Abstract
HIV-1 infection results in the development of a diverging quasispecies unique to each infected individual. Envelope (Env)-specific neutralizing antibodies (NAbs) typically develop over months to years after infection and initially are limited to the infecting virus. In some subjects, antibody responses develop that neutralize heterologous isolates (HNAbs), a phenomenon termed broadening of the NAb response. Studies of co-crystalized antibodies and proteins have facilitated the identification of some targets of broadly neutralizing monoclonal antibodies (NmAbs) capable of neutralizing many or most heterologous viruses; however, the ontogeny of these antibodies in vivo remains elusive. We hypothesize that Env protein escape variants stimulate broad NAb development in vivo and could generate such NAbs when used as immunogens. Here we test this hypothesis in rabbits using HIV Env vaccines featuring: (1) use of individual quasispecies env variants derived from an HIV-1 subtype A-infected subject exhibiting high levels of NAbs within the first year of infection that increased and broadened with time; (2) motif optimization of envs to enhance in vivo expression of DNA formulated as vaccines; and (3) a combined DNA plus protein boosting regimen. Vaccines consisted of multiple env variants delivered sequentially and a simpler regimen that utilized only the least and most divergent clones. The simpler regimen was as effective as the more complex approach in generating modest HNAbs and was more efficient when modified, motif-optimized DNA was used in combination with trimeric gp140 protein. This is a rationally designed strategy that facilitates future vaccine design by addressing the difficult problem of generating HNAbs to HIV by empirically testing the immunogenicity of naturally occurring quasispecies env variants.
1. Introduction
The generation of potent, broad neutralizing antibodies (NAbs) effective against HIV-1 from diverse clades remains a key objective for HIV vaccines. Numerous Envelope (Env) immunization studies have resulted in NAbs of limited potency and breadth [reviewed in [1, 2]] [3-5], and these observations are found both for subtype A and B Envs [6]. The extreme variability of this protein renders empirical searches for an ideal Env immunogen virtually impossible; thus a bioinformatics-based approach may be an attractive alternative [7]. Much progress has been made in developing and validating predictive T- and B cell peptide epitopes for HIV [8]. Furthermore, some improvements in NAb induction have been achieved using rationally designed immunogens that display neutralization epitopes [5, 9]. Recent studies have shown that extremely broad NmAbs typically undergo a high degree of affinity maturation [1, 10-13]. Using bioinformatics tools to compare the env genes in the evolving quasispecies population in subjects who develop HNAbs [14-17] may reveal key mutations involved in Env escape and increasing affinity. This knowledge could guide the choice of variants that are more effective in generating broad NAbs.
HIV-1 Env is a membrane-bound trimer, rendering it technically challenging to produce an authentic Env vaccine. A theoretical advantage of DNA vaccines expressing Env is the in vivo expression of trimers that more closely mimic the native structure present on the virion surface [18-20], and these vaccines can be delivered repeatedly, with no anti-vector immunity. Despite limited immunogenicity in humans, DNA vaccines have elicited strong immune responses in small mammals [19, 21] and modest responses in non-human primates [22] [23]. Codon-optimization of DNA from non-mammalian sources increases immunogenicity, and motif-optimization further addresses the problem by optimizing short nucleotide motifs differentially found in viral and host genomes [24]. Immunogenicity of DNA can also be enhanced by combining it with viral vectors [25] or proteins in prime-boost strategies [26] [13].
We recently reported that Env quasispecies antigens derived from a SHIV-infected macaque that developed moderate neutralization breadth partially replicated the response observed in that animal [27]. The vaccine was a codon-optimized DNA-based immunization delivered in the order that recapitulated the appearance of the natural variants. Here, we describe a vaccine that incorporates naturally occurring env variants isolated from a Clade A-infected human subject who developed HNAbs within the first year of infection, and who continued to broaden and increase in potency over the next several years [17, 28]. We characterized the mutational pathway of these envs and selected key variants to recapitulate the order of presentation for vaccination. We compared the immunogenicity of vaccines delivered to rabbits as a DNA prime followed by simultaneous protein plus DNA boosts. Vaccines consisted of env variants delivered sequentially and a simpler regimen that utilized only the least and most divergent clones. The simpler regimen was as effective as the more complex approach in generating modest HNAbs and was more efficient when modified, motif-optimized DNA was used.
2. Materials and Methods
2.1 Motif-Optimization of Env genes
The motif optimized (MO) HIV env sequences were generated through an application of the information theoretic motif-finding Robins-Krasnitz algorithm [29]. Briefly, motifs of seven or fewer nucleotides are identified which are either under or over-represented in a subset of genes from any organism’s genome, controlling for amino acid order and codon usage, and these are ranked by degree of bias. The algorithm is iterative and has been mathematically proven to converge. To generate the MO sequences, we applied the Robins-Krasnitz algorithm to the set of highly expressed genes in rabbit genome. As a proxy, we took the rabbit genes homologous to the 500 most highly expressed human genes. The top 100 under and over-represented motifs were identified and ranked. The env sequence was replaced with the set of sequences that maximizes the motif weights.
2.2. Sequence analysis
Full length envelope sequences from subject QA255, obtained from blood at multiple times post-infection from 189 through 1729 days were described elsewhere [28]. In silico analyses were performed on a total of 25 full-length envelope variants. Nucleotide sequences were aligned using HIVAlign (http://www.hiv.lanl.gov/content/sequence/HMM/HmmAlign.html) and manually annotated on Geneious Pro software 5.4.4 (Biomatters, Ltd). Sequences were aligned after the signal sequence and labeled according to HXB2 Env numbering. Potential N-linked glycosylation sites (PNGs) were identified using N-Glycosite (http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html). We tested for the effects of directional selection pressure, estimating the ratio of nonsynonymous to synonymous nucleotide substitutions [ω=dN/dS (also denoted KA/KS)] using Datamonkey [30]. Positive selection is inferred when dN/dS > 1. Selecton Webserver (http://selecton.tau.ac.il/) was used to detect adaptive codon substitutions [31] using the HKY85 model of nucleotide substitution and performing a likelihood ratio test on the M8 + beta [32] versus M8a [33]. Lineage-specific analysis was performed using the GA-Branch program (http://www.datamonkey.org/) [30]. Diversity and Divergence analyses, and prediction of the Most Recent Common Ancestral (MRCA) sequence were performed using the DIVEIN program available from the University of Washington Center For AIDS Research (http://indra.mullins.microbiol.washington.edu/DIVEIN/) [34].
2.3 DNA and trimeric protein vaccines
MO vaccine clones (n=19) were created by performing site-directed mutagenesis on a synthesized MO consensus sequence (GenScript USA Inc. Piscataway, NJ) [27]. Wild type recombinant gp140 derived from the most divergent clone 1729O (LCONS) was prepared as described [35].
2.4. Immunizations
Twenty-four female New Zealand White rabbits (>2.5 kg) were housed at R&R Research, Marysville, WA. All procedures followed IACUC-approved protocols by the Oregon Health & Science University and R&R Research. DNA was delivered intradermally by Gene Gun (Bio-Rad, Hercules, CA) [27]. All animals were vaccinated with either MO or WT DNA at weeks 0, 4, 12, and 20. Boosts included 36 μg of 1729O (most divergent) plasmid DNA concurrently with 50 μg recombinant LCONS gp140 protein mixed with an equal volume of polyethylenimine (PEI, branched; Sigma-Aldrich, St. Louis, MO), were delivered intramuscularly by needle injection at weeks 29 and 35. Blood was collected two weeks after each immunization; serum was separated and heat inactivated (1-hour at 56°C).
2.4. Antibody Assays
Binding antibodies were quantified by kinetic ELISA using recombinant LCONS trimeric gp140 (rgp140) protein. Avidity Indices was measured against recombinant LCONS trimeric gp140 (rgp140) Env [12]. The IgG concentration/serum dilution necessary to block viral infectivity by 50% was reported as IC50/ID50 (inhibitory concentration/dilution). In some cases, percent neutralization achieved by single serum dilutions was recorded. A pool of pre-immune sera was used as a negative control. Values were calculated with respect to virus only wells [(virus only - cells only) - (serum - cells only)] / (virus - cells only).
2.12. Statistical Analyses
Statistical analysis was performed using SAS statistical software, version 9.2. Binding antibody titers were transformed using natural logarithm due to data skew. Repeated measures ANOVA was used to explore differences in log titers between groups. First order autoregressive covariance structure was used to account for within-subject correlation. Pre-planned comparisons were performed using contrast t-test. Bonferroni method for multiple comparison adjustment was applied to control overall type I error. Statistical significance was inferred if p < 0.05. The association of the immunization strategy with the heterologous variant neutralization was analyzed using an Area Under the Curve (AUC) approach, using the neutralization value achieved by single dilution sera (1:20) at each time point and computed using the trapezoid rule. The neutralization AUC of each subject was calculated using the formula below: where D(Ti) is the baseline neutralization value at time point i, and i=0, 2, 6, …, 37 (week) and negative data was considered zero. One- way ANOVA was used to explore if differences in AUC exist between groups. Pair-wise comparisons with Bonferroni method for multiple comparison adjustment were applied. For avidity analysis of rabbit sera, repeated measures ANOVA was used to explore for differences in avidity indices between groups.
3. Results
3.1. Rational selection of immunogens using in silico analyses
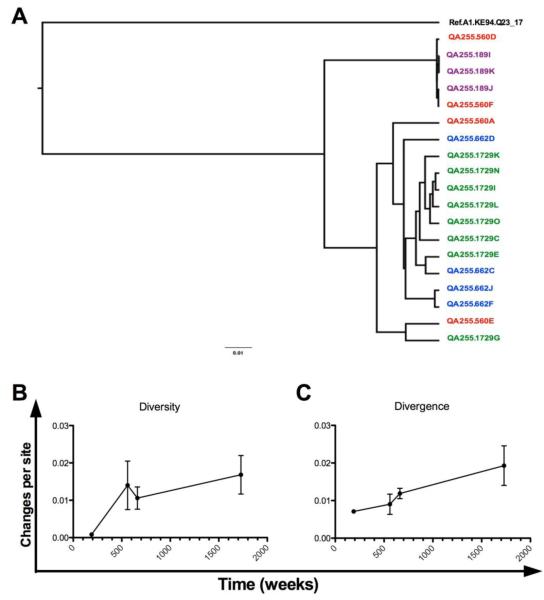
Subject QA255 was identified as having the greatest NAb breadth among 70 women at five years post-infection in a previous study [17]. We performed in silico analyses to identify patterns in the molecular evolution of HIV quasispecies envs derived from this subject [28]. Analysis of diversity, divergence, and potential N-linked glycosylation site (PNG) distribution between QA255 clones isolated at different time points showed that six amplicons from the earliest and latest time points shared similar sequence characteristics to each other, but not to other clones from their respective isolation times (data not shown). These six amplicons were excluded. Relatedness of the remaining 19 clones is shown in the phylogenetic tree (Figure 1a). These 19 clones and show a pattern of increasing diversity at each time point as well as increasing divergence based on cumulative changes from the theoretical most recent common ancestor (MRCA) (Figures 1b and c).
Figure 1. Phylogenetic analysis of QA255-derived Env quasispecies.
(A) Maximum Likelihood (ML) phylogeny illustrating the inferred relationships between env variants isolated from subject QA255 and chosen as vaccines. Wild type nucleotide sequences of the gp160 envelope were codon aligned, manually annotated, and used to reconstruct the phylogeny rooted using a Clade A sequence. Variants from different time points are color-coded as day 189: purple; day 560: red; day 662: blue; day 1729: green. (B) Genetic diversity between clones within each time point was calculated as number of nucleotide substitutions per site between each pair of sequences (i.e. pairwise distance) (C) Genetic divergence based on pairwise distance between clones from each time point and the theoretical Most Recent Common Ancestor illustrates the evolution of subject QA255’s quasispecies over time.
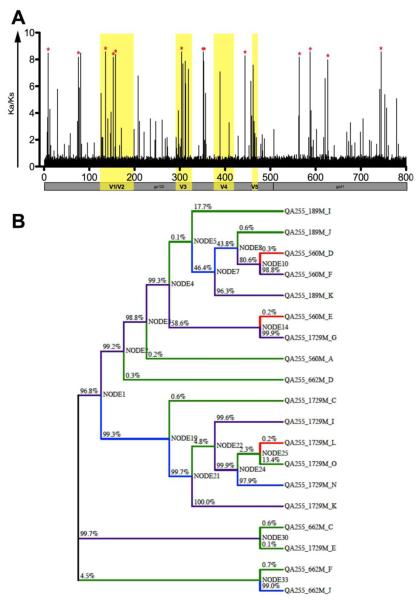
We next examined these clones to understand whether lineages evolved under differing categories or intensities of selective pressure (58). Reconstruction of the evolutionary history of dN/dS ratios along individual lineages showed a higher signal of response to diversifying selection imposed on amplicons collected from later time points. Two categories of diversifying and two categories of purifying selection were identified as the best fitting (Figure 2a). Positive selection was detected on 34% of branches that were segregated into two categories of dN/dS (dN/dS = 10000; dN/dS = 2.212). Similar to the ML phylogeny, most of the early amplicons clustered together with an ancestral branch (node 5) that showed neutral evolution. The branch leading to the cluster of late and intermediate late amplicons showed a high probability of positive selection (node 19), although some late amplicons were not positively selected after that point. We combined the lineage selection analysis with an analysis of the accumulation of non-synonymous mutations of each amplicon (Figure 3a) to infer a pattern of evolving neutralization escape, illustrated by increasing diversifying selection and accumulating non-synonymous mutations. From these analyses, clones 189J and 1729O were confirmed to be the least and most divergent amplicons. Several residues in env were evolving under strong positive selection, and these were predominantly concentrated in the V1/V2 loops and C3 regions of gp120, with a few occurring in the C1 and C4 regions, and in V3 and gp41 (Figure 2b).
Figure 2. QA255 quasispecies variants evolving under diversifying selection.
(A) Codon-specific analysis of positive selection. The red * indicates a posterior probability greater than 95% of dN/dS >1 (dN/dS described as Ka/Ks). Variable regions are highlighted in yellow. Boundary between gp120 and gp41 regions of gp160 is outlined beneath graph. (B) Phylogeny showing the distribution of estimated dN/dS per lineage. Four categories of dN/dS values (10000, 2.212, 0.633, and 0.219) were approximated according to the best-fitting model. Lineages are color-coded based on their assignment to each of these dN/dS categories and the percentage represents the posterior probability that dN/dS > 1.
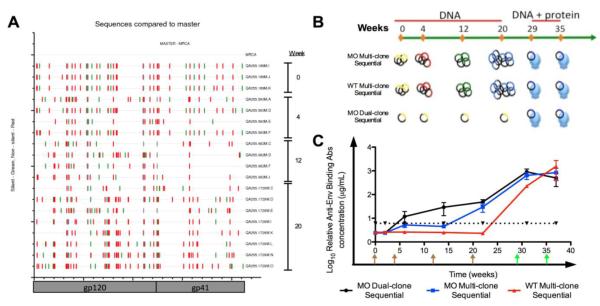
Figure 3. QA255 Env vaccine sequences, immunization approaches, and Env-specific binding antibodies.
(A) Highlighter plot illustrates accumulating non-synonymous changes in the sequence occurring longitudinally in the WT quasispecies variants incorporated in the various immunizations. Nucleotide differences from the Most Recent Common Ancestor sequence are indicated by tick marks (green, silent; red, non-silent; grey, gap). Codon location of the env gene is shown on the X-axis. Brackets to the right outline clones used for immunization on respective weeks. (B) Immunization Strategies. Rabbits were vaccinated with four either MO or WT DNA primes at weeks (brown arrows) 0, 4, 12, and 20, followed by two combination MO 1729O DNA plus LCONS protein boosts in the presence of PEI adjuvant (green arrows) on weeks 29 and 35. (C) LCONS Envelope-specific binding antibodies elicited by the immunization strategies. Relative antibody concentrations of serum samples collected two weeks after each immunization were measured by kinetic ELISA against trimeric LCONS gp140. MO Dual-clone Sequential (MO Dual-clone), black; MO Multi-clone Sequential (MO Multi-clone), blue; WT Multi-clone Sequential (WT Multi-clone), red. Dotted line represents 2.5× pre-immune titers.
3.2. Sequential inoculation of motif-optimized DNA elicits strong antibody responses
Three groups of eight rabbits were immunized with longitudinally isolated QA255 env genes administered sequentially to mimic the gradual divergence in QA255 quasispecies (Fig 3a). All rabbits received two immunizations with a combination of motif-optimized (MO) DNA (most divergent d1729O) and cognate protein (trimeric LCONS) (Figure 3b). Rabbits in the MO Multi-clone Sequential group were primed with the nineteen gp160 env genes given sequentially in four timepoint-specific subgroups, and then boosted with the DNA and protein combination described above. To assess the effects of motif optimization, we immunized a group of rabbits with non-optimized DNA (WT Multi-clone Sequential) using the same immunization regimen as the MO Multi-clone Sequential group. To determine the impact of the intermediate clones on the development of NAbs, rabbits in the MO Dual-clone Sequential group were primed with a single clone of least divergence (relative to the MRCA) isolated early in infection (d189) and boosted as described.
Binding antibodies were detected during the DNA primes using MO DNA only but not with WT prime (Figure 3c). Despite differences in responses to the prime, the first combined MO DNA plus protein boost increased antibody responses in all three groups to nearly comparable levels. The most notable increase between the final DNA prime (week 22) and first DNA plus protein boost (week 31) was in the WT Multi-clone Sequential group than either of the MO groups (p<0.0006 and p<0.0024 versus MO Dual-clone and MO Multi-clone Sequential, respectively), suggesting that the boosting strategy combining MO DNA and trimeric protein could greatly enhance the poor priming by WT DNA. Significant differences in binding antibody responses were only observed between the MO Dual-clone Sequential and WT Multi-clone Sequential group (p<0.0062); the difference between the two Sequential groups trended towards significance (p=0.0811, unadjusted p-value = 0.0270). Thus MO DNA was a more effective priming agent than WT, and two immunizations with combined MO DNA plus protein was as effective at raising Env-specific antibodies as four MO DNA primes followed by two combined MO DNA plus protein boosts. Immunizing with the least and most divergent clones led to similar levels of Env-specific binding Abs as immunizations with all clones given sequentially.
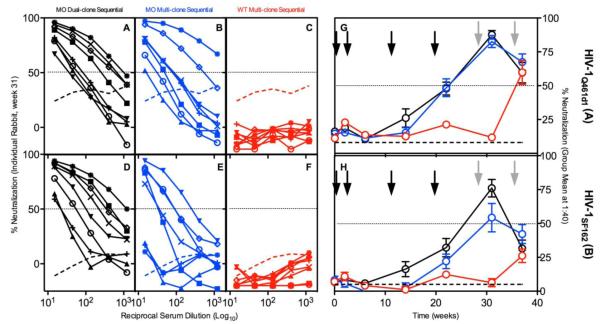
Modest heterologous NAbs were elicited against neutralization-sensitive Clade A and Clade B viruses and, to a lesser degree, against a subset of viruses from a more resistant heterologous panel. NAbs against HIV-1Q461d1 and HIV-1SF162 were detectable after the third DNA immunization and thereafter in the MO- but not WT-primed groups (Figure 4). The first combination MO DNA plus protein immunization boosted week 31 titers above 50% neutralization in 13/16 MO primed rabbits (Fig. 4A-F). The second combined MO DNA plus protein boost resulted in similar levels of NAbs in all immunization groups. MO Dual-clone- and MO Multi-clone Sequential immunization groups had stronger NAbs against Q461d1 than the WT Multi-clone Sequential group (p<0.0001 and p<0.001, respectively), thereby showing the advantage of using MO sequences. These groups also had significantly stronger SF162-NAbs than the WT Multi-clone Sequential (Table 1, P=0.0001 and P<0.05, respectively).
Figure 4. Heterologous rabbit humoral response against sensitive Clades A and B viruses.
Serial dilutions of sera from week 31 (post first DNA plus protein boost) was tested for neutralization against sensitive Clades A and B heterologous clones HIV-1Q461d1 (A-C) and HIV-1SF162 (D-F). Individual rabbit samples are shown with different symbols for inter-virus comparison. Thick broken line indicates pool pre-immune serum neutralization values. Thin dotted line demarks 50% neutralization. (G & H) Mean ± SEM neutralization values obtained at single dilution (1:40) of rabbit sera was plotted longitudinally. Columns: MO Dual-clone Sequential, black/left; MO Multi-clone Sequential, blue/middle; WT Multi-clone Sequential, red/right. Black arrows represent DNA immunizations, gray arrows represent DNA plus protein boosts.blue/middle; WT Multi-clone Sequential, red/right. Black arrows represent DNA immunizations, gray arrows represent DNA plus protein boosts.
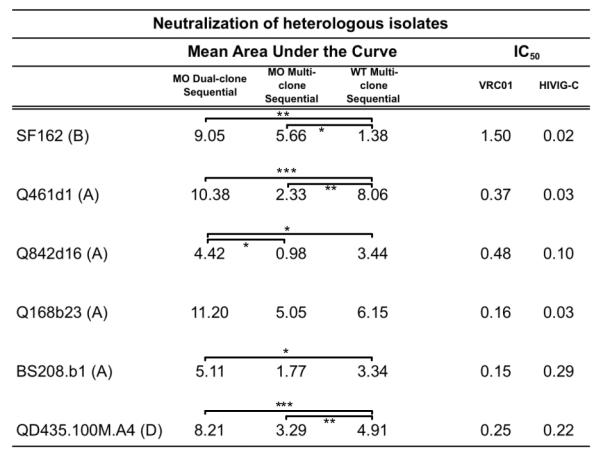
TABLE 1. Neutralization of selected heterologous isolates measured by area under the curve.
Area under the curve analysis was performed on longitudinal percent neutralization values taken from single dilution (1:20) rabbit sera. Numerical IC50 values for mNAb VRC01 and Clade C pooled polyclonal IgG from chronic infection are included for comparison. Letters in parentheses indicate clade of HIV-1 virus tested.
|
We did not detect neutralization ID50 titers against an extended panel of 16 heterologous viruses (data not shown). Although not reaching 50% neutralization, there was increasing neutralization activity in longitudinal samples of rabbit antisera. The area under the curve (AUC) of percent neutralization achieved by single-dilution (1:20) rabbit antisera against three subtype A and one subtype D viruses were calculated from week 0 to week 37 and shown in Table 1. This analysis shows significant differences between all three immunization groups against a subset of HIVIG-C sensitive clones [36]. The Dual-clone Sequential strategy was consistently the most effective when tested against this panel of heterologous viruses (Table 1), with higher AUC values in all cases.
3.3. MO DNA in Sequential inoculations elicit increasingly high avidity antibodies
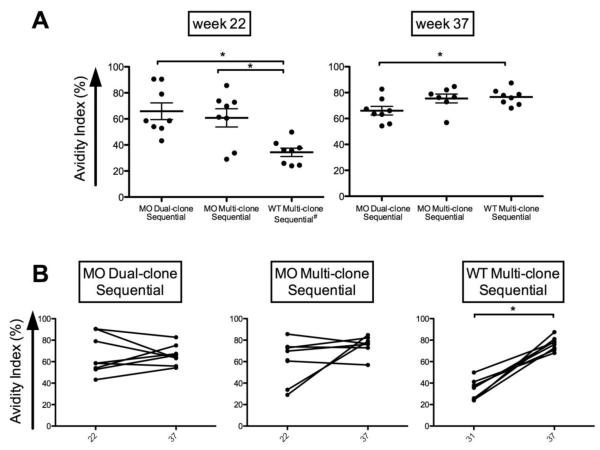
We compared the antibody avidity indices following the fourth MO DNA prime (week 22) and the second MO DNA plus protein boost (week 37) for MO Multi-clone and MO Dual-clone Sequential Groups (Figure 5a). Since no Env-specific antibodies were detectable after the fourth WT DNA prime for the WT Multi-clone Sequential Group, we compared responses following the first and second MO DNA plus protein boost (week 31 and 37). Both of the MO-primed groups have a higher avidity index at week 22 after only DNA than that of the WT-primed rabbits at week 31 after receiving DNA plus protein (p<0.005, Dual-clone Sequential; p<0.05, Sequential). However, no differences in avidity indices were identified between the two MO-primed groups after two DNA plus protein boosts. The avidity index of WT Multi-clone Sequential sera determined at week 37 increased significantly, surpassing that of MO Dual-clone Sequential group (p<0.01) (Figure 5b, top). A comparison of the change in avidity index between week 22 and 37 for MO-primed groups and week 31 and 37 for WT-primed groups shows that only the WT Multi-clone Sequential group increased significantly (p<0.001) (Figure 5b). Consistent with binding antibody and NAb data, combination DNA plus protein immunization induces high avidity after just two immunizations that is greater than that observed in the MO Dual-clone Sequential group.
Figure 5. Qualitative assessment of rabbit humoral responses.
(A) Avidity ELISA. Avidity indices against LCONS gp140 in samples collected after the final DNA prime for MO groups (week 22) and after the first DNA plus protein boost for WT Multi-clone Sequential (week 31) and after the final DNA plus protein boost (week 37). (B) Comparison of avidity indices between week 37 and week 22 (MO-primed group) and between week 37 and week 31 (WT-primed group) in samples as in top to determine affinity maturation. Asterisks indicate P<0.05 using tests described in the Methods.
4. Discussion
During HIV-1 infection, the appearance of broad NAbs follows that of the autologous response, which is a primary driving force in the divergence of the host quasispecies [37]. The specific amino acid sequences that accompany the broadening of this response are intensively studied in an effort to identify neutralization determinants [16, 38-43]. We hypothesize that one or more of these escape variants could effectively stimulate the development of heterologous NAbs. We thus chose multiple immunogens from the quasispecies of a subject who developed modest neutralization breadth. Selection of individual envs was guided by analyzing both lineage- and population-specific selection pressures, which showed that they represent a predominant population of the viral quasispecies.
The env genes selected by this strategy were highly immunogenic in rabbits when delivered as MO DNA vaccines, resulting in moderate levels of heterologous Tier 1 NAbs after DNA vaccines alone that were boosted when with combined MO DNA plus trimeric gp140 protein. These levels are similar to those obtained in vaccine studies with a poxvirus prime, protein boost approach using the most immunogenic of the subtype A early transmitted Envs [6]. Similar to that study, this sequential DNA and protein strategy was only effective in eliciting NAbs against sensitive heterologous Tier 1 viruses in the absence of significant autologous NAbs. The inherent neutralization resistance of these clones may explain these results; we confirmed that these variants were neutralization resistant, typical of recently isolated HIV-1 [44] and observed in prior vaccine studies [4]. Because these immunization regimens are not effective in generating NAbs that are as potent as those seen in infection, only responses against the most sensitive isolates can be measured. The MO Dual-clone Sequential approach was more immunogenic than the Multi-clone Sequential approach based on NAbs against four additional subtype A and D heterologous viruses. Both MO and WT Multi-clone Sequential vaccines resulted in higher avidity following the combination DNA plus protein boosting. Although the WT DNA did not elicit any NAbs, the strong boosting of binding antibodies by the MO DNA plus protein in all groups revealed the effectiveness of delivering these components concurrently. A caveat of this analysis is that trimeric gp140 on an ELISA plate may not recapitulate the Envelope spike on the virion; however, observing similar results in the neutralization activity of rabbit antisera confirms the benefit of co-immunizing with MO DNA and protein. We are currently examining these effects to understand the mechanisms involved.
This study shows the potential to exploit natural evolutionary variants to generate HNAb development in vivo. Moreover, a streamlined vaccine regimen using only two env clones to represent the quasispecies evolutionary divergence that occurred in vivo was as effective as the multi-clone presentation of groups of diverging envs for this subject. At this level of resolution, there is no apparent advantage to the multicomponent vaccine using these specific sequences. Other differences between these immunization strategies may also be occluded by the enhanced expression achieved by motif optimization of the vaccine clones. Taken together, our results indicate that modest NAbs can be elicited by a relatively simple vaccine strategy combining DNA and trimeric Env. Further work is clearly needed to predict the relationships between antigenicity and immunogenicity of Env vaccines in vivo and to understand the role of individual Env proteins in influencing B cell maturation and the development of NAbs. This strategy may hold significant promise when incorporated with Env immunogens from subjects with greater neutralization potency.
Highlights.
Immunization with naturally evolving env genes, given sequentially, is promising
Motif-optimized env sequences were superior to wild type for neutralizing antibodies
Simultaneous boosting with DNA and protein at different sites enhanced antibodies
Antibodies that neutralized heterologous HIV-1 isolates were obtained in rabbits
ACKNOWLEDGEMENTS
We thank Ann Hessell and Ilhem Messaoudi for their suggestions and comments on the manuscript. We are also grateful to William Sutton and Than-Phuong Chu for technical assistance. We thank Shelly Krebs, Dina Kovarik, and Pablo Jaworski for helpful discussions, Katie Bosch for generating the envelope clones used as a base for this study, and Travis Beckett for designing the consensus sequence. TZM-bl and 293T cell lines were obtained from the NIH AIDS Research and Reference Reagent Program. HIVIG-C was provided by David Montefiori. This work was supported by National Institutes of Health grants P01 AI087064 (H.R., N.L.H. and L.S.), P01 AI054564 (N.L.H., J.O. and L.S.), P51 OD011092 (N.L.H. and B.P.), HD058304 (J.O), NIH 5 T32 AI7472-17 (F.P.), and a Global Health Grand Challenges Explorations Grant from the Bill and Melinda Gates Foundation (N.L.H).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].McElrath MJ, Haynes BF. Induction of Immunity to Human Immunodeficiency Virus Type-1 by Vaccination. Immunity. 2010;33(4):542–54. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vaine M, Lu S, Wang S. Progress on the induction of neutralizing antibodies against HIV type 1 (HIV-1) BioDrugs. 2009;23(3):137–53. doi: 10.2165/00063030-200923030-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Smith DH, Winters-Digiacinto P, Mitiku M, O’Rourke S, Sinangil F, Wrin T, et al. Comparative immunogenicity of HIV-1 clade C envelope proteins for prime/boost studies. PLoS One. 2010;5(8):e12076. doi: 10.1371/journal.pone.0012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vaine M, Duenas-Decamp M, Peters P, Liu Q, Arthos J, Wang S, et al. Two Closely Related Env Antigens from the Same Patient Elicited Different Spectra of Neutralizing Antibodies against Heterologous HIV-1 Isolates. Journal of Virology. 2011;85(10):4927–36. doi: 10.1128/JVI.00081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zolla-Pazner S, Kong XP, Jiang X, Cardozo T, Nadas A, Cohen S, et al. Cross-clade HIV-1 neutralizing antibodies induced with V3-scaffold protein immunogens following priming with gp120 DNA. J Virol. 2011 Oct;85(19):9887–98. doi: 10.1128/JVI.05086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kraft Z, Strouss K, Sutton WF, Cleveland B, Tso FY, Polacino P, et al. Characterization of Neutralizing Antibody Responses Elicited by Clade A Envelope Immunogens Derived from Early Transmitted Viruses. Journal of Virology. 2008;82(12):5912–21. doi: 10.1128/JVI.00389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sekaly R, Pulendran B. Systems biology in understanding HIV pathogenesis and guiding vaccine development. Curr Opin HIV AIDS. 2012 Jan;7(1):1–3. doi: 10.1097/COH.0b013e32834e0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sirskyj D, Diaz-Mitoma F, Golshani A, Kumar A, Azizi A. Innovative bioinformatic approaches for developing peptide-based vaccines against hypervariable viruses. Immunol Cell Biol. 2011 Jan;89(1):81–9. doi: 10.1038/icb.2010.65. [DOI] [PubMed] [Google Scholar]
- [9].Moseri A, Tantry S, Sagi Y, Arshava B, Naider F, Anglister J. An optimally constrained V3 peptide is a better immunogen than its linear homolog or HIV-1 gp120. Virology. 2010 Jun 5;401(2):293–304. doi: 10.1016/j.virol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, et al. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Science Translational Medicine. 2011;3(81):81ra36–81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Richmond JF, Lu S, Santoro JC, Weng J, Hu SL, Montefiori DC, et al. Studies of the neutralizing activity and avidity of anti-human immunodeficiency virus type 1 Env antibody elicited by DNA priming and protein boosting. J Virol. 1998 Nov;72(11):9092–100. doi: 10.1128/jvi.72.11.9092-9100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sundling C, Forsell MNE, O’Dell S, Feng Y, Chakrabarti B, Rao SS, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. Journal of Experimental Medicine. 2010;207(9):2003–17. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vaine M, Wang S, Hackett A, Arthos J, Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine. 2010 Apr 9;28(17):2999–3007. doi: 10.1016/j.vaccine.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009 Jan;83(2):757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009 Jul;83(14):7337–48. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007 Jun;81(12):6187–96. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Piantadosi A, Panteleeff D, Blish CA, Baeten JM, Jaoko W, McClelland RS, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol. 2009 Oct;83(19):10269–74. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Doria-Rose N. DNA vaccine strategies: candidates for immune modulation and immunization regimens. Methods. 2003;31(3):207–16. doi: 10.1016/s1046-2023(03)00135-x. [DOI] [PubMed] [Google Scholar]
- [19].Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, et al. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006 May 22;24(21):4531–40. doi: 10.1016/j.vaccine.2005.08.023. [DOI] [PubMed] [Google Scholar]
- [20].Williams JA, Carnes AE, Hodgson CP. Plasmid DNA vaccine vector design: Impact on efficacy, safety and upstream production. Biotechnology Advances. 2009;27(4):353–70. doi: 10.1016/j.biotechadv.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001 Nov;75(22):10991–1001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barnett SW, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, et al. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001 Jun;75(12):5526–40. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mossman SP, Pierce CC, Watson AJ, Robertson MN, Montefiori DC, Kuller L, et al. Protective immunity to SIV challenge elicited by vaccination of macaques with multigenic DNA vaccines producing virus-like particles. AIDS Res Hum Retroviruses. 2004 Apr;20(4):425–34. doi: 10.1089/088922204323048177. [DOI] [PubMed] [Google Scholar]
- [24].Huang Y, Krasnitz M, Rabadan R, Witten DM, Song Y, Levine AJ, et al. A recoding method to improve the humoral immune response to an HIV DNA vaccine. PloS one. 2008;3(9):e3214. doi: 10.1371/journal.pone.0003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Doria-Rose NA, Ohlen C, Polacino P, Pierce CC, Hensel MT, Kuller L, et al. Multigene DNA priming-boosting vaccines protect macaques from acute CD4+-T-cell depletion after simian-human immunodeficiency virus SHIV89.6P mucosal challenge. J Virol. 2003 Nov;77(21):11563–77. doi: 10.1128/JVI.77.21.11563-11577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tomaras GD, Haynes BF. Strategies for eliciting HIV-1 inhibitory antibodies. Current Opinion in HIV and AIDS. 2010;5(5):421–7. doi: 10.1097/COH.0b013e32833d2d45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. Journal of virology. 2011 Jun;85(11):5262–74. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bosch KA, Rainwater S, Jaoko W, Overbaugh J. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology. 2010;398(1):115–24. doi: 10.1016/j.virol.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Robins H, Krasnitz M, Barak H, Levine AJ. A relative-entropy algorithm for genomic fingerprinting captures host-phage similarities. J Bacteriol. 2005 Dec;187(24):8370–4. doi: 10.1128/JB.187.24.8370-8374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Poon AFY, Frost SDW, Pond SLK. Detecting Signatures of Selection from DNA Sequences Using Datamonkey. 2009;537:163–83. doi: 10.1007/978-1-59745-251-9_8. [DOI] [PubMed] [Google Scholar]
- [31].Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W506–11. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000 May;155(1):431–49. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003 Jan;20(1):18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- [34].Deng W, Maust B, Nickle* D, Learn** G, Liu Y, Heath L, et al. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. BioTechniques. 2010;48(5):405–8. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, et al. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003 Oct;77(20):11244–59. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nixon DF, Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, et al. Anti-V3 Monoclonal Antibodies Display Broad Neutralizing Activities against Multiple HIV-1 Subtypes. PLoS One. 2010;5(4):e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Frost SDW. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proceedings of the National Academy of Sciences. 2005;102(51):18514–9. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moore PL, Gray ES, Morris L. Specificity of the autologous neutralizing antibody response. Current Opinion in HIV and AIDS. 2009;4(5):358–63. doi: 10.1097/COH.0b013e32832ea7e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009 Sep;5(9):e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bunnik EM, Lobbrecht MSD, van Nuenen AC, Schuitemaker H. Escape from autologous humoral immunity of HIV-1 is not associated with a decrease in replicative capacity. Virology. 2010;397(1):224–30. doi: 10.1016/j.virol.2009.11.009. [DOI] [PubMed] [Google Scholar]
- [41].Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous Neutralizing Humoral Immunity and Evolution of the Viral Envelope in the Course of Subtype B Human Immunodeficiency Virus Type 1 Infection. Journal of Virology. 2008;82(16):7932–41. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].van Gils MJ, Bunnik EM, Burger JA, Jacob Y, Schweighardt B, Wrin T, et al. Rapid Escape from Preserved Cross-Reactive Neutralizing Humoral Immunity without Loss of Viral Fitness in HIV-1-Infected Progressors and Long-Term Nonprogressors. Journal of Virology. 2010;84(7):3576–85. doi: 10.1128/JVI.02622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009 Sep;5(9):e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nkolola JP, Peng H, Settembre EC, Freeman M, Grandpre LE, Devoy C, et al. Breadth of Neutralizing Antibodies Elicited by Stable, Homogeneous Clade A and Clade C HIV-1 gp140 Envelope Trimers in Guinea Pigs. Journal of Virology. 2010;84(7):3270–9. doi: 10.1128/JVI.02252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]