SUMMARY
Hepatocyte Nuclear Factor (HNF)4α is a central regulator of gene expression in cell types that play a critical role in metabolic homeostasis, including hepatocytes, enterocytes, and pancreatic β-cells. Although fatty acids were found to occupy the HNF4α ligand-binding pocket and proposed to act as ligands, there is controversy about both the nature of HNF4α ligands as well as the physiological role of the binding. Here, we report the discovery of potent synthetic HNF4α antagonists through a high-throughput screen for effectors of the human insulin promoter. These molecules bound to HNF4α with high affinity and modulated the expression of known HNF4α target genes. Notably, they were found to be selectively cytotoxic to cancer cell lines in vitro and in vivo, although in vivo potency was limited by suboptimal pharmacokinetic properties. The discovery of bioactive modulators for HNF4α raises the possibility that diseases involving HNF4α, such as diabetes and cancer, might be amenable to pharmacologic intervention by modulation of HNF4α activity.
INTRODUCTION
HNF4α is a member of the nuclear receptor (NR) superfamily of transcription factors and binds as a homodimer to a relatively degenerate consensus DNA sequence consisting of two direct repeats separated by one or two nucleotides (Bolotin et al., 2010). It is expressed at high levels in hepatocytes, enterocytes, pancreatic epithelial cells (including β-cells), and renal tubular epithelial cells (Drewes et al., 1996; Jiang et al., 2003). In these cells, it sits at the heart of a transcriptional regulatory network that controls the expression of many genes, but particularly those involved in intermediary metabolism and maintenance of epithelial differentiation.
Because of the important role of HNF4α in regulating metabolic processes such as glucose and lipid homeostasis (Hayhurst et al., 2001; Odom et al., 2004; Stoffel and Duncan, 1997), and the high percentage of pharmaceuticals that target NR transcription factors, there has been a great interest in developing synthetic ligands for HNF4α (Hertz et al., 2001; Le Guevel et al., 2009; Sladek, 2011). However, this has proven to be difficult. While medium and long chain fatty acids (MCFAs and LCFAs, respectively) are invariably found bound in the HNF4α ligand binding pocket (LBP) in structural studies of HNF4α purified from bacteria (Dhe-Paganon et al., 2002; Duda et al., 2004), there has been little evidence that they modulate HNF4α activity, and attempts to study how different fatty acids affect the conformation of HNF4α have been stymied by the fact that ligand exchange in vitro is very poor (Dhe-Paganon et al., 2002; Wisely et al., 2002). This has raised questions about the extent to which HNF4α function is regulated by ligand binding, versus a model in which small molecules bound in the ligand binding pocket (LBP) play a structural role (Sladek, 2011).
Recently, linoleic acid was identified in the LBP of HNF4α purified from COS-7 cells. In vivo, linoleic acid was present in the LBP of HNF4α from mice that were in the fed state, yet was absent in fasted mice, suggesting that ligand binding is regulated (Yuan et al., 2009). A study in Drosophila using an HNF4α ligand-binding domain (LBD) sensor found that HNF4α LBD activation was highly modulated, although the nature of the ligand remained undetermined (Palanker et al., 2009). Because of the limited evidence indicating that ligand binding influences the state of transcriptional activity mediated by HNF4α (Yuan et al., 2009), studies of HNF4α have been restricted to genetic deletion (Chen et al., 1994; Duncan et al., 1997; Gupta et al., 2005; Hayhurst et al., 2001) or overexpression (Carter et al., 1993; Harnish et al., 1996; Inoue et al., 2002).
Previously, we described an assay for insulin promoter modulators based on a cell line derived from human fetal islets, T6PNE, which was engineered to express the β-cell transcription factors PDX-1, NeuroD1, and E47 (as a fusion protein with a modified estrogen receptor LBD to render it tamoxifen-inducible; E47MER)(Kiselyuk et al., 2010). Induction of E47 by tamoxifen resulted in dose-responsive expression of the insulin gene, as well as a number of other genes expressed in β-cells. T6PNE cells were adapted for high-throughput screening by transduction with a lentiviral vector expressing green fluorescent protein under the control of the human insulin promoter (Hao et al., 2006).
Here, as part of a continuing effort to discover novel regulators of the insulin promoter as tools to study diabetes, we used the T6PNE insulin promoter assay to screen a diverse synthetic chemical library. A screening hit, BIM5078, potently repressed insulin expression in that assay. BIM5078 bound to HNF4α with high affinity and modulated HNF4α target genes and metabolic processes controlled by HNF4α. Interestingly, BIM5078 and a related analog, BI6015, were selectively cytotoxic to transformed cells in vitro. In vivo, BI6015 induced apoptosis of a human hepatocellular carcinoma cell line. Further development of related compounds could lead to pharmacologic therapies for a variety of diseases, including diabetes and cancer, as well as provide a powerful tool for studying the physiological role of HNF4α.
RESULTS
Identification of a small molecule inhibitor of the insulin promoter: BIM5078
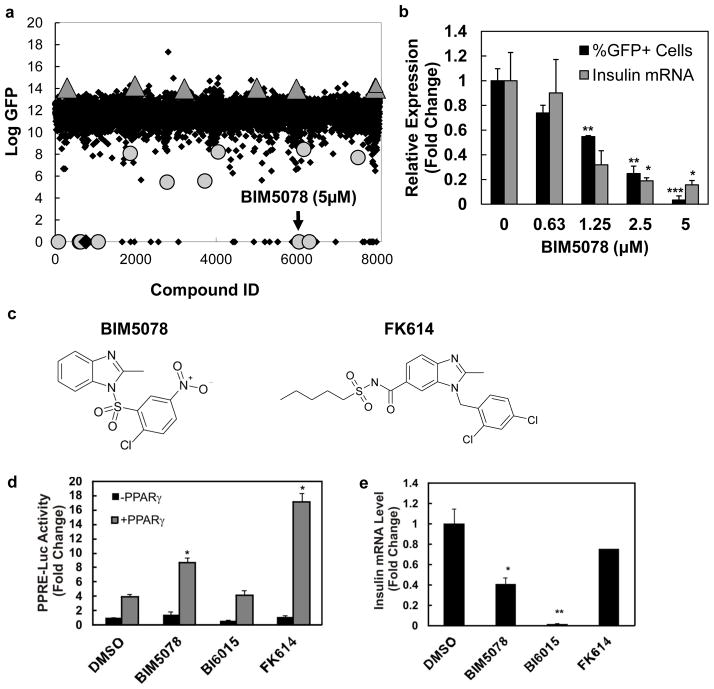
To identify molecules with novel mechanisms of action on the insulin promoter, a high-throughput screen of structurally diverse compounds, consisting of a subset of the ChemBridge DiverSet Library, was conducted for inhibition of the insulin promoter in T6PNE cells (Kiselyuk et al., 2010) (Fig. 1a, Supplementary Table 1). As previously described, primary hits were subjected to secondary counterscreens to assess potency on the endogenous insulin promoter and the modified estrogen receptor (Kiselyuk et al.) (Supplementary Fig. 1). From the screen, we identified one small molecule, 1-(2′-chloro-5′-nitrobenzenesulfonyl)-2-methylbenzimidazole, hereafter referred to as BIM5078, that passed the counterscreen for lack of tamoxifen-like activity on the modified estrogen receptor (Kiselyuk et al., 2010), and exhibited dose-responsive inhibition of endogenous insulin expression, with an IC50 = 930 nM (Fig. 1b).
Figure 1. Identification of BIM5078, a novel regulator of insulin gene transcription.
(a) High-throughput screen for compounds that modulate insulin promoter activity in T6PNE. A subset of the Chembridge DiverSet library of chemically diverse small molecules was used in a screen of T6PNE cells for compounds that modulated the activity of a human insulin promoter-GFP transgene. (b) BIM5078-mediated repression of insulin promoter activity is dose-dependent. T6PNE cells were exposed to the indicated concentration of BIM5078 for 48 hours. Effects of BIM5078 on the exogenous insulin promoter in T6PNE is reported as percent GFP+ cells, as determined by imaging the green channel and normalizing to the total number of cells per well. Endogenous insulin promoter activity was probed through quantitative real-time PCR (Q-PCR) for insulin and GAPDH mRNA. Insulin mRNA values are reported as normalized to GAPDH to control for nonspecific compound effects. Values represent the mean ± SE, n=3. *p<0.05, **p<0.01, ***p<0.001. (c) Structural homology between BIM5078 and PPARγ agonist FK614. (d) PPRE activation. HeLa cells were co-transfected with the PPRE reporter plasmid PPRE-Luc in the absence and presence of a PPARγ expression vector. BIM5078 and FK614, but not BI6015, activated PPRE-Luc in HeLa cells when co-transfected with a PPARγ expression vector. Values represent the mean ± SE, n=3. *p<0.005. (e) BIM5078 and BI6015 repressed endogenous insulin mRNA (normalized to 18S rRNA). FK614 had no effect on endogenous insulin expression. Values represent the mean ± SE, n=3. *p<0.05, **p<0.005, NS = no significance.
BIM5078 is structurally similar to FK614, a PPARγ agonist
A major problem with cell-based phenotypic as opposed to biochemical high-throughput screens is the difficulty in identifying the precise molecular target of small molecules that have the desired effect in the assay. To examine potential targets for BIM5078, we conducted a chemoinformatic analysis of BIM5078 that revealed structural similarity to an atypical PPARγ agonist, FK614, formerly evaluated as a therapeutic for type II diabetes (Fujimura et al., 2005) (Fig. 1c). Accordingly, BIM5078 and FK614 were tested in a PPAR response element (PPRE)-luciferase reporter assay. Consistent with their structural similarity, both BIM5078 and FK614 had activity as PPARγ agonists (Fig. 1d). BIM5078 activated the PPRE 2-fold, and activation was enhanced nearly 4-fold by co-transfection with a PPARγ expression vector (Fig. 1d). While both BIM5078 and FK614 were potent PPARg agonists, FK614 had no effect on insulin promoter activity (Fig. 1e), suggesting that PPARg activation was not responsible for the effect of BIM5078 on the insulin promoter. In addition, no change in luciferase activity was found when PPARα and PPARδ were co-transfected in the PPRE-luciferase reporter assay in the presence of BIM5078. Finally, as described in more detail below, BI6015, a compound structurally related to BIM5078, potently inhibited the insulin promoter, but lacked activity as a PPARγ agonist (Fig. 1d, e). Taken together, these data suggested that the repressive effect of BIM5078 on the insulin promoter was due to a target other than the PPARs.
BIM5078 bound directly to HNF4α and interacted with the LBP in situ
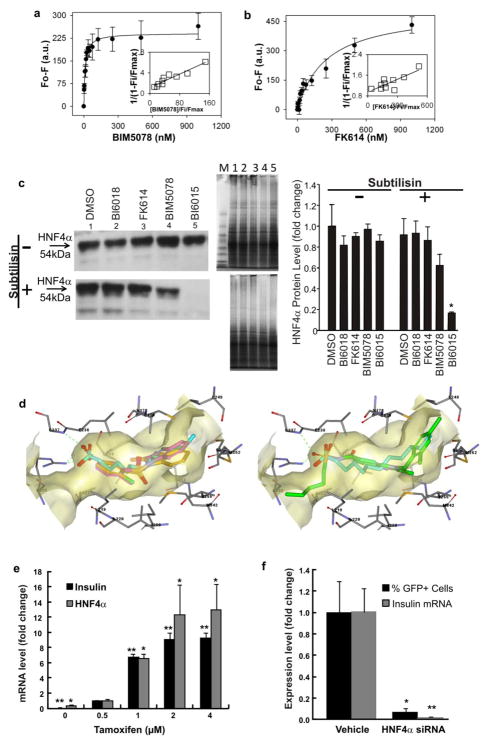
The only transcription factor known to interact with PPREs other than PPARs and their binding partner RXR is HNF4α (Nakshatri and Bhat-Nakshatri, 1998; Nicolas-Frances et al., 2000; Winrow et al., 1994), another member of the NR superfamily. Accordingly, we measured the direct binding of BIM5078 and FK614 to HNF4α. Binding was determined using full-length HNF4α and monitoring the intrinsic fluorescence of its aromatic amino acids Tyr/Trp (Petrescu et al., 2002).
The resultant EC50 ± standard errors were 11.9 ± 3.1 nM for BIM5078 (Fig. 2a) and 254 ± 56 nM for FK614 (Fig. 2b). Additional analysis of the data using the Hill equation demonstrated that the Hill coefficient for BIM5078 was 0.9 ± 0.3, i.e. close to unity, consistent with a single binding complex between the compound and HNF4α. The value of the Hill coefficient for FK614 was 0.6 ± 0.1, suggesting that FK614 was likely to have an additional binding mode characterized by high-micromolar affinity. To support this notion, we observed that the amplitude of the fluorescence change (Fmax) in the FK614 binding curve was almost double that of BIM5078, suggesting that some additional tryptophans might be affected upon FK614 binding. It is possible that hydrophobic groups present on FK614 resulted in non-specific binding elsewhere on the surface of HNF4α.
Figure 2. BIM5078 and BI6015 repress insulin promoter activity through HNF4α antagonism.
(a, b) Direct binding based on quenching of HNF4α aromatic amino acid fluorescence emission. Full-length HNF4α protein (100nM) in 2mL PBS was titrated with increasing concentrations of BIM5078 (a) or FK614 (b). The data are presented as the change in fluorescence intensity (Fo – F) plotted as a function of ligand concentration. Values represent the mean ± SE, n=3. Insets, linear plot of the binding curve from each panel. (c) DARTS assay. HepG2 cells were treated with DMSO (lane 1), BI6018 (lane 2), FK614 (lane 3), BIM5078 (lane 4) or BI6015 (lane 5) at a concentration of 20μM for 24hr. Total cell protein was extracted and each sample was split into three aliquots for proteolysis without (−) or with (+) subtilisin (left panels) or for Coomassie (InstantBlue) staining (middle panels) as a control to ensure that the compounds did not induce nonspecific proteolysis. Lane M-MW markers. Western blots were quantified using ImageJ software, demonstrating a statistically significant effect of BI6015 on subtilisin sensitivity. Values represent the mean ± SE of 3 biological replicates. *p<0.05. (d) Docking of BIM5078 (tan, left panel) BI6015 (violet, left panel), and FK614 (green, right panel) in the LBD of HNF4α with linoleic acid (cyan) crystallized. The nitro group of BIM5078 and BI6015 is predicted to form hydrogen bonds with Arg226 and Gly237. In this pose, the aryl chloro (BIM5078) and methyl (BI6015) groups occupy a hydrophobic pocket lined with Leu219, Leu 200 and Met342. The benzimidazole core occupies the same hydrophobic pocket as the carbon chain of the fatty acid. (e) HNF4α and insulin gene expression were upregulated when E47 was induced by tamoxifen in T6PNE which has a low level of insulin (Kiselyuk et al., 2010) and HNF4α expression at baseline. Differences in expression were measured relative to 0.5μM tamoxifen by Student’s t-test (*p<0.05, **p<0.001). (f) HNF4α siRNA repressed both exogenous and endogenous insulin promoter activity in T6PNE cells. Differences in expression between vehicle (DMSO or scrambled siRNA) and HNF4α siRNA were measured by Student’s t-test (*p<0.05, **p<0.01). Insulin and HNF4α mRNA levels in e and f were normalized to GAPDH.
The x-ray crystal structures of the human and rat HNF4α LBDs were reported some time ago, and it was noted that free fatty acids (FFAs) were always bound in the absence of exogenously added ligand (Dhe-Paganon et al., 2002; Duda et al., 2004). Using the crystal structure (PDB Code 1m7w), BIM5078 was docked into the LBD of HNF4α using the GOLD docking algorithm (Jones et al., 1997)(Fig. 2d, tan molecule in left panel). The high GoldScore (43.6) suggested that it is reasonable for BIM5078 to bind in the LBP in a position similar to that of the putative endogenous ligand (i.e., FFA). However, FK614, the PPARγ agonist that does not affect insulin promoter activity, did not dock well in the LBP (GoldScore 20.8)(Fig. 2d, green molecule in right panel). The primary contributing factor to the poor docking was that significant internal torsional strain was required for it to fit into the LBP.
BIM5078 mediated insulin promoter repression through HNF4α
For HNF4α to be the target of BIM5078, it must be expressed in T6PNE cells. To test this prediction, we conducted quantitative RT-PCR for HNF4α in T6PNE cells. While expressed at a very low level at baseline, it was induced more than 40-fold by E47 through tamoxifen administration in a saturable manner (Fig. 2e).
A further prediction of a model in which HNF4α acts on the insulin promoter is that direct inhibition of HNF4α expression by siRNA should inhibit insulin promoter activity. As predicted, transfection of HNF4α siRNA into T6PNE cells led to potent inhibition of insulin promoter activity both from the endogenous promoter as well as from the human insulin promoter-eGFP transgene (Fig. 2f). Taken together, these data support the notion that endogenous HNF4α may have a role in maintaining the activation state of the human insulin promoter in T6PNE cells.
BIM5078 affected the expression of known HNF4α target genes
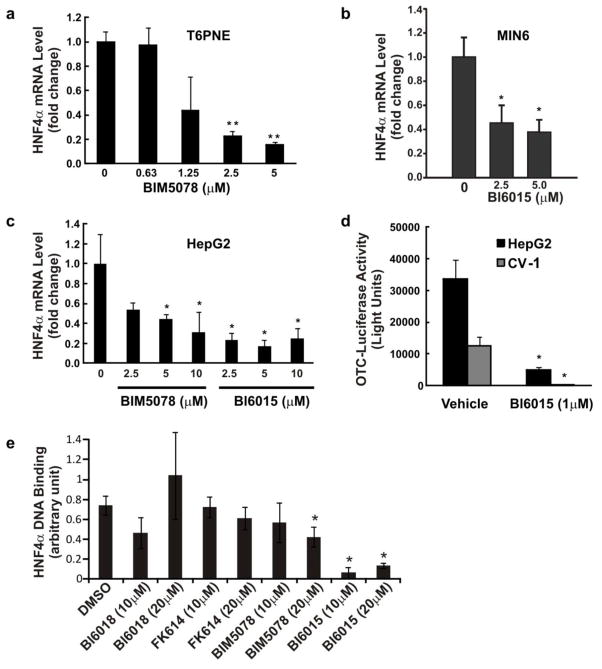
To provide additional evidence that BIM5078 inhibited HNF4α, we examined its effect on known HNF4α target genes. HNF4α autoregulates its own transcription through a complex feedback loop between HNF4α and HNF1α, part of which involves direct binding of HNF4α to its own promoter (Bailly et al., 2001). As predicted, BIM5078 potently repressed HNF4α expression in T6PNE (Fig. 3a), the murine insulinoma cell line MIN6 (Fig. 3b), and in the HepG2 hepatoma line (Fig. 3c), which has exceptionally high levels of HNF4α expression.
Figure 3. BIM5078 and BI6015 affect the expression of known HNF4α target genes.
(a, b and c) HNF4α antagonists BIM5078 and BI6015, repress HNF4α gene expression. T6PNE (a), MIN6 (b) and HepG2 (c) cells were cultured for 5 (b) or 48 (a, c) hours in the presence of BIM5078 or BI6015. HNF4α mRNA levels were normalized to 18S rRNA (a, c) or GAPDH (b) to control for nonspecific compound effects. Differences in expression were measured by Student’s t-test (*p<0.05, **p<0.002). (d) Effect of BI6015 on the OTC promoter. pGL3/mOTC-235, a plasmid containing the HNF4α-responsive OTC promoter driving the firefly luciferase gene, was co-transfected into HepG2 and CV-1 cells with a plasmid encoding full length human HNF4α. Cells were treated with DMSO or 1μM BI6015 for 48 hours. HepG2 cells had greater baseline stimulation of the OTC-luciferase transgene than CV-1 cells and BI6015 potently reduced this baseline activity in both cell lines. Values represent the mean ± SE, n=3. Differences in activity were measured by Student’s t-test (*p<0.05, **p<0.001). (e) Effect of compounds on HNF4α DNA binding. DNA binding was assessed using the HNF TransAM ELISA assay (Active Motif, #46296). Compounds were incubated with HepG2 cells overnight at the indicated concentration, a condition that had no effect on HNF4α protein levels (Fig. 1c). Nuclear protein extracts (5μg) were then assayed for HNF4α DNA binding activity using an anti-HNF4α specific antibody. Values represent the mean ± SD, n=3 biological replicates. Differences in activity were measured by Student’s t-test (*p<0.05).
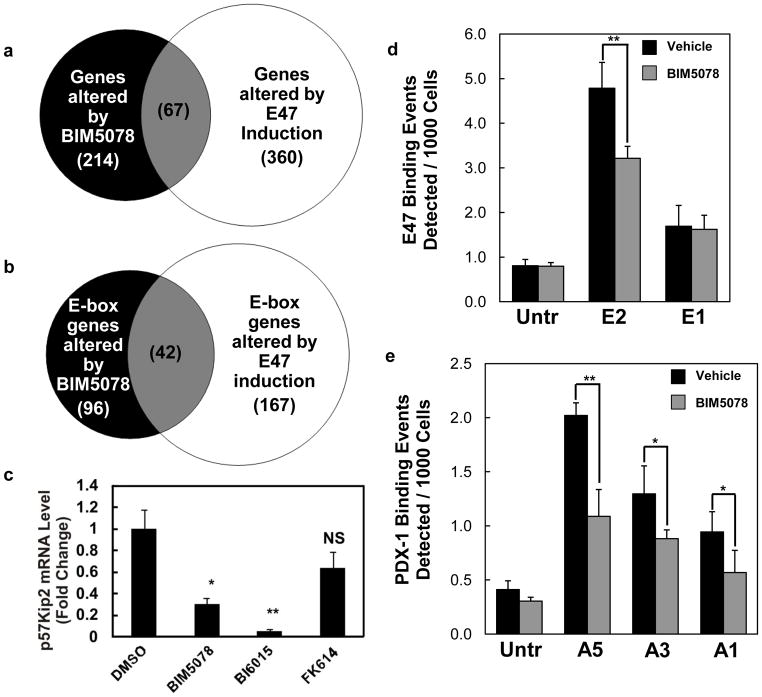
We expanded the analysis of the effects of BIM5078 on gene expression by performing global gene expression profiling of T6PNE cells in the presence and absence of BIM5078 (GSE 33432). We previously showed that T6PNE cells exhibit a pattern of gene expression similar to that of human pancreatic islets (GSE 18821) (Kiselyuk et al., 2010). Accordingly, we conducted a bioinformatic analysis to compare genes affected by BIM5078 in T6PNE cells (GSE 33432) and genes affected by genetic deletion of HNF4α in mouse islets (Gupta et al., 2007). Of the 156 identifiable genes altered in HNF4α-deleted islets compared to normal islets, 20 were exact matches to genes affected by BIM5078. An additional 36 genes were closely related to genes altered by HNF4α knockout, (e.g., cyclinD1 and Rab3a versus cyclinD2 and Rab3b), respectively. Using this analysis, 36% of the genes affected by genetic deletion of HNF4α were either identical or closely related to genes affected by pharmacologic inhibition of HNF4α. A Chi-square analysis showed that T6PNE cells treated with BIM5078 were statistically similar to the transcriptional profile of HNF4α genetic deletion (36% intersection, p<0.0001) when compared with 50,000 trials of randomly generated gene sets of the same size selected from genes expressed in T6PNE cells (11% intersection predicted). The concordance between the genes affected by genetic deletion and treatment with BIM5078 is particularly striking given that the analysis was done in different species (human versus mouse), different cell types (the cell line T6PNE cultured in vitro versus primary mouse islets), and genetic deletion in murine β-cells occurred during embryonic development, as soon as the insulin promoter became active. In summary, the gene expression data support that BIM5078 induces pharmacologic antagonism of HNF4α.
BIM5078 also affected a subset of genes with no known HNF4α binding sites
To further examine the effects of BIM5078 on gene expression, we utilized the web-based systems biology software NextBio to compare lists of genes containing cis-regulatory motifs with the list of genes modulated by BIM5078. We restricted our analysis to the Molecular Signatures Database (MSigDB) containing motif gene sets annotated by the Broad Institute to enable us to associate changes in our microarray studies with conserved, putative cis-regulatory elements (Xie et al., 2005). The most significantly correlated regulatory motifs were the E12, AP4 and MYOD binding sites, all of which contained the core CANNTG E-box sequence to which the critical insulin promoter transactivator E47 binds. It is interesting to note that there was a significant association with “HNF4α binding site geneset 1” (containing the motif VTGAACTTTGMMB) but not 4 other binding genesets representing alternate consensus binding sites for HNF4α, raising the possibility that BIM5078 may affect the activity of HNF4α at some sites preferentially over others, similar to the effect of other NR ligands on their target receptors (Hermanson et al., 2002).
Given the effect of BIM5078 on genes containing E-boxes in their promoter, we compared the genes altered by BIM5078 (GSE 33432) to a set of previously published genes altered by E47 induction in T6PNE cells (Kiselyuk et al., 2010). Of the 214 genes altered by BIM5078, 67 (31%, p<0.001) were also altered by E47 induction (Fig. 4a). This association was enhanced when only the genes containing E-boxes were compared. Specifically, of the 96 E-box containing genes altered by BIM5078, 42 (p<0.001) were also altered by E47 induction (Fig. 4b). Of the 67 genes altered by both BIM5078 and E47 induction in T6PNE, only one (SERPINI1) has been previously shown to bind HNF4α directly in ChIP-chip studies performed in islets (Odom et al., 2004).
Figure 4. BIM5078 and BI6015 affect the expression and binding of E47-responsive genes.
(a) Microarray data was analyzed using GeneSpring GX11. Of 214 genes whose expression changed with BIM5078 treatment, 67 (p<0.001) were also altered by induction of E47 in T6PNE cells. (b) Analysis in (a) was restricted to genes with promoter regions [−2kb, 2kb] around transcription start sites containing E-boxes (CANNTG). (c) HNF4α antagonists, BIM5078 and BI6015, reduced Kip2 mRNA in T6PNE cells. FK614 had no effect on Kip2 expression. Kip2 mRNA values are reported as normalized to 18s rRNA to control for nonspecific compound effects. Differences in expression between vehicle (DMSO) and BIM5078 or BI6015 were measured by Student’s t-test (*p<0.05, **p<0.01, NS = no significance). (d and e) BIM5078 decreases association of E47 and PDX-1 to the human insulin promoter. T6PNE cells were treated with 1μM tamoxifen and DMSO or 5μM BIM5078 for 48 hours followed by ChIP with anti-E47 antibody and anti-PDX-1 antibody. Q-PCR was performed with primers targeting E-box and A-box elements in the insulin promoter. (d) At 48 hours, treatment with BIM5078 significantly decreased binding of E47 to the distal E-box (E2), but not to the proximal E-box (E1). (e) Similarly, BIM5078 decreased binding of PDX-1 to all A-box elements tested (A1, A3, and A5) on the insulin promoter. Differences in binding between vehicle (DMSO) and BIM5078 were measured by Student’s t-test (*p<0.05, **p<0.01).
The effect of BIM5078 on E-box binding in the insulin promoter prompted us to examine the impact of BIM5078 on other genes regulated by E-boxes. We have previously shown that the cyclin-dependent kinase inhibitor p57Kip2 is directly regulated by E47 occupation of a particular E-box within the p57Kip2 promoter (Kiselyuk et al., 2010). Although p57Kip2 promoter does not contain an HNF4α binding site, BIM5078, but not FK614, which does not act on HNF4α, potently decreased Kip2 expression in T6PNE cells (Fig. 4c). These results suggest that an important effect of HNF4α on gene expression occurs through E-box sequences in the promoters of genes that may or may not contain HNF4α binding sites.
BIM5078 disrupted binding of E47 and PDX-1 to the human insulin promoter
While there is a well-characterized binding site for HNF4α in the rodent insulin promoter (Bartoov-Shifman et al., 2002), the cognate region of the human insulin promoter varies in sequence, and HNF4α has not been detected in ChIP-Chip studies as being directly bound to the human insulin promoter (Odom et al., 2004). However, there is a well-characterized HNF1α site in the human insulin promoter (Okita et al., 1999), suggesting one possible route through which HNF4α can act. Because of the evidence that HNF4α does not bind directly to the human insulin promoter, we further investigated the mechanism by which BIM5078 inhibits insulin gene expression. One possibility was that HNF4α acts indirectly, through effects on other insulin promoter transactivators. To test this hypothesis, ChIP assays were used to probe the binding of transcriptional activators E47 and PDX-1 to their regulatory sequences on the insulin promoter (i.e., E-box [CANNTG] and A-box [TAAT] motifs, respectively) in the presence and absence of BIM5078. After 48 hours, treatment with 5μM BIM5078 significantly decreased the association of E47 with the distal (E2) but not the proximal (E1) E-box of the insulin promoter (Fig. 4d). Furthermore, association of PDX-1 was significantly decreased at all tested A-boxes (Fig. 4e). These results are consistent with the hypothesis that BIM5078-mediated repression of insulin promoter activity occurs indirectly through disrupted binding of critical transcription factors.
Fatty acids bound to HNF4α preferentially inhibited the insulin promoter
Because of uncertainty in the literature about the role of natural or synthetic ligands in controlling HNF4α functional activity, we sought to determine whether compounds that have previously been reported to bind HNF4α had potency in our T6PNE assay. In the absence of exogenously added ligand, MCFAs and LCFAs were found in the HNF4α ligand-binding pocket in structural studies of HNF4α purified from bacteria (Dhe-Paganon et al., 2002; Duda et al., 2004) consistent with fatty acids being the natural ligands for HNF4α. Recently, linoleic acid was found to be bound in the LBP of HNF4α in COS-7 cells, as well as in the livers of fed but not fasted mice, suggesting that it might be a natural ligand (Yuan et al., 2009). Furthermore, linoleic acid binding to HNF4α in mammalian cells was slowly reversible (Yuan et al., 2009), while ligand exchange in vitro was shown to be irreversible. The relatively poor ligand exchange has raised questions about whether fatty acids are functionally relevant ligands or whether they play some other role, perhaps stabilizing the structure of the protein.
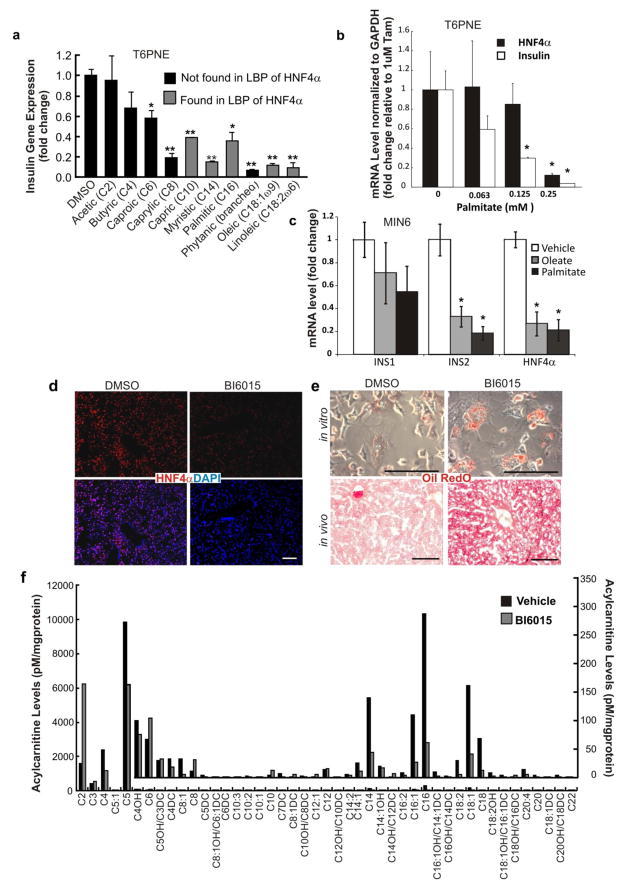
Based on the widely accepted mechanism of NR regulation, one would expect binding of the endogenous ligand (linoleic acid) to its native receptor (HNF4α) to induce changes in HNF4α functional activity. To test this, a broad spectrum of fatty acids was assessed for effects on insulin promoter activity. We found that MCFAs and LCFAs, including the polyunsaturated fatty acid linoleic acid, inhibited the insulin promoter as measured by quantitative RT-PCR (Fig. 5a), while short chain fatty acids (SCFA), such as acetic and butyric fatty acids, which have not been isolated in the ligand-binding pocket of HNF4α, did not have an effect on insulin gene expression. The same pattern of functional activity was observed using GFP+ cells as a measure of insulin promoter activity and very long chain fatty acids had no effect on the insulin promoter using that method (Supplementary Fig. 2). The correlation between fatty acids bound to HNF4α and those that have a repressive effect on the insulin promoter assay suggests a functional role for fatty acids in regulating HNF4α activity and is consistent with a role for HNF4α in β-cell lipotoxicity.
Figure 5. Metabolic effects of fatty acids and HNF4α antagonists.
(a, b) T6PNE cells were treated with 1μM tamoxifen and either fatty acid or vehicle (DMSO) for 48 hours, at which time cells were harvested for RNA isolation. Medium and long chain fatty acids, but not short chain fatty acids, inhibited insulin gene expression (a) (*p<0.01, **p<0.001). Palmitate induced a dose-dependent decrease in both insulin and HNF4α mRNA level (b) (*p<0.05). (c) MIN6 cells were treated with either 0.08mM Oleate or 0.02mM Palmitate for 110 days, at which time insulin and HNF4α mRNA levels were measured. mRNA levels were normalized to 18s rRNA (a) or GAPDH (b, c) to control for nonspecific compound effects. Values represent the mean ± SE, n=3, *p<0.05. (d) BI6015 decreased expression of HNF4α (red) in the liver. (e) BI6015 induced hepatic steatosis in murine hepatocytes in vitro and in vivo. Primary murine hepatocytes were exposed to DMSO or BI6015 (5μM) for 3 days, followed by fixation and staining with Oil Red O. BI6015 was injected IP once per day at either 10 or 30 mg/kg/day. Mice were sacrificed for analysis of organ histology after 5 days. Liver sections were stained with H&E and Oil Red O. (f) Acylcarnitine analysis by GC/MS was performed on cellular extracts of T6PNE cells. Biological duplicates were analyzed. Values represent the mean. Inlay depicts the same data beginning with C4-OH on the x-axis on a different scale to show less abundant species. Scale bars, d, e = 100 μm.
If fatty acids act to inhibit HNF4α function, they should affect the expression of HNF4α target genes, including HNF4α itself. Because the transcriptional role of the putative natural ligands to HNF4α, fatty acids, has remained unclear, we investigated their ability to modulate HNF4α expression. In T6PNE cells, where HNF4α gene expression is induced by tamoxifen simultaneously with the addition of the putative fatty acid ligand, we found that 48 hour treatment with palmitate significantly inhibited HNF4α mRNA levels (Fig. 5b). In MIN6 cells, oleate and palmitate inhibited the INS2 and HNF4α genes, but not INS1 (Fig. 5c). Interestingly, this effect occurred only in MIN6 cells subjected to prolonged fatty acid exposure, consistent with the much higher levels of insulin and HNF4α expression in MIN6 compared with T6PNE, more closely mimicking normal β-cells and the known long time course of β-cell lipotoxicity. Overall, these data are consistent with fatty acids being weak HNF4α antagonists compared with the more potent synthetic antagonists described here.
In addition to fatty acids, several other compounds have been proposed to be HNF4α ligands, including bezafibrate, acyl-CoA thioesters, 3,3,14,14-tetramethylhexadecanedioic acid (Medica-16), and 3-methyl-2-nitronaphthofuran (“Compound 5”) (Hertz et al., 2001; Le Guevel et al., 2009). It has been suggested that acyl-CoA thioesters, which are the best studied of the putative ligands, are too large for the HNF4α LBP (Bogan et al., 2000; Wisely et al., 2002). However, resolving whether fatty acids or their activated CoA thioester represent the immediate ligands regulating HNF4α is complicated by fatty acyl CoA thioester instability in the aqueous media used for HNF4α isolation/crystallization (Schroeder et al., 2005) as well as a report that HNF4α itself exhibits thioesterase activity (Hertz et al., 2005). In our hands, none of these compounds had consistent and robust effects on insulin promoter activity (Supplementary Fig. 3).
Development and characterization of BIM5078 analogs
Due to its promising behavior in vitro, pharmacokinetic studies were performed to evaluate the potential of BIM5078 for in vivo studies. BIM5078 was found to have relatively low plasma stability, only moderate microsomal stability (8% remaining after 3 h and 32% after 1.25 h, respectively), high binding to plasma proteins (98% bound after 4 h), and low solubility (0.17 μg/ml after 18 h).
As a result of its unfavorable pharmacokinetic properties, structural analogs of BIM5078 were examined to overcome some of these limitations. Replacement of the chloro group of BIM5078 with a methyl group resulted in BI6015 (2-Methyl-1-(2-methyl-5-nitrophenylsulfonyl)-1H-benzo[d]imidazole)(Supplementary Fig. 4). Additionally, we examined changes to the sulfonamide linker of BIM5078. Replacement of the -SO2- linker with a methylene gave BI6018, in which the chloro was also replaced with a hydroxy group (Supplementary Fig. 4). This compound was found to be inactive in the insulin promoter assay.
BI6015 reduced endogenous insulin gene expression 50-fold in T6PNE cells (Fig. 1e), and strongly repressed HNF4α gene expression in MIN6 (Fig. 3b) and HepG2 cells (Fig. 3c). The ornithine transcarbamoylase (OTC) promoter has been well characterized as responsive to HNF4α in transient transfection assays (Inoue et al., 2002). BI6015 inhibited luciferase expression driven by that promoter in both HepG2 cells and CV-1 cells (Fig. 3d). Interestingly, BI6015, unlike BIM5078, was found not to be a PPARγ agonist (Fig. 1d), supporting the identification of HNF4α rather than PPARg as the relevant target for the effects of the compounds. Thus, despite structural similarity between synthetic and natural ligands that bind PPARγ (Tontonoz and Spiegelman, 2008) and those that bind HNF4α, we found that a structural modification as subtle as the substitution of a methyl for a chloro group can dissociate their effects, thereby reducing off-target effects. BI6015 docked well into the HNF4α LBP (Fig. 2d, violet molecule in left panel), with a GoldScore of 42.1, very similar to that of BIM5078. It should be noted that the antagonists also dock in the LBP of the closely related NR HNF4γ (Gerdin et al., 2006)(Supplementary Fig. 5).
While gene expression studies and the high degree of structural homology of BI6015 with BIM5078 were consistent with BI6015 also interacting directly with HNF4α as its primary mechanism of action, it was important to demonstrate this directly. To accomplish that, we employed a novel assay termed drug affinity target stability (DARTS) (Lomenick et al., 2009). This assay takes advantage of the conformation change in a protein target induced by ligand binding. The conformation change is detected by a consequent change in the sensitivity to proteolysis. We employed this technique with the HNF4α antagonists BI6015 and BIM5078, as well as FK614 and BI6018, which were inactive in all assays that were responsive to HNF4α modulation. As expected for actual ligands, BI6015 and BIM5078 induced altered HNF4α protease sensitivity, with BI6015, the more potent compound, having a much greater effect, while BI6018 and FK614 had no effect (Fig. 2c).
NR ligands typically affect the binding of the receptor to DNA. Thus, we studied the effect of the compounds on HNF4α DNA binding with an ELISA assay in which an oligonucleotide containing an HNF4α binding site is attached to a plate well. Nuclear extracts from cells incubated with a particular compound were added to the wells, and an antibody against HNF4α was then used to detect binding of HNF4α to the oligonucleotide. Consistent with the gene expression and biochemical studies, BI6015 potently repressed HNF4α DNA binding. BIM5078 was active but less potent, while BI618 and FK614 were inactive (Fig. 3e).
Given the in vitro evidence that BI6015 was a potent HNF4α antagonist, we tested it for in vivo efficacy. In vivo PK studies of BI6015 in mice revealed a half-life of approximately 90 minutes on delivery by oral gavage or intraperitoneal (IP) injection, with moderate plasma levels (AUC = 1.6 μg·min/mL after a 30 mg/kg IP injection). In addition, high levels of BI6015 were found in the liver (3.1 μM at 24 hr after a 30 mg/kg IP injection in mice). Thus, in addition to improved specificity for HNF4α relative to other NRs, BI6015 has a more favorable stability profile over its predecessor, BIM5078, making it suitable for in vivo studies. In addition, a Ricerca hit profiling panel of 41 receptors/enzymes, including major cytochrome P450s and a number of NRs, including RXR, showed significant cross reactivity with only one CYP (2C19), and one L-type calcium channel (Supplementary Data).
BI6015 induced loss of HNF4α expression and hepatic steatosis in vivo
Injection of BI6015 into the peritoneum of mice once per day for 5 days at a dose of 30 mg/kg was well tolerated. A VetScan Liver Panel revealed that blood chemistries, including ALT, were unaffected by BI6015 treatment, suggesting an absence of hepatocellular death in the normal liver (Supplementary Fig. 6). Because HNF4α is highly expressed in the liver, hepatic tissue was harvested and examined, along with the intestine and kidney (other sites of high HNF4α expression).
Consistent with the effects of BIM5078 on HNF4α expression in vitro, both in human and murine derived cell lines, BI6015 induced a loss of HNF4α protein in the liver (Fig. 5d), but not in the intestine or kidney (Supplementary Fig. 7). It is possible that the lack of effect in the intestine and kidney was due to a high degree of hepatic first pass metabolism following IP administration, as hepatic microsomal stability studies on BI6015 showed that only 22% of the compound remained after a 60-minute incubation in vitro. Furthermore, no difference in the number of cells expressing the proliferation marker Ki67 was observed in liver, intestine or kidney with BI6015, as compared with vehicle-treated animals.
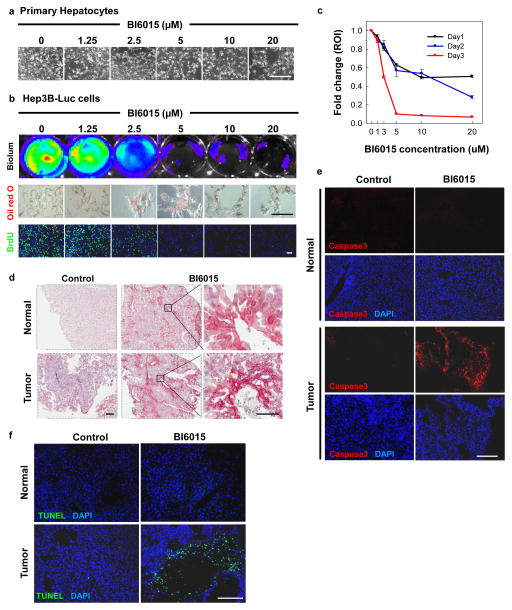
Although there was no evidence of hepatocellular death, the hepatocytes from livers of mice injected with BI6015 exhibited marked fat accumulation (steatosis)(Fig. 5e, 6d), similar to the effect of genetic deletion of HNF4α in the liver (Hayhurst et al., 2001). Steatosis and vesicular changes were dose-dependent between 10 and 30 mg/kg of BI6015 administered daily or every other day, as evidenced by increased fat staining with Oil Red O, but steatosis was limited to regions around vessels, again consistent with extensive hepatic metabolism limiting the area of the liver that was exposed to active compound. In vitro, BI6015 induced steatosis in primary mouse hepatocytes, again without evidence of cell death (Fig. 5e).
Figure 6. HNF4α antagonists are cytotoxic to hepatocellular carcinoma.
(a) Toxicity was not observed in primary hepatocytes treated with BI6015 in vitro. (b) BI6015 was potently toxic to Hep3B-Luc cells, as measured by bioluminescence. Fat accumulation, measured by Oil Red O staining (red), was enhanced in Hep3B-Luc cells treated with BI6015, while cell division, measured by BrdU uptake (green), was blocked in a dose-dependent manner. (c) Bioluminescence of Hep3B-Luc treated with BI6015 was quantified at 24, 48 and 72 hours after treatment. Decreased bioluminescence correlated directly with decreased cell counts. (d) Hep3B-Luc cells were injected into the liver parenchyma of nude mice, hereafter referred to as HCC mice. Animals were injected IP with 30 mg/kg of BI6015 daily or every other day, as tolerated. Treatment with BI6015 resulted in marked induction of Oil Red O staining (red) in both the normal liver and tumor region, as compared with DMSO. (e and f) Apotosis was evaluated by immunostaining for cleaved caspase-3 (red) and a TUNEL assay (green). High levels of cleaved caspase-3 expression were found in the tumor regions, but not in the normal liver of BI6015-treated HCC mice as compared with the DMSO control. A similar phenomenon was observed with TUNEL positive cells, which were only found in tumor regions of BI6015-treated HCC mice, predominantly in regions surrounding the necrotic core of the tumor. Scale bars, a,d, e,f=200 μm (inset for d=50 μm), b=100 μm.
BI6015 administration resulted in changes in fatty acid oxidation
Given the known role of HNF4α in both fatty acid biosynthesis and utilization (Odom et al., 2004; Palanker et al., 2009), we studied the effect of BI6015 on fatty acid profiles. Acylcarnitine profiles of T6PNE cells treated with BI6015 revealed decreased levels of longer chain acylcarnitines, particularly C18, C18:1, C16, and C14 (Fig. 5f) with a concurrent increase in acetylcarnitine. This is consistent with BI6015 acting as an HNF4α antagonist and with HNF4α antagonism increasing fatty acid oxidation (Degrace et al., 2004). However, we also observed that BI6015 induced hepatic steatosis. Together, these results indicate that hepatic steatosis resulting from BI6015 administration and supposed HNF4α inhibition cannot be explained by impaired fatty acid oxidation. A similar phenomenon has been reported in mice fed the conjugated trans-10, cis-12-isomer of linoleic acid, in which lipid accumulation is observed despite increases in fatty acid oxidation (Degrace et al., 2004).
BI6015 is cytotoxic to human hepatocellular carcinoma (HCC)
While studying BIM5078 and BI6015 in vitro, we unexpectedly noted that they were toxic to a number of different tumor cell lines but not to cultured primary cells. To further characterize this phenomenon, we examined the effects of BI6015 on a variant of the human hepatoma cell line Hep3B that was engineered to express a luciferase transgene under the control of the CMV promoter, in parallel with primary hepatocytes. BI6015 treatment was markedly toxic to Hep3B cells (Fig. 6b, c) but spared primary hepatocytes (Fig. 6a). These observations coincided with cell cycle arrest, as evidenced by decreased BrdU (green) incorporation after 48 hours of treatment (Fig. 6b). Consistent with what we and others have observed with respect to HNF4α antagonism, perturbations to hepatocytes by BI6015 resulted in characteristic steatosis in the absence of cell death (Hayhurst et al., 2001) (Fig. 6d). To determine the scope of tumor cytotoxicity, BI6015 was submitted to the developmental therapeutics program (DTP) at the NCI/NIH, where it was screened in vitro against a panel of 60 human tumor cell lines. Slowed growth and/or toxicity were observed with BI6015 treatment on a wide range of cancer cell lines (Supplementary Fig. 8). Similarly, siRNA to HNF4α also resulted in toxicity to HepG2 cells and to a greater extent to T6PNE cells.
To extend the in vitro finding of cytotoxicity to tumor cells in vivo, we studied the effects of BI6015 treatment in a human orthotopic xenograft model, in which luciferase-expressing Hep3B cells were injected directly into the liver parenchyma (Yao et al., 2003). Once the tumor was established, as defined by a doubling of luciferase counts on 3 consecutive reads by bioluminescent imaging, the animals were injected IP with 30 mg/kg of BI6015 daily or every other day, as tolerated. After 20–57 days of treatment, the tumors and normal liver samples were harvested and analyzed. There was a marked induction of Oil Red O staining in both the normal liver and tumor samples, although the background level of Oil Red O in the tumor cells was high (Fig. 6d). Interestingly, the distribution of the steatotic cells was limited, being restricted to areas around the hepatic blood vessels (Fig. 6d).
Within the tumor, but not in the normal liver, there were regions with high levels of apoptotic cells positive for TUNEL and cleaved caspase-3 (Fig. 6e, f). While some treated animals exhibited decreased luciferase counts, this did not occur in all animals. Higher doses of BI6015 could not be delivered due to limited solubility of the compound.
DISCUSSION
HNF4α is an attractive target for pharmacologic manipulation. Not only is it at the center of multiple complex feedback loops that maintain differentiated function in the pancreas and liver, but HNF4α has also been implicated in a number of disease states, including diabetes, inflammatory bowel disease, cancer, and others. Here, we report the discovery of small molecule inhibitors of HNF4α that help interrogate and clarify complex processes driven by HNF4α-dependent pathways.
Until about a decade ago, HNF4α was considered to be an orphan receptor. Structural studies identified tightly bound fatty acids in the LBP of HNF4α, which existed in a mixture of active and inactive conformations, suggesting the fatty acids were playing a structural role, rather than inducing a specific conformational change (Dhe-Paganon et al., 2002; Wisely et al., 2002). Affinity isolation followed by mass spectrometry (AIMS) was used more recently to reveal that linoleic acid (LA, C18:2ω6) was bound to HNF4α in the livers of fed but not fasted mice, suggesting that ligand binding could be reversible (Yuan et al., 2009). These results were also consistent with findings in Drosophila showing that a GAL4-dHNF4 ligand sensor could be activated by starvation or administration of exogenous long chain fatty acids (Palanker et al., 2009). However, the study by Yuan et al. did not find evidence of a significant effect of ligands on HNF4α transactivation. In contrast, we found that MCFAs and LCFAs, including the polyunsaturated fatty acid linoleic acid, antagonized the insulin promoter, while shorter chain fatty acids, which have not been found in the LBP of HNF4α (Dhe-Paganon et al., 2002), did not. Our ability to observe effects of fatty acids on HNF4α activity in T6PNE cells may be because HNF4α expression is induced in those cells by tamoxifen simultaneously with the addition of fatty acids.
The observed correlation between fatty acids that have been found to be bound to HNF4α and those that have a repressive effect on the insulin promoter assay, together with evidence that HNF4α expression is inhibited by fatty acids, suggests that fatty acids are weak HNF4α antagonists. This is consistent with the finding that linoleic acid inhibited HNF4α protein expression (Yuan et al., 2009). The finding that fatty acids antagonize HNF4α activity may provide insight into the mechanism by which fatty acids exert their biological effects, e.g., in βcell lipotoxicity as well as other disorders in which there are high circulating levels of fatty acids.
Based on the data presented, we believe that BIM5078 and BI6015 act as HNF4α antagonists by binding directly in the LBP. This thinking is supported by our biochemical data showing tight binding, although we can’t use those results to differentiate orthosteric from allosteric interactions. BIM5078 and BI6015 are structurally similar to FK614, a known PPARγ agonist, and we show that BIM5078 retains PPARγ agonist activity, strongly indicating that the compound binds within the PPARγ LBP. FK614 bound poorly to HNF4α and was inactive in all of the assays for effects on HNF4α activity. Of particular interest, PPARγ and HNF4α, both of which are affected by BIM5078, share fatty acids as their natural ligands. This provides support that BIM5078 and BI6015 are HNF4α ligands. Consistent with that, our docking experiment shows that BIM5078 and BI6015 have binding poses that are similar to the fatty acids observed in HNF4α crystal structures. The effect of BIM5078, acting as an antagonist of HNF4α and an agonist for PPARγ, is similar to that of fatty acids that potently activate PPARγ, while we and others found them to inhibit HNF4α (Yuan et al., 2009). Regardless, the atomic details of BIM5078 and BI6015 interaction with HNF4α are yet to be revealed and depend on the development of ligands with improved solubility to enable future structural work.
Genetic deletion of HNF4α in pancreatic β-cells did not result in loss of insulin gene expression (Gupta et al., 2007), in contrast to our results with T6PNE cells that displayed high sensitivity to small molecule modulation of HNF4α activity and enabled discovery of viable antagonists in the short time frame in which HTS is performed. We show that HNF4α not only appears to act on a number of target genes through effects on E47, which we induced at a submaximal level in T6PNE for the screening purposes, but it is also induced by E47 expression, a previously unknown mechanism of HNF4α transcriptional regulation. This suggests that HNF4α and E47 form a multicomponent regulatory network involving complex feedback loops, similar to what has been described for the interaction between HNF4α and HNF1α (Odom et al., 2004). We think that it is this relationship between E47 and HNF4α that renders the insulin promoter in T6PNE cells susceptible to HNF4α modulation, affording us the opportunity to discover compounds that antagonized HNF4α activity.
The effect of HNF4α antagonists on E-box containing genes was unexpected, as the vast majority of these genes are not known to contain HNF4α binding sites and do not bind to HNF4α in ChIP-Chip assays (Odom et al., 2004). However, a similar phenomenon was reported for the human intestinal cell line Caco-2, in which two-thirds of the genes that bound HNF4α in their ChIP-Chip assay had no discernable HNF4α binding site (Boyd et al., 2009). Classically, NRs exert their effects by binding to highly conserved DNA binding elements, but it is known that NRs can also act indirectly via tethering to target genes (Adler et al., 1988). NR tethering has been reported to occur through a number of DNA binding transcription factors, including members of the bHLH class including E47 (Murayama et al., 2004) and the homeodomain class (Stender et al., 2010). On the human insulin promoter, which does not contain an HNF4α binding site and does not bind to HNF4α in ChIP-Chip assays (Odom et al., 2004), we found that binding of both E47 and PDX1 was inhibited by BIM5078, which we propose to be an HNF4α antagonist. On the p21 promoter, HNF4α was shown to act through a DNA binding independent mechanism involving binding to the bHLH factor c-myc (Hwang-Verslues and Sladek, 2008). Altogether, these results suggest a model in which HNF4α is recruited to a transcriptional complex bound to DNA in a ligand-dependent manner, but might not necessarily involve direct binding of HNF4α to DNA.
While studying the effect of BIM5078 and BI6015 in vitro, we noticed marked toxicity in a variety of tumor cell lines but not in cells cultured from primary tissue. HNF4α has previously been described to have a role in tumor pathogenesis, but the studies are conflicting, with both upregulation and downregulation of HNF4α expression being reported in association with tumor progression. Knockdown of HNF4α mRNA by siRNA has been shown to inhibit growth and proliferation of colorectal cancer cells in vitro (Schwartz et al., 2009). This is consistent with reports that have shown that HNF4α is upregulated in human hepatocellular carcinoma (Xu et al., 2001). However, others have shown downregulation of HNF4α promotes tumorigenesis in hepatocellular and other cancers (Ning et al.).
Several lines of evidence suggest that the effects of BI6015 on transformed cells are mediated through HNF4α. First, we detected marked toxicity with selective HNF4α knockdown by siRNA. Furthermore, the concentration at which BI6015 induced cytotoxicity was very similar to that at which it affected the expression of downstream targets of HNF4α. This would not be expected if the effects on cancer cells were caused by an off-target effect.
In vivo, BI6015 caused dose-dependent hepatic steatosis in normal hepatocytes and in Hep3b xenografted cells. This is consistent with the effects of genetic ablation of HNF4α (Hayhurst et al., 2001). Importantly, the induction of steatosis provided a biomarker for where the compound was acting, which appeared to be restricted to regions surrounding blood vessels. Apoptosis was induced in Hep3b cells but not in primary hepatocytes, consistent with the in vitro results. Of note, apoptotic cells also were distributed in a perivascular pattern, mimicking that of steatosis. The lack of activity of BI6015 distal to the vessels suggests either poor tissue penetration, as has been shown for a number of chemotherapeutics including doxorubicin (Minchinton and Tannock, 2006), or extensive hepatic metabolism that limits the amount of active compound to regions surrounding vessels. The in vitro pharmacology with hepatic microsomes suggests that extensive hepatic metabolism is likely to be occurring in vivo, supporting the need for additional medicinal chemistry to develop more stable molecules. We think that the limited in vivo efficacy in the Hep3B orthostatic xenograft model could have been due to a high degree of hepatic metabolism, thereby restricting the amount of drug that penetrated into the tumor, which would be consistent with steatosis in both normal liver and tumor samples being localized to the region around blood vessels. Nonetheless, our data indicate that BIM5078 and BI6015 might be promising agents, and HNF4α a promising target for cancer therapy. In addition, these compounds provide powerful tools for studying the function of HNF4α. Additional studies will be required to determine whether an appropriate balance between the anti-tumor effects of HNF4α and the side effects of inhibiting HNF4α, e.g., hepatic steatosis, can be achieved. The fact that mice administered BI6015 for up to a month have tolerated it well raises the hope that this will be the case.
SIGNIFICANCE
NF4α is a member of the NR class of transcription factors. This family has 48 members in humans and characterized by being regulated by small molecule ligands to control a wide range of processes in which it is desirable to have a small molecule regulate a gene expression program. As such, NRs have been highly desirable pharmacologic targets. However, many have yet to be characterized in terms of the nature of the endogenous ligand and/or the development of synthetic ligands that can be used pharmacologically. One of those has been HNF4α, which plays a central role in controlling gene expression in the liver, pancreas, kidney, and intestine. Here, we describe two compounds, BIM5078 and BI6015, and present evidence that they are HNF4α antagonists. As expected, the antagonists repressed known HNF4α target genes. Surprisingly, the compounds were found to be selectively cytotoxic to transformed cells, raising the possibility that they might be useful for cancer therapy.
EXPERIMENTAL
Cell culture, compound library screening, and confirmatory assays, including RT-PCR, are described in (Kiselyuk et al., 2010). Detailed methods are presented in the Supplementary Material.
Supplementary Material
Supplementary Figure 1. BIM5078 passed confirmatory and counter screens.(a) Compounds found to modulate the insulin-eGFP transgene in the primary screen were confirmed in dose-response assays. Three potential activators and four potential repressors were identified. (b) T6PNE contains E47MER, which translocates to the nucleus and becomes transcriptionally active in the presence of tamoxifen. Compounds found to reproducibly increase activity of the insulin promoter driving eGFP in the secondary screen (a) were tested for their ability to bind a multimerized E-box and drive luciferase expression. Compound 5157189 was found be estrogenic. (c) Similarly, compounds found to decrease the functional activity of the insulin-eGFP transgene (a) were tested for their ability to prevent the translocation of E47MER. Compounds 5867913, 5849200, 5359535 and 5816458 were found to be estrogenic antagonists. (d) Compounds that did not interact with E47MER were tested for their ability to modulate endogenous insulin mRNA. BIM5078, but not 5544524 or 5120083, had activity on the endogenous insulin promoter.
Supplementary Figure 2. Fatty acids bound to HNF4α preferentially inhibit exogenous insulin promoter activity. T6PNE cells were treated with 0.12 mM fatty acids for 48 hours. Effects on the exogenous insulin promoter in T6PNE is reported as percent GFP+ cells, as determined by imaging the green channel and normalizing to the total number of cells per well. Values represent the mean ± SE, n=8.
Supplementary Figure 3. Effect of reported HNF4α ligands on insulin promoter activity. T6PNE cells were treated with bezafibrate, Medica-16 or a nitro-naphthofuran derivative (“Compound 5”) for 48 hours in the presence of either 0.5 μM (a) or 1 μM (b) tamoxifen. Effects on the exogenous insulin promoter transgene in T6PNE are reported as percent GFP+ cells. Values represent the mean ± SE, n=3.
Supplementary Figure 4. Structures of BI6015 and BI6018.
Supplementary Figure 5. Docking of BI6015 (purple) in the LBD of HNF4γ with linoleic acid (cyan) crystallized. The nitro group of BI6015 forms hydrogen bonds with the backbone N of Gly197 and the side chain guanadinium group of Arg 186. The remaining parts of BI6015 primarily make hydrophobic interactions with the binding pocket, which is for the most part hydrophobic in nature. The empirical docked score (GOLD Fitness score) for this pose is 40.06, similar to that for HNF4α.
Supplementary Figure 6. Blood chemistry of mice treated with BI6015. Mice (NONcNZO10/LtJ or ICR) were injected IP with vehicle (DMSO) or BI6015 once daily for 5 days. Prior to sacrifice, blood was drawn and analyzed using a VetScan blood analyzer, measuring alkaline phosphatase (ALP, IU/L), alanine aminotransferase (ALT, IU/L), gamma glutamyl transferase (GGT, IU/L), bile acids (BA, μmol/L), total bilirubin (TBIL, mg/dL), albumin (ALB, g/dL), blood urea nitrogen (BUN, mg/dL), and cholesterol (CHOL, mg/dL). Five groups of mice were studied: normal mice injected with DMSO (Normal DMSO, n=4), normal mice injected with BI6015 at a dose of 30 mg/kg/day (Normal BI6015H, n=4) or 10 mg/kg/day (Normal BI6015L, n=4), mice injected with the hepatocellular carcinoma (HCC) cell line Hep3B and treated with DMSO (HCC DMSO, n=5) for 13–29 days, and mice injected with Hep3B and treated with BI6015 at a dose of 30 mg/kg/2day (HCC BI6015, n=4) for 29–36 days.
Supplementary Figure 7. HNF4α expression does not change in the intestine or kidney in mice receiving IP injection of BI6015. HNF4α expression was assessed as described in the Methods. Scale bar, 100 μm.
Supplementary Figure 8. NCI Panel. Data were generated by the NCI Developmental Therapeutics Program, as previously described (Shoemaker, 2006).
Supplementary Table 1. Small molecule screening data
Supplementary Table 2. List of genes affected both by genetic deletion of HNF4α in vivo and pharmacologic inhibition of HNF4α in vitro. 36% of the genes altered by genetic deletion of HNF4α in the mouse in vivo are either identical or closely related to genes modulated by pharmacologic inhibition of HNF4α in the human cell line, T6PNE.
Supplementary Table 3. List of genes affected by both pharmacologic inhibition of of HNF4α and induction of E47. Analysis was restricted to genes altered at least two-fold. The expression of 214 genes were significantly altered by BIM5078 at least two-fold. Of these, 67 were also modulated by E47 induction through 4-hydroxytamoxifen. This association was enhanced when only the genes containing E-boxes were compared. 96 E-box containing genes were altered by BIM5078. 42 of these were also altered by E47 induction
HIGHLIGHTS.
HNF4α modulators identified by high-throughput screening
Fatty acids act as HNF4α antagonists in an HNF4α-responsive assay Synthetic
HNF4α antagonists are selectively cytotoxic to transformed cells
Acknowledgments
The OTC-luciferase reporter plasmid was a kind gift from Professor Frank J. Gonzalez (National Institute of Health). Roy Williams (SBMRI Informatics Core) and Kang Liu (SBMRI Genomics Core) provided support for the microarray studies. Ed Sergienko (Conrad Prebys Center for Chemical Genomics) aided in the analysis of the fluorescence quenching assay. Sumeet Salaniwal (Conrad Prebys Center for Chemical Genomics) aided in the docking studies. AK, SHL, SFK, FL designed experiments. AK, SHL, SFK, SA, TC, HH, LH, BS, JH, MR-L, performed experiments. PB, PIA, and MM assisted with HTS. JC synthesized FK614. MY, AP, and MID performed medicinal chemistry. HAH and FS designed and carried out binding assays. ADR performed metabolic flux studies. LS performed pharmacokinetic analysis. MZ and YC designed and carried out tumor xenograft studies. AK, SHL, HF, SFK, FL analyzed data. AK, SHL, and FL wrote the paper. All the authors read, revised and approved the manuscript. This work was supported by the Sanford Children’s Health Research Center, the Kieckhefer Foundation, the UCSD Genetics Training Grant, the UC Systemwide Biotechnology Research and Education Program Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Program, the Juvenile Diabetes Research Foundation, an American Diabetes Association Clinical Scientist Training Award, and USPHS, National Institutes of Health grants DK41402 (F.S.) and DK77573 (H.A.H.). High-throughput screening and some medicinal chemistry was carried out in the SBMRI’s Conrad Prebys Center for Chemical Genomics.
Footnotes
There are no conflicts of interest.
References
- Adler S, Waterman ML, He X, Rosenfeld MG. Steroid receptor-mediated inhibition of rat prolactin gene expression does not require the receptor DNA-binding domain. Cell. 1988;52:685–695. doi: 10.1016/0092-8674(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC. An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Research. 2001;29:3495–3505. doi: 10.1093/nar/29.17.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoov-Shifman R, Hertz R, Wang H, Wollheim CB, Bar-Tana J, Walker MD. Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4 alpha. J Biol Chem. 2002;277:25914–25919. doi: 10.1074/jbc.M201582200. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Dallas-Yang Q, Ruse MD, Jr, Maeda Y, Jiang G, Nepomuceno L, Scanlan TS, Cohen FE, Sladek FM. Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. Journal of molecular biology. 2000;302:831–851. doi: 10.1006/jmbi.2000.4099. [DOI] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd M, Bressendorff S, Moller J, Olsen J, Troelsen JT. Mapping of HNF4alpha target genes in intestinal epithelial cells. BMC Gastroenterol. 2009;9:68. doi: 10.1186/1471-230X-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Gulick T, Raisher BD, Caira T, Ladias JA, Moore DD, Kelly DP. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J Biol Chem. 1993;268:13805–13810. [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Dawson MI, Ye M, Cao X, Farhana L, Hu QY, Zhao Y, Xu LP, Kiselyuk A, Correa RG, Yang L, et al. Derivation of a retinoid X receptor scaffold from peroxisome proliferator-activated receptor gamma ligand 1-Di(1H-indol-3-yl)methyl-4-trifluoromethylbenzene. ChemMedChem. 2009;4:1106–1119. doi: 10.1002/cmdc.200800447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrace P, Demizieux L, Gresti J, Chardigny JM, Sebedio JL, Clouet P. Hepatic steatosis is not due to impaired fatty acid oxidation capacities in C57BL/6J mice fed the conjugated trans-10, cis-12-isomer of linoleic acid. J Nutr. 2004;134:861–867. doi: 10.1093/jn/134.4.861. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- Drewes T, Senkel S, Holewa B, Ryffel GU. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda K, Chi YI, Shoelson SE. Structural basis for HNF-4alpha activation by ligand and coactivator binding. J Biol Chem. 2004;279:23311–23316. doi: 10.1074/jbc.M400864200. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(-/-) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Sakuma H, Konishi S, Oe T, Hosogai N, Kimura C, Aramori I, Mutoh S. FK614, a novel peroxisome proliferator-activated receptor gamma modulator, induces differential transactivation through a unique ligand-specific interaction with transcriptional coactivators. J Pharmacol Sci. 2005;99:342–352. doi: 10.1254/jphs.fp0050578. [DOI] [PubMed] [Google Scholar]
- Gerdin AK, Surve VV, Jonsson M, Bjursell M, Bjorkman M, Edenro A, Schuelke M, Saad A, Bjurstrom S, Lundgren EJ, et al. Phenotypic screening of hepatocyte nuclear factor (HNF) 4-gamma receptor knockout mice. Biochem Biophys Res Commun. 2006;349:825–832. doi: 10.1016/j.bbrc.2006.08.103. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Gao N, Gorski RK, White P, Hardy OT, Rafiq K, Brestelli JE, Chen G, Stoeckert CJ, Jr, Kaestner KH. Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev. 2007;21:756–769. doi: 10.1101/gad.1535507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Vatamaniuk MZ, Lee CS, Flaschen RC, Fulmer JT, Matschinsky FM, Duncan SA, Kaestner KH. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao E, Tyrberg B, Itkin-Ansari P, Lakey JR, Geron I, Monosov EZ, Barcova M, Mercola M, Levine F. Beta-cell differentiation from nonendocrine epithelial cells of the adult human pancreas. Nat Med. 2006;12:310–316. doi: 10.1038/nm1367. [DOI] [PubMed] [Google Scholar]
- Harnish DC, Malik S, Kilbourne E, Costa R, Karathanasis SK. Control of apolipoprotein AI gene expression through synergistic interactions between hepatocyte nuclear factors 3 and 4. J Biol Chem. 1996;271:13621–13628. doi: 10.1074/jbc.271.23.13621. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- Hertz R, Kalderon B, Byk T, Berman I, Za’tara G, Mayer R, Bar-Tana J. Thioesterase activity and acyl-CoA/fatty acid cross-talk of hepatocyte nuclear factor-4{alpha} J Biol Chem. 2005;280:24451–24461. doi: 10.1074/jbc.M500732200. [DOI] [PubMed] [Google Scholar]
- Hertz R, Sheena V, Kalderon B, Berman I, Bar-Tana J. Suppression of hepatocyte nuclear factor-4alpha by acyl-CoA thioesters of hypolipidemic peroxisome proliferators. Biochemical pharmacology. 2001;61:1057–1062. doi: 10.1016/s0006-2952(01)00578-0. [DOI] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol. 2008;22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ. Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4alpha (HNF4alpha). HNF4alpha regulates ornithine transcarbamylase in vivo. Journal of Biological Chemistry. 2002;277:25257–25265. doi: 10.1074/jbc.M203126200. [DOI] [PubMed] [Google Scholar]
- Jiang S, Tanaka T, Iwanari H, Hotta H, Yamashita H, Kumakura J, Watanabe Y, Uchiyama Y, Aburatani H, Hamakubo T, et al. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4alpha (HNF4alpha) isoforms in human and rats. Nuclear receptor. 2003;1:5. doi: 10.1186/1478-1336-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of molecular biology. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kiselyuk A, Farber-Katz S, Cohen T, Lee SH, Geron I, Azimi B, Heynen-Genel S, Singer O, Price J, Mercola M, et al. Phenothiazine neuroleptics signal to the human insulin promoter as revealed by a novel high-throughput screen. J Biomol Screen. 2010;15:663–670. doi: 10.1177/1087057110372257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guevel R, Oger F, Lecorgne A, Dudasova Z, Chevance S, Bondon A, Barath P, Simonneaux G, Salbert G. Identification of small molecule regulators of the nuclear receptor HNF4alpha based on naphthofuran scaffolds. Bioorganic & medicinal chemistry. 2009;17:7021–7030. doi: 10.1016/j.bmc.2009.07.079. [DOI] [PubMed] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. Target identification using drug affinity responsive target stability (DARTS) Proc Natl Acad Sci U S A. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- Murayama A, Kim MS, Yanagisawa J, Takeyama K, Kato S. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 2004;23:1598–1608. doi: 10.1038/sj.emboj.7600157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nakshatri H, Bhat-Nakshatri P. Multiple parameters determine the specificity of transcriptional response by nuclear receptors HNF-4, ARP-1, PPAR, RAR and RXR through common response elements. Nucleic Acids Res. 1998;26:2491–2499. doi: 10.1093/nar/26.10.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas-Frances V, Dasari VK, Abruzzi E, Osumi T, Latruffe N. The peroxisome proliferator response element (PPRE) present at positions -681/-669 in the rat liver 3-ketoacyl-CoA thiolase B gene functionally interacts differently with PPARalpha and HNF-4. Biochem Biophys Res Commun. 2000;269:347–351. doi: 10.1006/bbrc.2000.2249. [DOI] [PubMed] [Google Scholar]
- Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640–7651. doi: 10.1158/0008-5472.CAN-10-0824. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Yang Q, Yamagata K, Hangenfeldt KA, Miyagawa J, Kajimoto Y, Nakajima H, Namba M, Wollheim CB, Hanafusa T, et al. Human insulin gene is a target gene of hepatocyte nuclear factor-1alpha (HNF-1alpha) and HNF-1beta. Biochem Biophys Res Commun. 1999;263:566–569. doi: 10.1006/bbrc.1999.1412. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu AD, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Ligand specificity and conformational dependence of the hepatic nuclear factor-4alpha (HNF-4alpha) J Biol Chem. 2002;277:23988–23999. doi: 10.1074/jbc.M201241200. [DOI] [PubMed] [Google Scholar]
- Schroeder F, Huang H, Hostetler HA, Petrescu AD, Hertz R, Bar-Tana J, Kier AB. Stability of fatty acyl-coenzyme A thioester ligands of hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Lipids. 2005;40:559–568. doi: 10.1007/s11745-005-1416-y. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Algamas-Dimantov A, Hertz R, Nataf J, Kerman A, Peri I, Bar-Tana J. Inhibition of colorectal cancer by targeting hepatocyte nuclear factor-4alpha. Int J Cancer. 2009;124:1081–1089. doi: 10.1002/ijc.24041. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2011;334:3–13. doi: 10.1016/j.mce.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci U S A. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Winrow CJ, Marcus SL, Miyata KS, Zhang B, Capone JP, Rachubinski RA. Transactivation of the peroxisome proliferator-activated receptor is differentially modulated by hepatocyte nuclear factor-4. Gene expression. 1994;4:53–62. [PMC free article] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y, Ji Y, Wu X, Han Z, Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]
- Yamasaki N, Imito T, Murai Y, Hiramura T, Oku T, Sawada K. In: Preparation of benzimidazole derivatives as drugs. Appl PI, editor. Fujisawa Pharmaceutical Co., Ltd; 1997. [Google Scholar]
- Yao X, Hu JF, Daniels M, Yien H, Lu H, Sharan H, Zhou X, Zeng Z, Li T, Yang Y, et al. A novel orthotopic tumor model to study growth factors and oncogenes in hepatocarcinogenesis. Clin Cancer Res. 2003;9:2719–2726. [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. BIM5078 passed confirmatory and counter screens.(a) Compounds found to modulate the insulin-eGFP transgene in the primary screen were confirmed in dose-response assays. Three potential activators and four potential repressors were identified. (b) T6PNE contains E47MER, which translocates to the nucleus and becomes transcriptionally active in the presence of tamoxifen. Compounds found to reproducibly increase activity of the insulin promoter driving eGFP in the secondary screen (a) were tested for their ability to bind a multimerized E-box and drive luciferase expression. Compound 5157189 was found be estrogenic. (c) Similarly, compounds found to decrease the functional activity of the insulin-eGFP transgene (a) were tested for their ability to prevent the translocation of E47MER. Compounds 5867913, 5849200, 5359535 and 5816458 were found to be estrogenic antagonists. (d) Compounds that did not interact with E47MER were tested for their ability to modulate endogenous insulin mRNA. BIM5078, but not 5544524 or 5120083, had activity on the endogenous insulin promoter.
Supplementary Figure 2. Fatty acids bound to HNF4α preferentially inhibit exogenous insulin promoter activity. T6PNE cells were treated with 0.12 mM fatty acids for 48 hours. Effects on the exogenous insulin promoter in T6PNE is reported as percent GFP+ cells, as determined by imaging the green channel and normalizing to the total number of cells per well. Values represent the mean ± SE, n=8.
Supplementary Figure 3. Effect of reported HNF4α ligands on insulin promoter activity. T6PNE cells were treated with bezafibrate, Medica-16 or a nitro-naphthofuran derivative (“Compound 5”) for 48 hours in the presence of either 0.5 μM (a) or 1 μM (b) tamoxifen. Effects on the exogenous insulin promoter transgene in T6PNE are reported as percent GFP+ cells. Values represent the mean ± SE, n=3.
Supplementary Figure 4. Structures of BI6015 and BI6018.
Supplementary Figure 5. Docking of BI6015 (purple) in the LBD of HNF4γ with linoleic acid (cyan) crystallized. The nitro group of BI6015 forms hydrogen bonds with the backbone N of Gly197 and the side chain guanadinium group of Arg 186. The remaining parts of BI6015 primarily make hydrophobic interactions with the binding pocket, which is for the most part hydrophobic in nature. The empirical docked score (GOLD Fitness score) for this pose is 40.06, similar to that for HNF4α.
Supplementary Figure 6. Blood chemistry of mice treated with BI6015. Mice (NONcNZO10/LtJ or ICR) were injected IP with vehicle (DMSO) or BI6015 once daily for 5 days. Prior to sacrifice, blood was drawn and analyzed using a VetScan blood analyzer, measuring alkaline phosphatase (ALP, IU/L), alanine aminotransferase (ALT, IU/L), gamma glutamyl transferase (GGT, IU/L), bile acids (BA, μmol/L), total bilirubin (TBIL, mg/dL), albumin (ALB, g/dL), blood urea nitrogen (BUN, mg/dL), and cholesterol (CHOL, mg/dL). Five groups of mice were studied: normal mice injected with DMSO (Normal DMSO, n=4), normal mice injected with BI6015 at a dose of 30 mg/kg/day (Normal BI6015H, n=4) or 10 mg/kg/day (Normal BI6015L, n=4), mice injected with the hepatocellular carcinoma (HCC) cell line Hep3B and treated with DMSO (HCC DMSO, n=5) for 13–29 days, and mice injected with Hep3B and treated with BI6015 at a dose of 30 mg/kg/2day (HCC BI6015, n=4) for 29–36 days.
Supplementary Figure 7. HNF4α expression does not change in the intestine or kidney in mice receiving IP injection of BI6015. HNF4α expression was assessed as described in the Methods. Scale bar, 100 μm.
Supplementary Figure 8. NCI Panel. Data were generated by the NCI Developmental Therapeutics Program, as previously described (Shoemaker, 2006).
Supplementary Table 1. Small molecule screening data
Supplementary Table 2. List of genes affected both by genetic deletion of HNF4α in vivo and pharmacologic inhibition of HNF4α in vitro. 36% of the genes altered by genetic deletion of HNF4α in the mouse in vivo are either identical or closely related to genes modulated by pharmacologic inhibition of HNF4α in the human cell line, T6PNE.
Supplementary Table 3. List of genes affected by both pharmacologic inhibition of of HNF4α and induction of E47. Analysis was restricted to genes altered at least two-fold. The expression of 214 genes were significantly altered by BIM5078 at least two-fold. Of these, 67 were also modulated by E47 induction through 4-hydroxytamoxifen. This association was enhanced when only the genes containing E-boxes were compared. 96 E-box containing genes were altered by BIM5078. 42 of these were also altered by E47 induction