. Below pH 7.4 the polymersome transitioned to cylindrical micelles, spherical micelles, and finally unimers as the pH was decreased. The pH-induced structural transition of mPEG2kDa-b-(polyHis29kDa)2 nanostructures may be caused by the increasing hydrophilic weight fraction of mPEG2kDa-b-(polyHis29kDa)2 and can help to disrupt the endosomal membrane through proton buffering and membrane fusion of mPEG2kDa-b-(polyHis29kDa)2. In addition, a hydrophilic model dye, 5(6)-carboxyfluorescein encapsulated into the aqueous lumen of the polymersome showed a slow, sustained release at pH 7.4 but greatly accelerated release below pH 6.8, indicating a desirable pH sensitivity of the system in the range of endosomal pH. Therefore, this polymersome that is based on a biocompatible histidine-based miktoarm polymer and undergoes acid-induced transformations could serve as a drug delivery vehicle for chemical and biological drugs.
Keywords: Polymersome, pH sensitivity, Poly(L-histidine), Structural transition, Miktoarm polymer
1. Introduction
Phospholipids self-assemble in the body, forming various biological vesicles and membranes. Liposomes, prepared using natural or synthetic lipids, mimic these naturally occurring vesicles and are attractive as delivery vehicles because of their unique ability to encapsulate hydrophilic molecules inside the aqueous lumen as well as hydrophobic drugs between the hydrophobic tails of the lipids.1, 2 However, the lack of chemical flexibility and physical stability of conventional liposomes has limited their applications in drug delivery. These issues have led to a great deal of interest in polymer-based delivery approaches.3, 4 Polymer-based vesicles are referred to as “polymersomes” and are attracting rapidly growing interest in the field of drug delivery owing to their enhanced stability, controllable molecular weights, and easy modification.5, 6 Much effort has been devoted to the development of various polymersomes with the goal of clinically efficacious drug delivery through mimicking natural structures, introducing stimulus-responsiveness, and providing biocompatibility.7
Most polymersomes reported thus far have been fabricated from amphiphilic linear diblock copolymers, which are essentially combinations of a hydrophilic block and a hydrophobic block.8, 9 However, these diblock copolymers do not completely mimic the natural architecture of phospholipid bilayer vesicles. In addition, the weight/volume fraction of these polymers must be well-controlled to avoid the formation of other structures such as spherical or tubular micelles. Therefore, more complex polymers such as miktoarm polymers have attracted significant attention as building blocks for self-assembling materials, owing to their ability to mimic the characteristics of phospholipid structures. In particular, the unique properties of the structures that assemble from these polymers in solution have led to their application in drug delivery.10–12 In our recent study, amphiphilic AB2 type miktoarm polymers exhibited a greater ability to form vesicles than did typical diblock copolymer structures.13
Intelligent polymersome carriers are designed to release their payloads in a controlled manner upon arrival at the target site in response to a specific external or internal stimulus, which can significantly improve therapeutic efficacy and reduce side effects. Of all of the potential stimuli, a pH change is a particularly useful intrinsic signal for tumor-specific drug delivery because of the acidity in the tumor extracellular matrix and in endocytic organelles in contrast to the constant physiological blood pH of 7.4.14 To construct a pH-sensitive polymersome, biodegradable linkages or proton-buffering components have been introduced into vesicle-forming block copolymers. The former approach has been used with hydrophobic segments such as poly(lactic acid) and poly(ε-caprolactone), and the acid-catalyzed hydrolysis induces vesicle rupture, resulting in pH-triggered drug release. These polymersome carriers showed some encouraging in vitro/vivo results.15–17 However, their slow hydrolysis rates limited their potential for drug delivery.7 The proton-buffering approach involves the incorporation of proton-buffering groups such as weakly acidic (carboxyl) or basic (amino) pendant groups; it also allows pH-responsiveness, because of the pH-dependent ionization (protonation/deprotonation) of these groups, and then allows sharp structural transitions (soluble/insoluble). These characteristics lead to rapid drug release in response to a small change in pH18, 19 and an enhanced therapeutic efficacy owing to the low degree of sequestration of drug inside acidic endosomes/lysosomes.
Even if polymersomes are constructed as natural mimetics and the loaded drugs are released in a pH-sensitive manner, to be clinically applicable their components must be biocompatible. Among natural sources, poly(amino acid)-based polymers are a candidate material because of their versatile functional groups, high biocompatibility, low toxicity, and potential biodegradability.20, 21 Thus far, a number of pH-sensitive polymersomes prepared from poly(L-lysine),22, 23 poly(L-glutamate),24, 25 and poly(L-arginine)26 have been studied; however, none of these has shown desirable pH-sensitivity for intracellular drug delivery.
Thus, this study sought to develop a novel pH-sensitive polymersome carrier that exhibits phospholipid-mimicking characteristics, pH sensitivity, and biocompatibility, by using a poly(amino acid)-based AB2 miktoarm polymer. Biocompatible poly(ethylene glycol) (PEG) was used as the hydrophilic A arm to avoid polymersome uptake by the reticuloendothelial system (RES).27 Biocompatible, amphoteric poly(L-histidine) (polyHis)28 was used for both of the hydrophobic B arms to confer pH sensitivity on the vesicle carrier. The vesicle formation, pH-triggered morphological transitions, and cytotoxicity of this 3-miktoarm polymer were investigated to assess the suitability of this material as a drug delivery vehicle.
2. Experimental
2.1. Materials
Nα-Boc-N(im)-2,4-dinitrophenyl-L-histidine (Boc-His(DNP)-OH) isopropanol solvate was purchased from BACHEM Co. (Torrance, CA). Thionyl chloride, propargylamine, methyl acrylate, ethylenediamine, methoxy poly(ethylene glycol) (mPEG, Mn 2 kDa), p-toluenesulfonyl chloride, ethylenediaminetetraacetic acid (EDTA) tetrasodium salt hydrate, copper (II) sulfate pentahydrate, sodium L-ascorbate, 2-mercaptoethanol, pyrene, RPMI1640 medium, 4-(2-hydroxy-ethyl)-1-piperazine (HEPES), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), doxorubicin (DOX or adriamycin (ADR)), recombinant human insulin, L-glutamine, D-glucose, acetone, dimethylsulfoxide (DMSO), dimethylformamide (DMF), methanol, 1,4-dioxane, diethyl ether, phosphate buffered saline (PBS), branched polyethyleneimine (bPEI; Mw 25 kDa and Mn 10 kDa), poly(L-lysine) hydrogen bromide (PLL·HBr; Mw (viscosity) 23 kDa), 5(6)-carboxyfluorescein (CF), and Triton X-100 were purchased from Sigma-Aldrich Co. (St. Louis, MO). Fetal bovine serum (FBS), penicillin-streptomycin antibiotics, and trypsin-EDTA solution were purchased from Invitrogen Co. (Carlsbad, CA).
2.2. Synthesis and characterization of block and core components
To synthesize azide-functionalized mPEG for the A block (Fig. S1a), mPEG was modified by a 2-step synthetic procedure via tosylation and then azide functionalization. As the first step, mPEG (2 g, 1 mmol) in anhydrous pyridine (10 mL) was reacted with p-toluenesulfonyl chloride (1.93 g, 10 mmol) for 24 h at room temperature (RT). The reaction mixture was centrifuged and precipitated into diethyl ether (80 mL). The obtained monotosylated mPEG powder (1.7 g) was dried in vacuo at RT. In the second step, the monotosylated mPEG (1.7 g) in DMF (10 mL) was reacted with sodium azide (0.65 g) for 24 h at RT under a N2 atmosphere, and then dichloromethane (25 mL) was added into the reaction mixture. The resultant mixture was sequentially washed with cold deionized water (DIW), cold 6 M HCl, and then cold DIW again. The organic layer was dried over anhydrous magnesium sulfate and then precipitated in diethyl ether. The identity of the resultant mPEG-N3 (1.35 g) was confirmed by 1H-nuclear magnetic resonance (1H-NMR) spectroscopy and Fourier Transform-Infrared (FT-IR) spectroscopy.
To synthesize N(im)-2,4-dinitrophenyl-L-histidine-N-carboxyanhydride hydrochloride (DNP-His-NCA·HCl) as a cyclic monomer of the B blocks (Fig. S1b), thionyl chloride (2.5 mL) was added dropwise into Boc-His(DNP)-OH isopropanol solvate (2.5 g) in anhydrous 1,4-dioxane (20 mL) under inert N2 gas and slow stirring. The reaction mixture was stirred for 1 h and was then poured into anhydrous diethyl ether (100 mL) to remove excess thionyl chloride and precipitate the product (DNP-His-NCA·HCl). The precipitate was filtered and dried in vacuo at RT. After further purification by recrystallization in a mixture of acetone-diethyl ether, DNP-His-NCA·HCl (1.9 g) was obtained and characterized by 1H-NMR spectroscopy.
The core component, a polyamidoamine (PAMAM) type tri-functional dendron (D1) with an alkyne group and two primary amine groups was synthesized as previously reported (Fig. S1c).29 Briefly, propargylamine (D0; 0.1 g, 1.8 mmol) in methanol (2 mL) was added dropwise into methyl acrylate (0.57 mL, 6.2 mmol) in methanol (2 mL) over 30 min in an ice-water bath. The reaction mixture was stirred for 30 min at 0 °C and then for an additional 24 h at RT under a N2 atmosphere. After evaporating the reaction solution, the residual methyl ester-terminated dendron (D0.5; 0.33 g) was dried in vacuo. Then, D0.5 (0.33 g) in methanol (2 mL) was further added dropwise into redistilled ethylenediamine (1 mL) in methanol (2 mL) over 1 h in an ice-water bath. The reaction mixture was stirred for 1 h at 0 °C and then for an additional 48 h at RT under a N2 atmosphere. The reaction solution was evaporated, and then the residue was dried in vacuo at 35 °C to obtain the amino-terminated D1 (0.29 g). D0.5 and D1 were characterized by 1H-NMR spectroscopy.
2.3. Synthesis and characterization of 3-miktoarm block copolymers
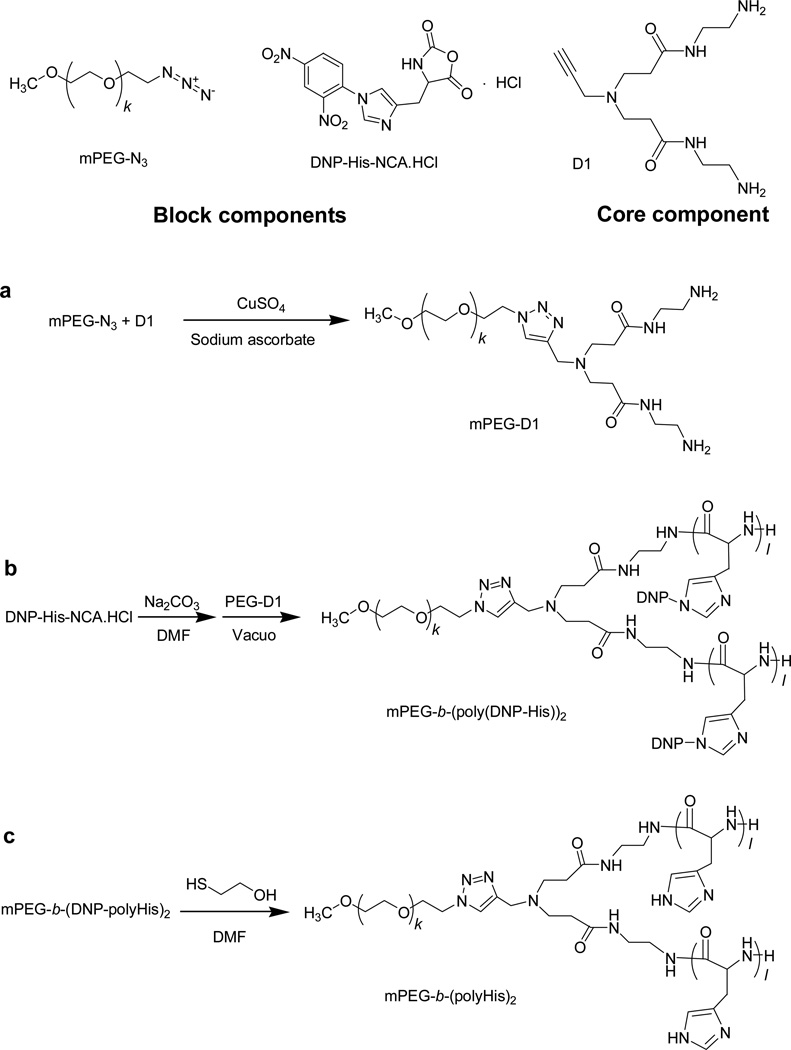
Synthesis of the mPEG-dendron macroinitiator (mPEG-D1) was carried out via click chemistry (Fig. 1a). mPEG-N3 (1 g, 0.5 mmol) in DIW (10 mL) was mixed by adding D1 (0.2 g, 0.7 mmol), CuSO4·5H2O (0.25 g, 1 mmol), and sodium ascorbate (0.59 g, 3 mmol) under stirring. The resultant solution was stirred vigorously for 48 h at RT. Then, EDTA sodium salt was added to the reaction solution, and the resultant mixture was dialyzed against DIW for 48 h in a dialysis membrane with a 1 kDa molecular weight cut-off (MWCO) to remove Cu2+ ions and the excess reactants. The purified solution was then extracted with dichloromethane and the organic layer was finally precipitated in diethyl ether to obtain mPEG-D1. To remove any water residue in the macroinitiator, the product was further treated by an azeotropic distillation in toluene for 2 h at 120 °C in an N2 atmosphere. The completion of the click chemistry reaction was confirmed by the disappearance of the azide group in FT-IR spectroscopy, and the chemical structure of mPEG-D1 was characterized by 1H-NMR spectroscopy.
Fig. 1.
Synthetic procedure for synthesis of the 3-miktoarm star copolymer.
Two primary amine groups of mPEG-D1 initiated a ring-opening polymerization (ROP) of DNP-His-NCA·HCl to form the AB2-type miktoarm polymer (Fig. 1b) In brief, sodium carbonate (0.48 g, 4.5 mmol) was added into DNP-His-NCA·HCl (1.15 g, 3 mmol) in anhydrous DMF (10 mL) under N2 gas and stirred for 1 h to remove the hydrogen chloride attached to the cyclic His derivative. The reaction solution was mixed with mPEG-D1 (0.12 g, 0.06 mmol) and then the resultant solution was stirred for 72 h at RT in vacuo. To remove unreacted monomers, the reaction mixture was poured into diethyl ether (500 mL). The resultant precipitate, mPEG-b-(poly(DNP-His))2, was filtered and dried in vacuo at RT. The chemical structure and molecular weight distribution of the precipitate were characterized by 1H-NMR spectroscopy and gel permeation chromatography (GPC), respectively.
Removal of the DNP protecting groups (Fig. 1c) was performed by the addition of 2-mercaptoethanol into mPEG-b-(poly(DNP-His))2 (0.5 g) in DMF (10 mL). After 6 h of vigorous stirring, the resultant mixture was poured into diethyl ether (200 mL) to precipitate the product. The precipitate was filtered and dried in vacuo at RT. The crude product was dissolved in DIW pre-adjusted to pH 3 with 1 M HCl. Then, using a dialysis membrane (MWCO 2 kDa), the solution was dialyzed against acidic water (pH 3) for 24 h and then neutral water for another 48 h. Finally, the dialysate was lyophilized to obtain the purified product mPEG-b-(polyHis)2. The chemical structure of the product was confirmed by 1H-NMR spectroscopy.
For all synthesized components and polymers, 1H-NMR spectra were recorded on a Varian 400 spectrometer in deuterated solvents. Their FT-IR spectra were recorded from 400 to 4000 cm−1 on a Bruker Tensor 37 IR spectrophotometer (± 1 cm−1) using KBr pellets. Gel permeation chromatography (GPC) measurements of polymers were performed on an Agilent 1100 Series high performance liquid chromatography (HPLC) system equipped with a TSKgel G3000HHR GPC column equilibrated at 30°C and a refractive index detector. DMF with 10 mM LiBr solution was used as the eluent at a flow rate of 1 mL/min, and a series of polystyrene standards with Mp (peak top molecular mass) ranging from 1 kDa to 40 kDa were used to calibrate the molecular weight.
The proton buffering capacity of the synthesized 3-miktoarm polymer was evaluated by a typical acid-base titration.28, 30, 31 Briefly, the polymer (5 mg in 10 mL 150 mM NaCl) was adjusted to pH 2 with 1 M HCl. The resultant solution was titrated with 0.1 M NaOH, and the pH was monitored with a pH meter. The average value from three titrations was plotted, and the buffering capacity was calculated as follows:
where ΔVNaOH is the volume of 0.1 M NaOH, CNaOH is the concentration of NaOH solution (0.1 M), and mpolymer is the mass of polymer (mPEG-b-(polyHis)2).
2.4. Preparation and physicochemical characterization of polymersomes
mPEG-b-(polyHis)2 (5 mg) was dissolved in DMSO (10 mL), and then 5 M NaOH (5 µL) was added into the polymer solution to neutralize residual charges. The solution was vigorously stirred overnight and was then transferred into a dialysis membrane (MWCO 2 kDa) and dialyzed against 10 mM borate buffer (pH 9.0) with 140 mM NaCl. The outer buffer phase was replaced with fresh solution at 2, 4, and 12 h, and the polymer solution inside the membrane was recovered after 24 h. The solution was adjusted to a predetermined pH with 1 N HCl and was further equilibrated at RT under gentle stirring for 48 h prior to characterization. The surface charge of polymersomes prepared at pH 7.0, pH 7.4, and pH 9.0 were measured with a Zetasizer 3000HS (Malvern Instrument, Inc, Worcestershire, UK) at a wavelength of 677 nm and a constant angle of 90° at RT.
Light scattering (LS) experiments were carried out on a Brookhaven Instruments Corp. system consisting of a BI-200SM goniometer and a BI-9000AT autocorrelator. The temperature was controlled at 25°C with a circulating water bath unless otherwise specified. Dust was removed from the sample by filtration with a 0.80 µm disposable membrane filter immediately prior to the measurement. In dynamic light scattering (DLS) measurements, the measured second order autocorrelation function G2(t) was converted to the normalized first order autocorrelation function g1(t)32, which was further analyzed by the constrained regularized CONTIN algorithm33 to yield information on the normalized distribution of relaxation time G(tR). In the case of simple diffusion, the angular dependence of the relaxation time is described by Dapp=1/(tRq2), where Dapp is the apparent translational diffusion coefficient and q is the magnitude of the scattering wave vector. This magnitude is given by q=4πnsin(θ/2), where n is the refractive index of the solvent, λ is the wavelength of the incident beam in vacuo (632.8 nm) and θ is the scattering angle. For a dilute solution with qRg~1, the intrinsic translational diffusion coefficient D0 is correlated to the angular dependent Dapp via the equation: Dapp=D0(1+CRg2q2), where Rg is the radius of gyration and C is a characteristic parameter of the molecular architecture. D0 is obtained from the intercept of a plot of Dapp vs. q2 extrapolated to the zero angle. The hydrodynamic radius Rh is related to D via the Stokes-Einstein equation: D=kT/6πηRh, where k is the Boltzmann constant and η is the viscosity of water at temperature T. The polydispersity index (PDI) of the sample is calculated from the relative variance of the intensity-weighted distribution at 90° by the integrated software.
Static light scattering (SLS) measurements were carried out at angles from 30° to 120°. The excess intensity of scattered light Iex(q) is related to the radius of gyration (Rg) via the partial Zimm equation: 1/Iex(q) = P(1+Rg2q2/3), where q is the magnitude of the scattering wave vector and P is an experimental parameter. Rg is determined from the slope and intercept of a plot of 1/Iex(q) vs. q2.
Transmission electron microscopy (TEM) observations were performed on a Philips TECNAI T12 electron microscope. To prepare the sample, one drop of the aqueous solution was deposited onto a Formvar-carbon coated copper grid. After 5 min, the excess solution was wicked away by touching a filter paper to the edge of the grid. The sample was then observed directly by TEM without staining.
A fluorescence study was performed on a SHIMADZU RF-5301PC spectrofluorometer using pyrene as a hydrophobic florescent probe. Pyrene solution (2 µL) in ethanol was added to 3 mL of polymer solution to achieve a final pyrene concentration of 6×10−7 M, and the resultant solution was stirred for 48 h before measurement. Excitation and emission bandwidths were 5 and 3 nm, respectively. The intensity ratios (I1/I3) of the first band (372 nm) to the third band (383 nm) in the pyrene emission spectra were analyzed as a function of polymer concentration and pH. The critical association concentration (CAC), which indicates the onset of polymer association, was estimated as the concentration above which the I1/I3 value decreased.
2.5. Polymersome encapsulation and in vitro release of the hydrophilic dye 5(6)-carboxyfluorescein
CF loaded polymersomes were prepared by mixing 0.5 mL of the polymer dissolved in DMSO (4 mg/mL) into 2 mL of borate buffer solution (10 mM, pH 9.0) containing 150 mM CF in a glass vial, followed by vigorous stirring for 2 h to obtain a homogenous solution. The solution was then transferred into a dialysis bag (MWCO 15 kDa) and dialyzed against 1 L of borate buffer (10 mM, pH 9.0) containing 140 mM NaCl. The outer buffer phase was replaced twice with fresh buffer and the solution inside the membrane was recovered after 4 h. Afterwards, the CF containing polymersomes were separated from free CF by gel filtration with a Sephadex G-25 column (1.5 cm×40 cm) using the borate buffer as the eluent at a flow rate of 0.5 mL/min. Fifty µL of the gel filtrated solution was mixed with 50 µL 2 wt.% Triton-X 100 and the mixture was further diluted to 5 mL by adding the pH 9.0 borate buffer, and the solution was briefly sonicated for 10 min to trigger complete release of the encapsulated dye. The fluorescence intensity (λex=490 nm and λem=520 nm) of the resulting solution was measured with a SPECTRAmax M2 spectrofluorometer. The amount of encapsulated CF was calculated from a linear regression equation relating the fluorescence intensity (I) to the CF concentration (C, ng/mL) by using a series of CF standard solutions (5–200 ng/mL) in pH 9.0 borate buffer: C= 0.0265×I−0.70 (correlation coefficient R2=0.999). The encapsulated volume is given as 1000×(mEV)/(mFmP), where mE and mF are amount in mg of the encapsulated and feed CF, respectively; mP is amount in mg of the polymer used; V is volume in mL of the CF-loaded polymersome solution.
CF self-quenches when trapped within a vesicle at high concentration but is readily detected by fluorescence when it is released from the vesicle.34 Right after gel filtration, 0.2 mL of the CF-loaded polymersomes was mixed with 1.8 mL of PBS pre-adjusted to different pH in a glass vial, and then the vial was immediately immersed into in a 37°C water bath equipped with a mechanic shaker at a speed of 100 RPM. At predetermined intervals, 0.1 mL of the mixture was withdrawn, and then the fluorescence intensity (It) was measured. The percentage of released CF was calculated from: (It-I0)/(Itot-I0)×100, where I0 is the fluorescence intensity right after the gel filtration and Itot is the fluorescence intensity after disrupting the vesicle and completely releasing the dye upon addition of Triton X-100.
2.6. Polymersome cytotoxicity testing
This study used two multidrug resistant (MDR) cell lines: MCF7/ADR-RES cells (a DOX-induced multidrug resistant subline of human breast adenocarcinoma MCF7 cells) and A2780/ADR (a DOX-induced multidrug resistant subline of human ovarian carcinoma A2780 cells). The cells were cultured in culture medium (i.e., RPMI1640 medium) supplemented with glucose (2 g/L) and 10% heat-inactivated FBS under humidified air containing 5% CO2 at 37°C. For MCF7/ADR-RES cells, insulin (4 mg/L) was also added to the culture medium. To maintain the MDR characteristics, the cells were treated with DOX (400 ng/mL) weekly.
For the MTT-based cell viability assay, the cells were seeded at a density of 5×103 cells/well in 96-well plates. The seeded cells were cultured for 24 h prior to adding various concentrations of polymersomes and representative polycations (i.e., bPEI and PLL). After a 3 d incubation period, the culture medium was replaced with fresh medium. MTT solution (10 µL; 5 mg/mL) was added to the cells (0.1 mL of culture medium) on day 5 of incubation. After an additional 4 h incubation, the MTT-containing medium was removed. Formazan crystals produced by living cells were dissolved in DMSO, and their absorbance was measured at 570 nm using a microplate reader.
3. Results and Discussion
3.1. Synthesis and characterization of block and core components for 3-miktoarm block copolymers
As shown in Fig. 1, a histidine-based 3-miktoarm block copolymer was synthesized via a “core-first” strategy combining click chemistry and ring-opening polymerization (ROP) of α-amino acid N-carboxyanhydrides (NCA).35 The A and B blocks and the core component were synthesized and prepared individually. For the A block, the hydroxyl group of mPEG was tosylated in the presence of p-toluenesulfonyl chloride and the identity of the resultant monotosylated mPEG (mPEG-TOS, 85% yield) was confirmed by the observation of the corresponding peaks of the TOS group in the 1H-NMR spectrum: δ 2.41 for -O-SO2-C6H4-CH3 and δ 7.37 and δ 7.8 for -O-SO2-C6H4-CH3 in CDCl3 (Fig. S2a). In the presence of sodium azide, the monotosylated mPEG was converted to azide-functionalized mPEG (mPEG-N3). The chemical structure of the resultant mPEG-N3 was confirmed by the disappearance of several peaks (-O-SO2-C6H4-CH3 at δ 2.41, 7.37, and 7.8 and -O-CH2CH2-TOS at δ 4.18) and the appearance of one new peak (-O-CH2CH2-N3 at δ 3.4) in the 1H-NMR spectrum (CDCl3) (Fig. S2b). Further confirmation of the newly generated azide functionality was provided by the appearance of a unique band at 2105 cm−1 in the FT-IR spectrum (Fig. S2c), and the mPEG-N3 (A block) was obtained at a yield of 85%.
For the B blocks, the cyclic monomer DNP-His-NCA·HCl was prepared from Boc-His(DNP)-OH (Fig. S1b). In the presence of thionyl chloride, the carboxylic acid group and the Boc-protected amine group of Boc-His(DNP)-OH isopropanol reacted and cyclized. The resultant DNP-His-NCA·HCl was confirmed by a peak corresponding to –NH-CO-O- in NCA at δ 8.58 in the 1H-NMR spectrum (d6-DMSO) (Fig. S3), and the pale yellow product was obtained at a yield of 95%.
A PAMAM-type tri-functional dendron (D1) with an alkyne group and two primary amine groups as the core component was designed to link with the A block and the B blocks, respectively (Fig. S1c). First, two methyl acrylate molecules were reacted with primary amines of propargylamine (D0), resulting in a dendron (D0.5) with two methyl esters (80% yield). Ethylenediamine further modified D0.5 into D1 (70% yield). 1H-NMR spectra in CDCl3 confirmed the conversion of D0.5 to D1 by the disappearance of –CO-O-CH3 at δ 3.68 and the newly introduced –CO-NH-CH2CH2NH2 at δ 1.45 (Fig. S4).
3.2. Synthesis and physicochemical characterization of 3-miktoarm block copolymers
An alkyne group of the core component (D1) was first reacted with the azide group (N3) of the A block component via click chemistry, resulting in mPEG-D1 (72% yield; Fig. 1a). The success of this synthesis was confirmed in two ways. First, the azide groups at 2105 cm−1 disappeared from the FT-IR spectrum of mPEG-D1 (Fig. S5a). Second, triazole protons appeared as a singlet at 8.02 ppm in the 1H-NMR spectrum of mPEG-D1 (Fig. S5b).
Then, after desalting DNP-His-NCA·HCl, ROP of DNP-His-NCA was initiated by the two primary amine groups of mPEG-D1. This synthesis yielded the polymer mPEG-b-(poly(DNP-His))2 (75 % yield), and its chemical structure was confirmed by 1H-NMR spectrum (Fig. S6a). Based on the mPEG (Mn 2 kDa) of mPEG-b-(poly(DNP-His))2, the degree of polymerization (DP) of poly(DNP-His) in mPEG-b-(poly(DNP-His))2 was calculated as 42 according to the 1H-NMR spectrum, with an average DP for each poly(DNP-His) block of 21. The polydispersity (Mw/Mn) of the molecular weight of this polymer was measured as 1.14 using GPC, implying a narrow MWD of mPEG-b-(polyHis)2 (Fig. S6b).
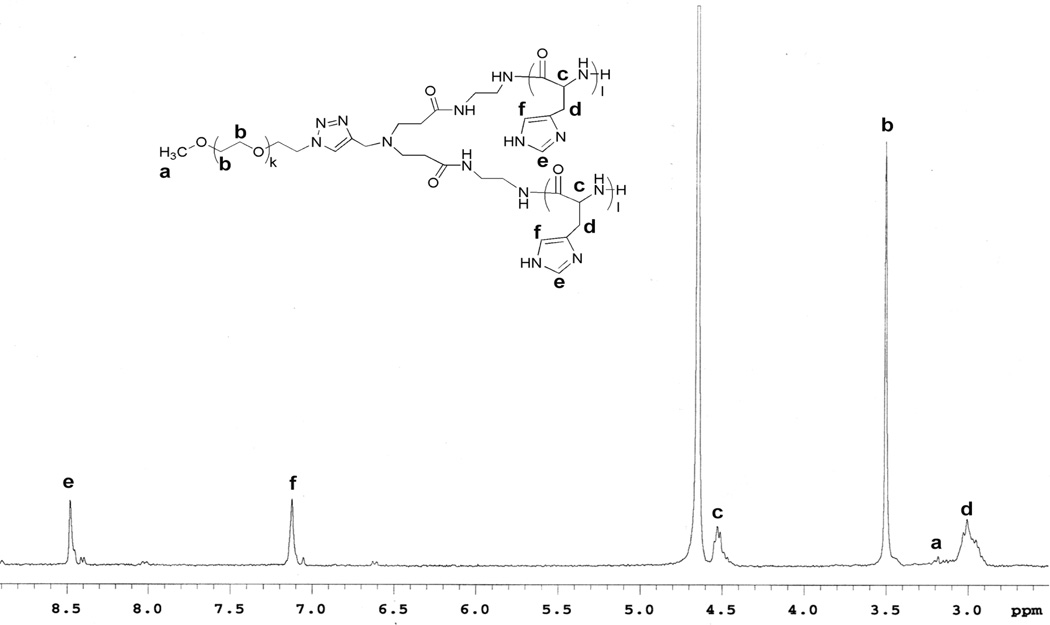
The protected DNP group of mPEG-b-(poly(DNP-His))2 was removed by mercaptoethanol, resulting in the designed 3-miktoarm block copolymer, mPEG-b-(polyHis)2. The chemical structure of the final polymer was confirmed by 1H-NMR spectroscopy in D2O with 0.1 wt.% DCl (Fig. 2), as indicated by the complete disappearance of the DNP protons. Based on the 1H-NMR spectrum, the k and l values of mPEG-b-(polyHis)2 were 45 and 21, respectively, and the weight fraction of mPEG in mPEG-b-(polyHis)2 was estimated as 0.25 (Fig. 1c). The polydispersity of mPEG-b-(polyHis)2 was estimated based on that of mPEG-b-(poly(DNP-His))2 and should be similarly narrow.
Fig. 2.
1H-NMR spectrum of mPEG-b-(polyHis)2 in D2O with 0.1 wt.% DCl.
These synthesis results indicate that the overall synthetic routes from the block and core components to 3-miktoarm block copolymers are well-designed and well-controlled, resulting in a very narrow molecular weight distribution (Mw/Mn=1.14) and that the synthesized mPEG-b-(polyHis)2 is a structural mimic of phospholipids. Moreover, the synthetic design allowed for a suitable mPEG weight fraction (25 wt.%) in the miktoarm copolymer for polymersome fabrication.13
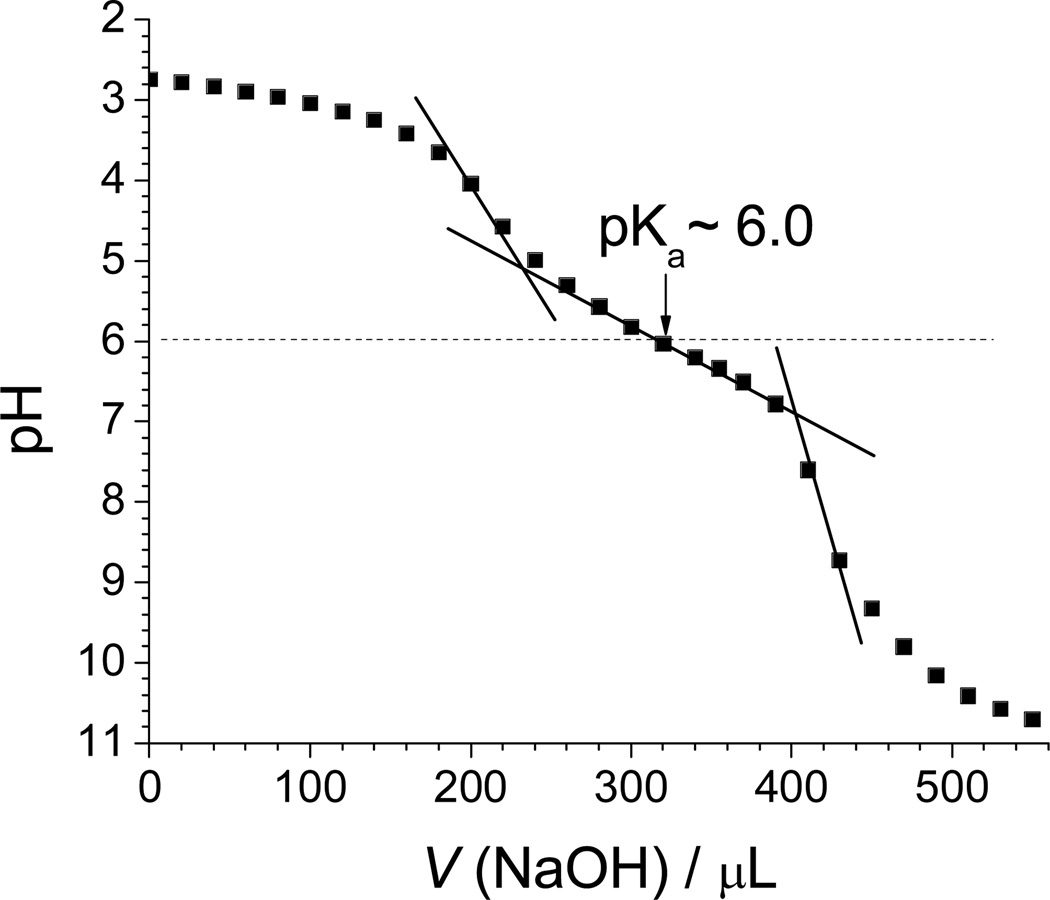
Histidine-based polymers and their derivatives are well-known pH-sensitive polymers and showed reasonable proton buffering.28 Therefore, we evaluated whether mPEG-b-(polyHis)2 has proton buffering capacity in the endolysosomal pH range and assessed the strength of this proton buffering. As shown in Fig. 3, the synthesized mPEG-b-(polyHis)2 exhibited a strong proton buffering capacity within the pH range from neutral to pH 5, and had an apparent pKa of 6.0. The results indicate that the 3-miktoarm copolymer may be able to escape from endosomes/lysosomes. When estimating the buffering power of mPEG-b-(polyHis)2 (namely, mPEG2kDa-b-(polyHis2.9kDa)2 when considering the molecular weights of each block) per unit mass within pH 4–7.4, the proton buffering capacity was found to be 4.2 µmol/mg compared with 3.78 µmol/mg for mPEG2kDa-b-polyHis3.1kDa.36 The histidine-based 3-miktoarm copolymer showed a slightly better proton buffering capacity than the histidine-based diblock copolymer. Considering the weight fractions of the buffering blocks (here, polyHis) in the block copolymers, the two polyHis2.9kDa blocks (i.e., 5.8 kDa of polyHis) in mPEG-b-(polyHis)2 represent 5.65 µmol/mg compared with 6.22 µmol/mg of polyHis3.1kDa in mPEG2kDa-b-polyHis3.1kDa.36 This result supports our previous findings that the proton buffering capacity of polyHis decreases as the molecular weight of polyHis increases.36
Fig. 3.
Acid-base titration of mPEG-b-(polyHis)2 in 150 mM NaCl. The solid lines were drawn to estimate the buffering range and the pKa value.
3.3. Polymersome preparation
To fabricate supramolecular nanostructures, mPEG-b-(polyHis)2 was first dissolved in dimethylsulfoxide (DMSO), a common solvent for both the mPEG and polyHis moieties and then dialyzed against a pH 9.0 borate buffer. Self-assembly of the polymer in aqueous solution was driven by hydrophobic interactions among the polyHis moieties due to the complete deprotonation of the imidazole groups.
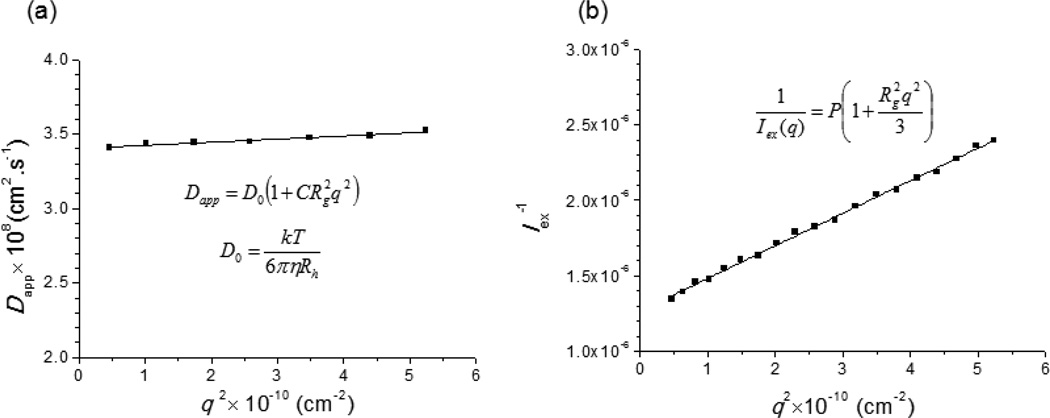
After dialysis, the nanostructures of mPEG-b-(polyHis)2 were characterized by LS-based approaches and confirmed by TEM images. First, LS techniques were used to measure both the hydrodynamic radius (Rh) and the radius of gyration (Rg) using DLS and SLS, respectively, and then to estimate the characteristic radius ratio (Rg/Rh), a parameter that describes the shape of the nanostructure and depends on the polymer architecture, chain conformation, and polydispersity.37, 38 Thus, after monitoring the normalized relaxation time distribution (G(tR)) using DLS mode (Fig. S7), a relationship between the angular dependent diffusion coefficient (Dapp) and the magnitude of scattering (q) of the polymersome was plotted to estimate the intrinsic translational diffusion coefficient (D0 = 3.4 × 10−8 cm2/s) (Fig. 4a). Using the Stokes-Einstein equation and the estimated D0, the Rh was calculated as 70.2 nm. In addition, DLS measurement determined that the size polydispersity index (PDI) was 0.05, indicating a narrow size distribution of the nanostructure. In addition, to calculate Rg, SLS was conducted from 30°~120°, and a partial Zimm plot was generated from the excess intensity of scattered light (Iex(q)) and the magnitude of the scattering (q) (Fig. 4b). The resultant Rg was determined as 71.9 nm. Based on the findings from DLS and SLS, the ratio of Rg to Rh (Rg/Rh) was determined to be 1.02 (71.9 nm/70.2 nm). This value suggests that the mPEG-b-(polyHis)2 nanostructures may be thin-walled hollow spheres (i.e., polymersomes), which generally have Rg/Rh values of unity.39, 40
Fig. 4.
(a) Angle dependence (DLS mode) and (b) a partial Zimm plot (SLS mode) of the mPEG-b-(polyHis)2 nanostructure in borate buffer (0.3 mg/mL, pH 9.0). Solid lines represent linear fits to the data points.
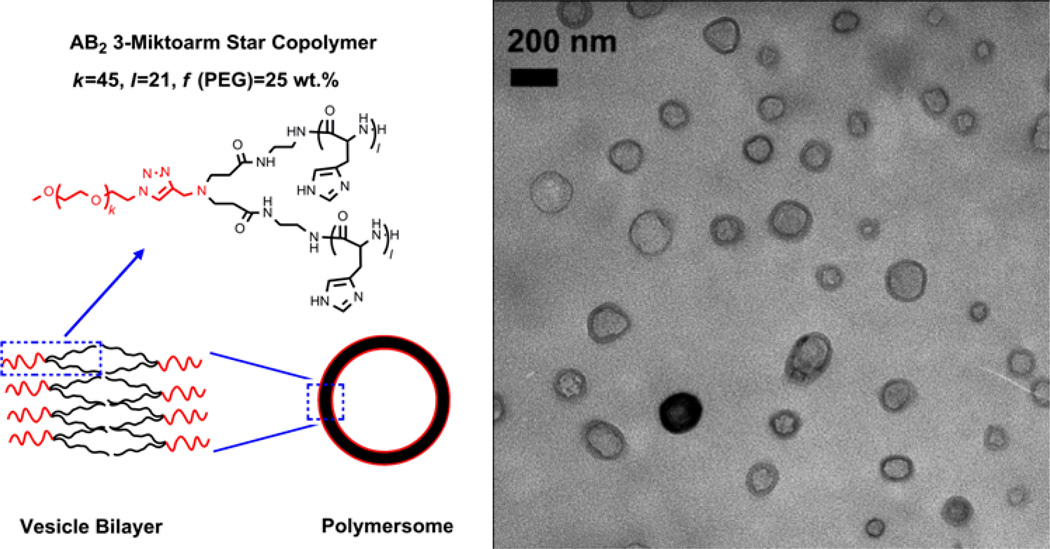
To support this theoretical estimation of the structure of mPEG-b-(polyHis)2 polymersomes, the nanostructures were visualized by TEM imaging to provide additional evidence. As shown in Fig. 5, spherical unilamellar vesicles with diameters of 100–200 nm were clearly observed, consistent with the size determined by DLS. As the polymer possesses a phospholipid-like structure,13 the polymer vesicles formed from mPEG-b-(polyHis)2 might be constructed in a similar way as a liposome. That is, the hydrophilic mPEG arms point “out” to the water on either side of the bilayer while the hydrophobic polyHis arms point “in” to the core of the bilayer (Fig. 5). These findings confirmed that mPEG-b-(polyHis)2 forms a polymersome, unlike the micellar nanostructure formed from its diblock counterpart (i.e., PEG-b-polyHis with a similar PEG fraction).28 These results may strengthen our previous report about the superior ability of the AB2-type architecture to form vesicles.13
Fig. 5.
Micrograph of polymersomes observed by TEM and a schematic illustration of a unilamellar vesicle formed by the miktoarm star copolymer. The degree of polymerization (DP) for ethylene glycol (k) and histidine (l) in each arm as well as the weight fraction (f) of PEG are also indicated.
Immediately after preparation, the surface charge of mPEG-b-(polyHis)2–based polymersomes in borate buffer saline at pH 9.0 was −17.65±1.24 mV. When the polymersomes were exposed to extracellular pHs (e.g., normal blood pH 7.4 and slightly acidic pathological pH 7.0), their negative surface charges were reduced to −4.84±0.09 mV at pH 7.4 and then - 1.42±0.10 mV at 7.0 because the polyHis blocks near the surface become protonated, increasing their cationic character as the pH decreases.
3.4. pH-induced structural transition of polymersomes
Cellular internalization of nanoparticulate drug delivery systems frequently occurs via endocytosis. Therefore, escaping from endosomes/lysosomes is an important step to enable the drugs loaded in the nanoparticles to achieve their therapeutic effects. Liposomes are well known to fuse with endosomal membrane components, resulting in endosomal lysis and release of the loaded drugs.41 Thus, although mPEG-b-(polyHis)2 exhibited a strong buffering capacity in the endolysosomal pH range (Fig. 3), the structural transition of the mPEG-b-(polyHis)2 polymersome was also investigated to determine whether interactions occur between endosomes and the 3-miktoarm block copolymer polymersomes owing to their structural mimicking of phospholipids and liposomes.
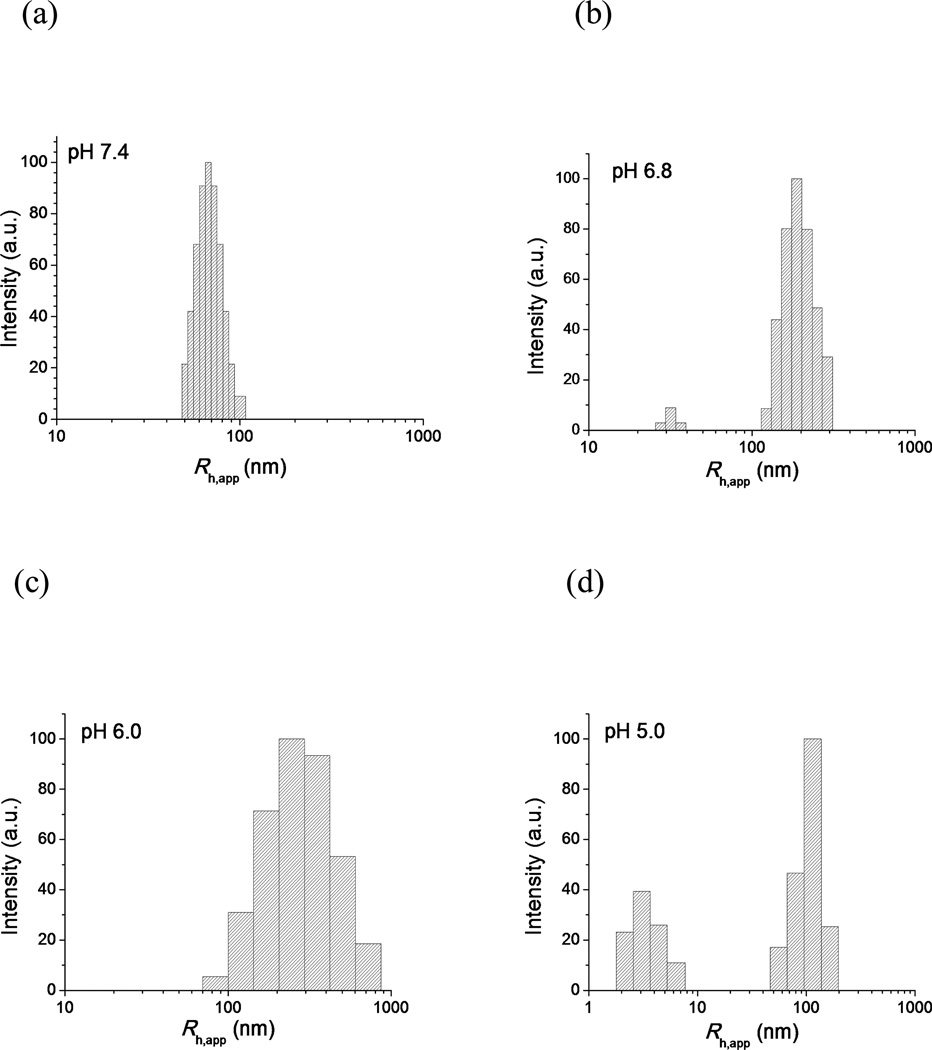
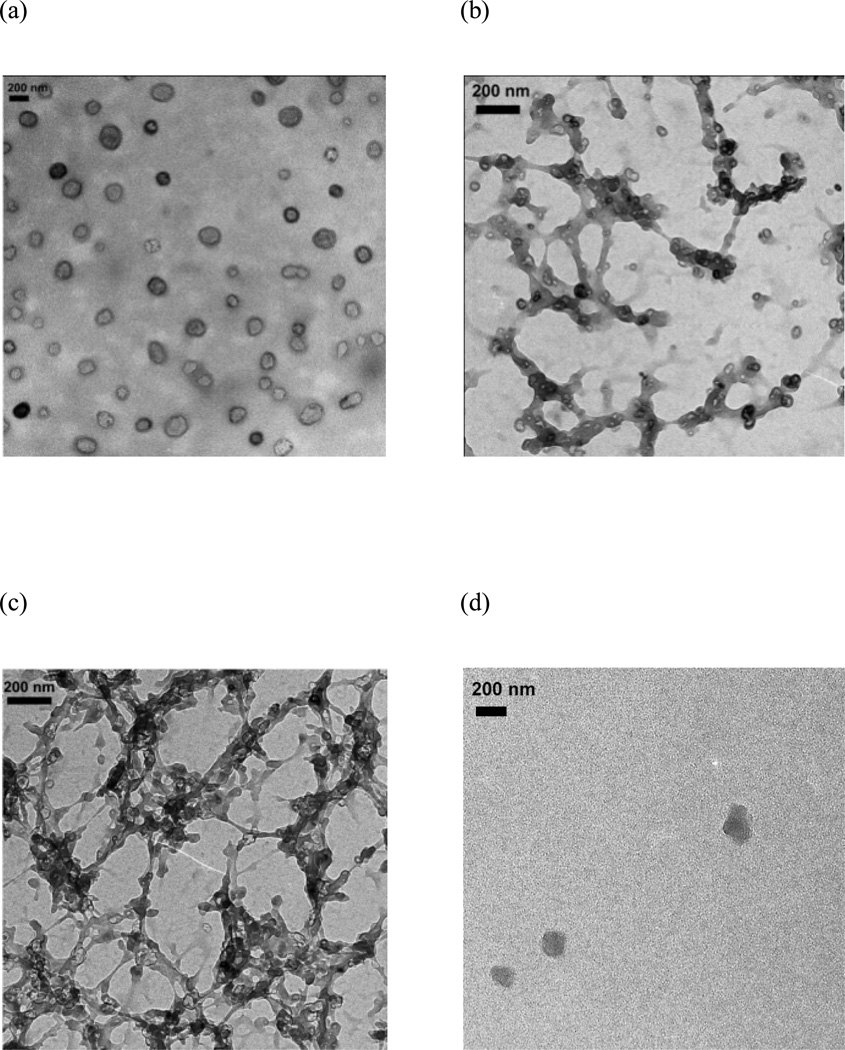
The mPEG-b-(polyHis)2 polymersomes were exposed to various pHs between pH 7.4 and pH 5.0 and their structural transitions were studied by DLS (Fig. 6) and TEM (Fig. 7). At pH 7.4, the DLS histogram of the nanostructure still exhibited a unimodal distribution similar to pH 9.0 (Fig. 6a), and the TEM micrograph also confirmed the presence of a stable vesicular structure (Fig. 7a). Interestingly, when the pH was decreased to 6.8, the polymersomes transformed into elongated and branched nanostructures with a few small spherical vesicles (Fig. 7b), resulting in a bimodal size distribution with apparent hydrodynamic radii (Rh,app) equivalent to ca. 30 nm and 200 nm, respectively (Fig. 6b). As the pH was decreased further to 6.0, small vesicles could hardly be detected among the elongated and branched nanostructures (Fig. 7c), and the DLS histogram showed a unimodal but quite broad Rh,app distribution from ca. 80 nm to 900 nm (Fig. 6c). As the pH was further decreased to 5.0, the branched structures disappeared and only a few micelle-like structures were observed (Fig. 7d), and a bimodal size distribution was detected once again by DLS with Rh,app values of approximately 2.5 nm and 100 nm (Fig. 6d). The larger size should be attributed to the micelles observed by TEM while the smaller radius of several nanometers may be correlated with single polymer chains,42 i.e., mPEG-b-(polyHis)2 unimers that were too small to be visualized by TEM.
Fig. 6.
DLS intensity-weighted apparent hydrodynamic radius (Rh,app) distribution at 90° for the polymer solution (0.3 mg/mL) at (a) pH 7.4, (b) pH 6.8, (c) pH 6.0, and (d) pH 5.0.
Fig. 7.
TEM micrographs of the polymer nanostructures at (a) pH 7.4, (b) pH 6.8, (c) pH 6.0, and (d) pH 5.0.
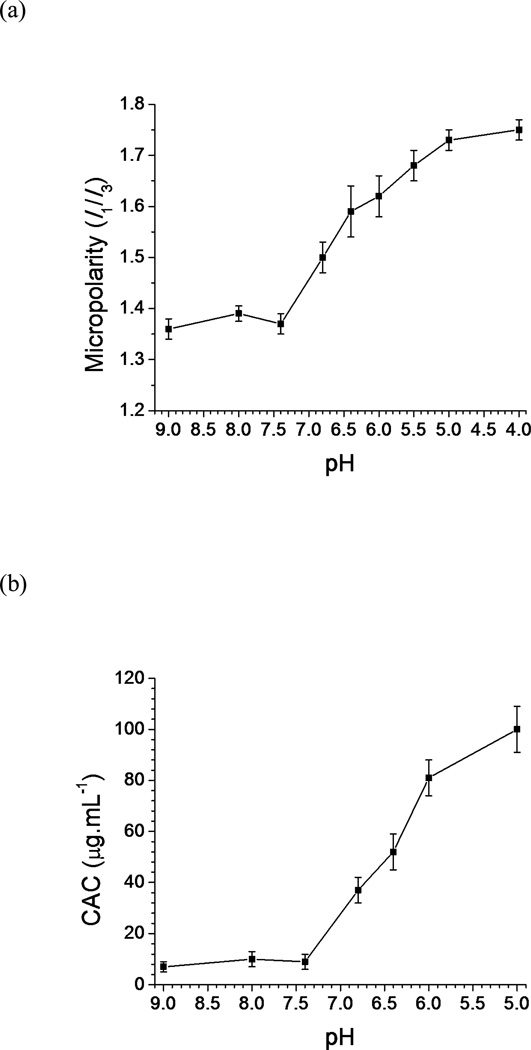
A fluorescence study was performed using pyrene as a hydrophobic fluorescent probe to investigate the physiochemical characteristics of the pH-induced nanostructural transition. As the fluorescence emission spectrum of solubilized pyrene is very sensitive to its environment,43 the ratio of the intensity of the first band to that of the third band (I1/I3, also referred as micropolarity) was monitored as a function of pH (Fig. 8a). No significant change in the I1/I3 value occurred between pH 9.0 and pH 7.4; however, when the pH was decreased to pH 6.8, a remarkable increase was observed in the I1/I3 value. The I1/I3 value continued to increase as the pH was further decreased until a relative plateau was reached below pH 5.0. The increase in I1/I3 from pH 6.8 to pH 5.0 suggested a corresponding increase in the polarity, in other words, a decrease in the hydrophobicity of the microenvironment surrounding pyrene molecules.44
Fig. 8.
Variation in (a) the micropolarity and (b) the critical association value (CAC) as a function of pH for the 0.3 mg/mL polymer solution (mean ± standard deviation; n=3).
The pyrene method was also used to determine the critical association concentration (CAC) of the polymer solution at different pH values (Fig. S8). As shown in Fig. 8b, no significant change in the CAC value was observed between pH 9.0 and pH 7.4. Similar to the trend of the micropolarity, the CAC value sharply increased to 37 µg/mL (4.74 µM) at pH 6.8 and continued to increase as the pH was decreased, until it reached as high as 100 µg/mL (12.8 µM) at pH 5.0. The increase in the CAC with acidification indicates that the mPEG-b-(polyHis)2 nanostructures become increasingly hydrophilic.
The pH-induced structural transition of the mPEG-b-(polyHis)2 nanostructure may be caused by the increasing hydrophilicity of mPEG-b-(polyHis)2 as the pH is decreased. At pH 7.4, the mPEG-b-(polyHis)2 existed as a polymersome with a hydrophilic mPEG block and a hydrophobic polyHis block, with the relatively low CAC (9 µg/mL or 1.15 µM)suggesting good thermodynamic stability during in vivo circulation, making it a desirable drug carrier. However, the micropolarity of pyrene indicates that the hydrophobicity of the polyHis moiety was greatly reduced below pH 6.8 as the imidazole groups were gradually ionized, which increased the electrostatic repulsion and hydration of the hydrophobic segment.45 The change in the hydrophilicity-hydrophobicity balance in mPEG-b-(polyHis)2 nanostructures caused interfacial instability in the vesicular structure. To stabilize hydrophobic/hydrophilic interface, structural transitions occurred from spherical vesicles (here, polymersomes) to cylindrical micelles, spherical micelles, and then finally soluble unimers.46 The mPEG-b-(polyHis)2 nanostructure at pH 6.8 could arise because of the coexistence of untransitioned polymersomes and transitioned cylindrical micelles, with one polymersome potentially acting as a core linking with several cylindrical micelles. As the pH was decreased further, all polymersomes disappeared and some cylindrical micelles transformed into spherical micelles. Thus, at pH 6.0, the mPEG-b-(polyHis)2 nanostructure may include interconnecting nanostructures of cylindrical micelles and spherical micelles. A further decrease in pH led to the coexistence of spherical micelles and unimers (at pH 5.0), but with continuing acidification, mPEG-b-(polyHis)2 became a water-soluble unimer. These structural transitions of mPEG-b-(polyHis)2 nanostructures in the endolysosomal pH range could indicate that mPEG-b-(polyHis)2 polymersomes can fuse with endolysosomal membranes and that therapeutic payloads delivered by these polymersomes can be released from the endolysosomes.
3.5. Hydrophilic drug encapsulation and release
A self-quenching fluorescent probe, 5(6)-carboxyfluorescein (CF), was used as a hydrophilic model dye47 to test the encapsulation capability of the mPEG-b-(polyHis)2 polymersome. CF was physically encapsulated into the aqueous lumen of the polymersome and the dye-loaded polymersomes were separated by gel filtration as described in the experimental section. Calculation of the encapsulation content and efficiency for vesicles was different from that of polymeric micelles and is not only dependent on the vesicle size but is also very dependent on drug and vesicle concentrations. Therefore, as another parameter, the encapsulation volume that is independent on the drug and vesicle concentrations is more commonly used to evaluate the encapsulation capacity of vesicles. The encapsulation efficiency, which is defined as the encapsulated volume per milligram of the polymer used,48 was determined to be 0.92±0.10 µL/mg and it is comparable to that of liposomes (1–3 µL/mg) with similar sizes.49
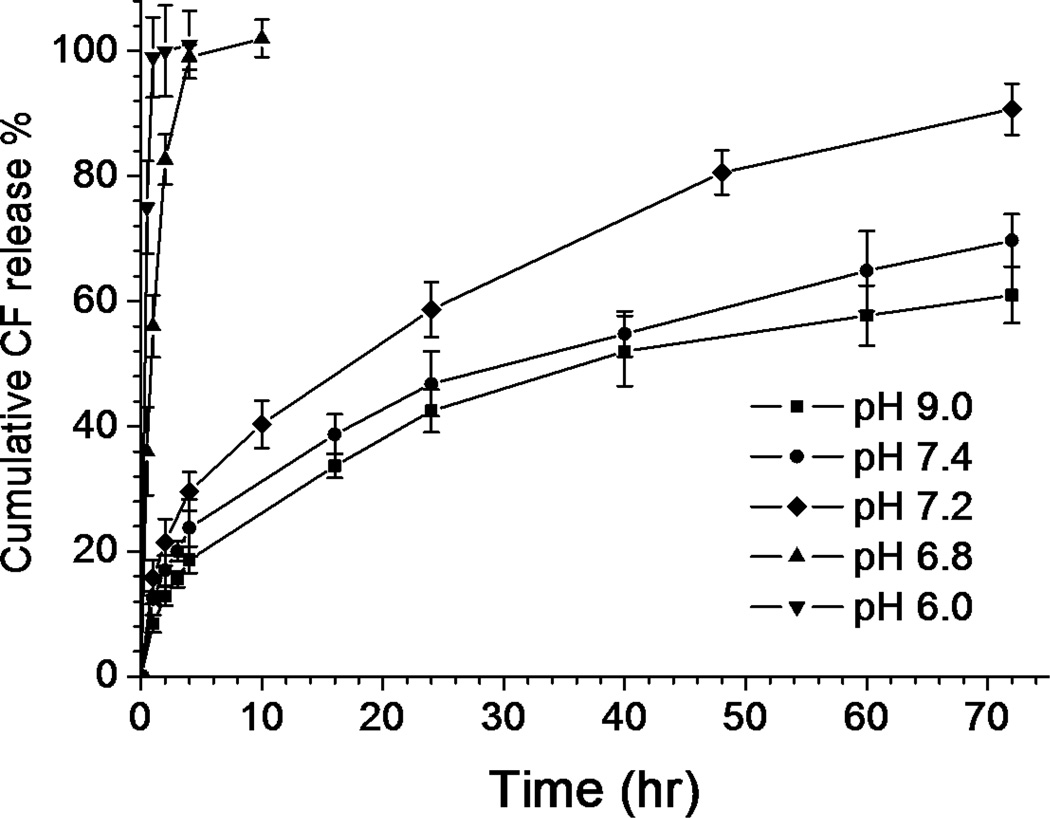
In vitro release of the encapsulated dye at different pHs upon dilution by a factor of ten was carried out under the conditions of T=37°C and 300 mOsm/L. In Fig. 9, the encapsulated CF showed sustained release profiles at pH 9.0, 7.4 and 7.2 within the studied time scale (72 h). At pH 7.4, around 30% of the dye remained inside the polymersome after 72 h. However, a significantly accelerated release was observed at pH 6.8 and the encapsulated dye was almost completely released after 4 h. The transition became more obvious as pH further decreased to 6.0, in which case 100% dye release was achieved after only 1 h. It is interesting to note that the transition in the release profile occurred simultaneously with the pH-induced morphological transition; therefore it is reasonable to consider that the enhanced dye release at the lower pHs was triggered by the destabilization of the polymersome bilayers as a result of the protonation of the polyHis moiety. More importantly, the release profile indicates a desirable pH sensitivity for intracellular delivery; the carrier is able to hold the majority of its cargo over an extended period of time during systemic circulation (pH 7.4) while rapid release of the payload can be triggered once in the acidic environment of endosomes (pH 5.5–6.5).50, 51
Fig. 9.
In vitro release profiles of the encapsulated CF in the polymersome at different pH (mean ± standard deviation; n=3).
3.6. Cytotoxicity of polymersomes
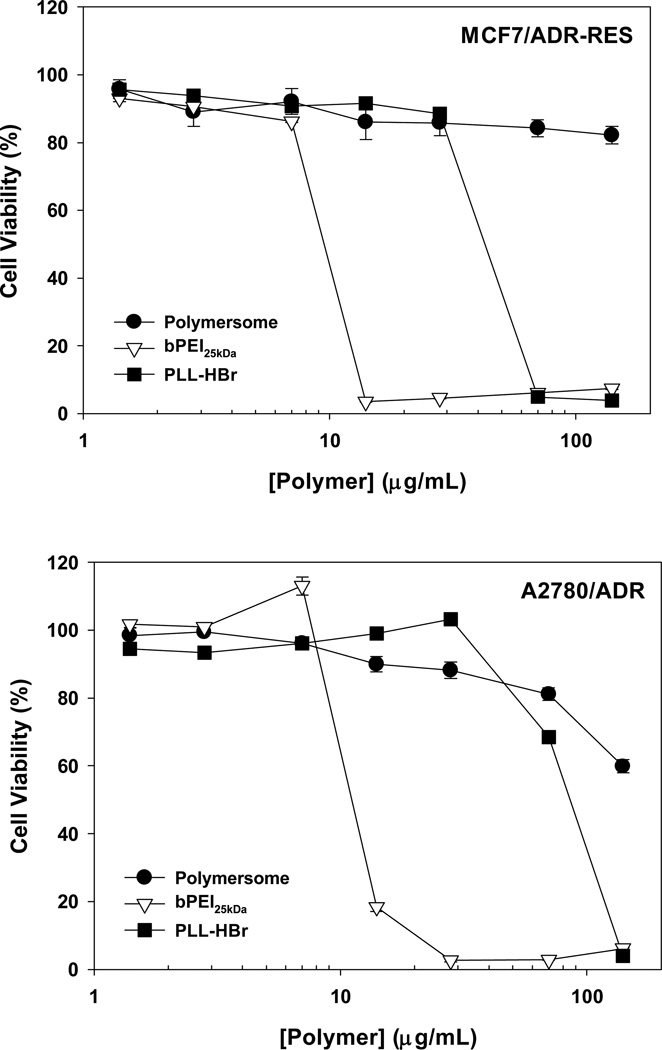
It is important to assess the biocompatibility of these pH-sensitive mPEG-b-(polyHis)2 polymersomes to support their application to in vitro and in vivo studies and eventually to clinical studies. Thus, two kinds of MDR cells (5000 cells seeded for each cell line) were exposed to the polymersomes for 5 d. Their viabilities were over 80% when the cells were treated with 70 µg/mL polymersomes (Fig. 9). In the case of MCF7/ADR-RES cells, mPEG-b-(polyHis)2 polymersomes at 140 µg/mL did not significantly harm the cell viability (> 80%). On the other hand, in MCF7/ADR-RES cells, representative polycations such as bPEI and PLL killed half of all cells that were dosed at ~10 µg/mL and ~26 µg/mL (42 µg/mL for PLL·HBr), respectively (Fig. 10). Also, for A2780/ADR cells, the IC50 concentrations were ~11 µg/mL for bPEI and ~52 µg/mL for PLL (85 µg/mL for PLL·HBr) (Fig. 10). In comparison, our results indicate that mPEG-b-(polyHis)2 polymersomes have negligible cytotoxicity.
Fig. 10.
Cytotoxicity of mPEG-b-(polyHis)2 polymersomes and representative polycations against MCF7/ADR-RES and A2780/ADR cells after a 5 d incubation (mean ± standard error; n=6).
The 3-miktoarm mPEG-b-(polyHis)2 copolymers formed polymeric vesicles, which mimic phospholipid liposomes and loaded comparable encapsulation volumes of a hydrophilic model drug. The polymersome nanostructures were transformed to micelles by a decrease in pH. The differential response to varying pH environments may provide more desirable release profiles, such as a slow drug release in blood or extracellular pHs but an accelerated drug release in intracellular endosomal compartments. However, the colloidal stability of polymersomes is still a very important consideration in blood and extracellular environments and should be investigated because serum proteins can interact with polymersomes and sequentially disrupt the polymersomal nanostructures, leading to unwanted drug release.
4. Conclusion
We designed and successfully synthesized 3-miktoarm mPEG-b-(polyHis)2 copolymers that mimic phospholipid structures. The miktoarm copolymer self-assembled in aqueous environments to form nano-sized polymersomes with a strong proton buffering capacity and a low cytotoxicity; these poymersomes were stable above pH 7.4 but underwent a vesicle-to-micelle transition between pH 6.8 and pH 5.0 as a result of the gradual protonation of the imidazole groups. Drug-loaded polymersomes can thus provide pH-dependent drug release. In conclusion, these biocompatible, pH-sensitive polymersomes represent promising drug carriers.
Supplementary Material
Acknowledgements
This work was supported by NIH grant CA101850. The authors are grateful to Kwonhyeok Yoon and Hana Cho for technical assistant and Darren Stirland and Joseph Nichols for careful editorial aid.
References
- 1.Goren D, Grob M, Lorenzoni P, Burger MM. Tumour Biol. 1997;18:341–349. doi: 10.1159/000218048. [DOI] [PubMed] [Google Scholar]
- 2.Andresen TL, Jensen SS, Jorgensen K. Prog Lipid Res. 2005;44:68–97. doi: 10.1016/j.plipres.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Discher DE, Eisenberg A. Science. 2002;297:967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- 4.Discher DE, Ortiz V, Srinivas G, Klein ML, Kim Y, Christian D, Cai S, Photos P, Ahmed F. Prog Polym Sci. 2007;32:838–857. doi: 10.1016/j.progpolymsci.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christian DA, Cai S, Bowen DM, Kim Y, Pajerowski JD, Discher DE. Eur J Pharm Biopharm. 2009;71:463–474. doi: 10.1016/j.ejpb.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine DH, Ghoroghchian PP, Freudenberg J, Zhang G, Therien MJ, Greene MI, Hammer DA, Murali R. Methods. 2008;46:25–32. doi: 10.1016/j.ymeth.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng F, Zhong Z, Feijen J. Biomacromolecules. 2009;10:197–209. doi: 10.1021/bm801127d. [DOI] [PubMed] [Google Scholar]
- 8.Lee JC, Bermudez H, Discher BM, Sheehan MA, Won YY, Bates FS, Discher DE. Biotechnol Bioeng. 2001;73:135–145. doi: 10.1002/bit.1045. [DOI] [PubMed] [Google Scholar]
- 9.Letchford K, Burt H. Eur J Pharm Biopharm. 2007;65:259–269. doi: 10.1016/j.ejpb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Lodge TP, Rasdal A, Li Z, Hillmyer MA. J Am Chem Soc. 2005;127:17608–17609. doi: 10.1021/ja056841t. [DOI] [PubMed] [Google Scholar]
- 11.Khanna K, Varshney S, Kakkar A. Polym Chem. 2010;1:1171–1185. [Google Scholar]
- 12.Soliman GM, Sharma R, Choi AO, Varshney SK, Winnik FM, Kakkar AK, Maysinger D. Biomaterials. 2010;31:8382–8392. doi: 10.1016/j.biomaterials.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 13.Yin H, Kang SW, Bae YH. Macromolecules. 2009;42:7456–7464. [Google Scholar]
- 14.Lee ES, Gao Z, Bae YH. J Control Release. 2008;132:164–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed F, Discher DE. J Control Release. 2004;96:37–53. doi: 10.1016/j.jconrel.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed F, Pakunlu RI, Srinivas G, Brannan A, Bates F, Klein ML, Minko T, Discher DE. Mol Pharmaceut. 2006;3:340–350. doi: 10.1021/mp050103u. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Meng F, Cheng R, Zhong Z. J Control Release. 2010;142:40–46. doi: 10.1016/j.jconrel.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Lomas H, Canton I, MacNeil S, Du J, Armes SP, Ryan AJ, Lewis AL, Battaglia G. Adv Mater. 2007;19:4238–4243. [Google Scholar]
- 19.Du J, Tang Y, Lewis AL, Armes SP. J Am Chem Soc. 2005;127:17982–17983. doi: 10.1021/ja056514l. [DOI] [PubMed] [Google Scholar]
- 20.Kopecek J. Eur J Pharm Sci. 2003;20:1–16. doi: 10.1016/s0928-0987(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 21.Adams ML, Lavasanifar A, Kwon GS. J Pharm Sci. 2003;92:1343–1355. doi: 10.1002/jps.10397. [DOI] [PubMed] [Google Scholar]
- 22.Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Nat Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Hernandez J, Lecommandoux S. J Am Chem Soc. 2005;127:2026–2027. doi: 10.1021/ja043920g. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Hayward RC. J Am Chem Soc. 2008;130:7496–7502. doi: 10.1021/ja801268e. [DOI] [PubMed] [Google Scholar]
- 25.Holowka EP, Pochan DJ, Deming TJ. J Am Chem Soc. 2005;127:12423–12428. doi: 10.1021/ja053557t. [DOI] [PubMed] [Google Scholar]
- 26.Holowka EP, Sun VZ, Kamei DT, Deming TJ. Nat Mater. 2007;6:52–57. doi: 10.1038/nmat1794. [DOI] [PubMed] [Google Scholar]
- 27.Moghimi SM, Hunter AC, Murray JC. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 28.Lee ES, Shin HJ, Na K, Bae YH. J Control Release. 2003;90:363–374. doi: 10.1016/s0168-3659(03)00205-0. [DOI] [PubMed] [Google Scholar]
- 29.Lee JW, Kim BK, Kim HJ, Han SC, Shin WS, Jin SH. Macromolecules. 2006;39:2418–2422. [Google Scholar]
- 30.Kang HC, Bae YH. Adv Funct Mater. 2007;17:1263–1272. [Google Scholar]
- 31.Kang HC, Kang HJ, Bae YH. Biomaterials. 2011;32:1193–1203. doi: 10.1016/j.biomaterials.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu B. Laser light scattering. New York: Academic Press, Inc.; 1990. [Google Scholar]
- 33.Provencher SW. Biophys J. 1976;16:27–41. doi: 10.1016/S0006-3495(76)85660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lelkes PI. In: Liposome Technology. Gregoriadis G, editor. Boca Raton: CRC Press; 1984. pp. 225–256. [Google Scholar]
- 35.Peng SM, Chen Y, Hua C, Dong CM. Macromolecules. 2009;42:104–113. [Google Scholar]
- 36.Park WR, Kim D, Kang HC, Na K, Bae YH. submitted. [Google Scholar]
- 37.Savin G, Burchard W, Luca C, Beldie C. Macromolecules. 2004;37:6565–6575. [Google Scholar]
- 38.Nie T, Zhao Y, Xie Z, Wu C. Macromolecules. 2003;36:8825–8829. [Google Scholar]
- 39.Zhou S, Burger C, Chu B, Sawamura M, Nagahama N, Toganoh M. Science. 2001;291:1944–1947. doi: 10.1126/science.291.5510.1944. [DOI] [PubMed] [Google Scholar]
- 40.Liu T, Diemann E, Li H, Dress AW, Muller A. Nature. 2003;426:59–62. doi: 10.1038/nature02036. [DOI] [PubMed] [Google Scholar]
- 41.Torchilin VP. Nat Rev Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 42.Vamvakaki M, Palioura D, Spyros A, Armes SP, Anastasiadis SH. Macromolecules. 2006;39:5106–5112. [Google Scholar]
- 43.Kalyanasundaram K, Thomas JK. J Am Chem Soc. 1977;99:2039–2044. [Google Scholar]
- 44.Chung JE, Yokoyama M, Okano T. J Control Release. 2000;65:93–103. doi: 10.1016/s0168-3659(99)00242-4. [DOI] [PubMed] [Google Scholar]
- 45.Yin H, Lee ES, Kim D, Lee KH, Oh KT, Bae YH. J Control Release. 2008;126:130–138. doi: 10.1016/j.jconrel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, Choi YJ, Lim YB. Expert Opin Drug Deliv. 2010;7:341–351. doi: 10.1517/17425240903559841. [DOI] [PubMed] [Google Scholar]
- 47.Meng F, Engbers GH, Feijen J. J. Control. Release. 2005;101:187–198. doi: 10.1016/j.jconrel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 48.Perkins WR, Minchey SR, Ahl PL, Janoff AS. Chem. Phys. Lipids. 1993;64:197–217. doi: 10.1016/0009-3084(93)90066-c. [DOI] [PubMed] [Google Scholar]
- 49.Zhang XM, Patel AB, de Graaf RA, Behar KL. Chem. Phys. Lipids. 2004;127:113–120. doi: 10.1016/j.chemphyslip.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 50.Grabe M, Oster G. J. Gen. Physiol. 2001;117:329–344. doi: 10.1085/jgp.117.4.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson P, Jones AT, Stephens DJ. Adv. Drug. Deliv. Rev. 2005;57:43–61. doi: 10.1016/j.addr.2004.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.