Abstract
The liver is a major organ of lipid metabolism, which is markedly changed in response to physiological nutritional demand; however, the regulation of hepatic lipogenic gene expression in early life is largely unknown. In this study, we show that expression of glycerol-3-phosphate acyltransferase 1 (GPAT1; Gpam), a rate-limiting enzyme of triglyceride biosynthesis, is regulated in the mouse liver by DNA methylation, an epigenetic modification involved in the regulation of a diverse range of biological processes in mammals. In the neonatal liver, DNA methylation of the Gpam promoter, which is likely to be induced by Dnmt3b, inhibited recruitment of the lipogenic transcription factor sterol regulatory element–binding protein-1c (SREBP-1c), whereas in the adult, decreased DNA methylation resulted in active chromatin conformation, allowing recruitment of SREBP-1c. Maternal overnutrition causes decreased Gpam promoter methylation with increased GPAT1 expression and triglyceride content in the pup liver, suggesting that environmental factors such as nutritional conditions can affect DNA methylation in the liver. This study is the first detailed analysis of the DNA-methylation–dependent regulation of the triglyceride biosynthesis gene Gpam, thereby providing new insight into the molecular mechanism underlying the epigenetic regulation of metabolic genes and thus metabolic diseases.
The liver is a major organ of lipid metabolism, which is physiologically changed during organ maturation (1,2). The rate of hepatic de novo lipogenesis (i.e., triglyceride [TG] biosynthesis) is very low during the suckling period, when fat intake is high from milk, but it rises with the onset of weaning, when glucose is used as a source of energy (1). Thus, hepatic gene expression may change markedly before and after weaning, which could be regulated in response to nutritional demand.
TG is the major storage form of energy in animals. TG biosynthesis begins with the acylation of glycerol-3-phosphate by glycerol-3-phosphate acyltransferase 1 (GPAT1; Gpam) to form lysophosphatidic acid; this is the rate-limiting step in the hepatic TG biosynthesis pathway (3). In the acylation process, fatty acids produced by stearoyl CoA desaturase 1 (SCD1; Scd1) and fatty acid synthase (FAS; Fasn) are used as acyl donors. Among the lipogenic enzymes, GPAT1 plays an important role in the regulation of hepatic TG biosynthesis (4,5). The lipogenic genes such as Gpam, Scd1, and Fasn are activated by sterol regulatory element–binding protein-1c (SREBP-1c), which is a transcription factor and master regulator of lipogenesis. Indeed, their promoter regions contain the SREBP-responsive elements (SREs) (6–8). Aberrant lipogenic gene regulation can contribute to fatty liver, which is associated with obesity, type 2 diabetes, and insulin resistance (9). However, the molecular mechanism involved in the regulation of lipogenic genes during early life remains largely unclear.
The methylation of cytosine residues in DNA is a major epigenetic modification, and its role is well studied in organ development and cell differentiation (10–12). In most instances, DNA methylation of the promoter regions causes suppression of gene expression (13). In mammals, three CpG DNA methyltransferases (Dnmt)—Dnmt1, Dnmt3a, and Dnmt3b—coordinately regulate DNA methylation in the genome. Dnmt1 promotes DNA methylation after DNA replication and plays a major role in the maintenance of methylation (14). Dnmt3a and Dnmt3b are required for the initiation of de novo DNA methylation (10).
DNA methylation may be affected by environmental factors, thereby regulating a variety of metabolic processes and diseases (15–18). Although the fetal and neonatal periods, which are highly plastic to environmental changes, should be under the epigenetic control, the role of DNA methylation in early life has not fully been addressed. This study is the first demonstration that the DNA methylation status of the Gpam promoter and its mRNA expression are inversely correlated during mouse liver maturation. This study highlights the role of DNA methylation in the regulation of lipogenic genes, thereby providing new insight into the molecular mechanism underlying epigenetic regulation of metabolic diseases.
RESEARCH DESIGN AND METHODS
Animals and the experiment with high-fat/high-sucrose diet–fed dams.
Pregnant female C57BL/6 mice were obtained from Japan SLC (Hamamatsu, Japan). The mice were fed ad libitum a standard rodent chow, CRF1 (Charles River Japan, Tokyo, Japan). Offspring at the indicated ages were used for tissue sampling. They were weaned at 25 days of age and thereafter fed CRF1 throughout the experiment.
The experiment with high-fat/high-sucrose (HF/HS) diet–fed dams was performed as follows. Six-week-old male and female C57BL/6 mice (Japan SLC) were crossed, and pregnant dams were used. Two weeks before the beginning of mating and throughout the experiment, dams were fed ad libitum either CRF1 (standard) or HF/HS diet (D12079B; Research Diets, New Brunswick, NJ). Schematic experimental design is shown in Supplementary Fig. 1. Five-day-old offspring were used for analysis. All animal experiments were approved by Institutional Animal Care and Use Committee of Tokyo Medical and Dental University (approval identification number 0090041).
Quantitative real-time PCR analysis.
Gene expression levels were measured as described (19). The primers used are shown in Supplementary Table 1.
Western blot analysis.
The nuclear fraction of cell lysate was prepared using a ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas, Glen Burnie, MD). Mitochondrial fraction of the liver was prepared as described (4). Western blot analysis was performed as described previously (19). Anti-GPAT1 (sc-161674; Santa Cruz Biotechnology, Santa Cruz, CA), anti-SCD1 (ab19862; Abcam, Cambridge, MA), anti-FAS (sc-48357; Santa Cruz Biotechnology), anti–SREBP-1 (ab3259; Abcam), anti-CoxIV (ab14774; Abcam), anti–α-tubulin (T9026; Sigma-Aldrich, St. Louis, MO), anti-Lamin a/c (sc-20681; Santa Cruz Biotechnology), anti-Dnmt3b (IMG184A; Imgenex, San Diego, CA), or histone H1 (sc-10806; Santa Cruz Biotechnology) was used as the primary antibody.
Bisulfite DNA methylation analysis.
Sodium bisulfite treatment of genomic DNA was performed with a BisulFast DNA modification kit (Toyobo, Tokyo, Japan) according to the manufacturer’s instructions. Sequential PCR amplification of the genes of interest was performed using specific primers (Supplementary Table 2). The PCR profile was as described in Supplementary Table 2. The amplified fragments were ligated into the vector pGEM-T-easy (Promega KK, Tokyo, Japan) and sequenced. At least 16 bacterial colonies were picked up per PCR amplification. A web-based tool, quantification tool for methylation analysis, was used for bisulfite sequencing analysis of CpG methylation (http://quma.cdb.riken.jp/) (20).
Quantification of DNA methylation of the Gpam promoter.
DNA digestion with methylation-sensitive HpaII and quantitative real-time PCR were performed as described previously (21), with the following primers: forward primer 5′-CCCTAAAACTGGCTCCGGA-3′ and reverse primer 5′-CAGCCAATCGAAAGCTTCAGA-3′. The forward primer contains the HpaII site of Gpam promoter (underlined).
Primary culture of mouse hepatocytes.
Primary hepatocytes were isolated as described previously (22,23).
Preparation of recombinant adenovirus.
The full-length mouse Dnmt3b1 cDNA was subcloned into the pShuttle vector provided in the BD Adeno-X Expression System (BD Biosciences, Franklin Lakes, NJ). BD Adeno-X enhanced green fluorescent protein (GFP) was used as a control (Ad-GFP; BD Biosciences). Ad–SREBP-1c (active-nuclear form) (24) was kindly provided from Dr. Hitoshi Shimano (Tsukuba University). Each recombinant adenovirus (Ad-Dnmt3b, Ad-SREBP-1c, or Ad-GFP) was added to the medium of primary cultured hepatocytes (1.8 × 107 infection-forming units in 500 μL).
TG/diacylglycerol synthesis in isolated liver and primary hepatocytes.
The liver was dissected from tendon to tendon and placed in modified Krebs-Henseleit buffer containing 4% fatty acid-free bovine serum albumin (Sigma-Aldrich), 5 mmol/L glucose, and 0.5 mmol/L palmitate, giving a palmitate-to-bovine serum albumin molar ratio of 1:1. After a 30-min preincubation period, liver strips were transferred to vials containing 0.5 μCi/mL [1-14C]palmitate (GE Healthcare Life Sciences, Buckinghamshire, U.K.) for 60 min. For primary hepatocytes, cells were incubated with [1-14C]palmitate in medium for 6 h. From the liver or cells, lipids were extracted with chloroform/methanol (2:1) and resolved by thin-layer chromatography (hexane/ethyl ether/acetic acid = 60:40:3) followed by photoimaging detection.
Transfection and luciferase assay with methylated plasmids.
A luciferase gene construct containing the Gpam promoter fragment (from −489 to +79, taking the first nucleotide of exon 1 as +1) was prepared. For in vitro DNA methylation, the construct was digested with Asp718/XhoI, and a fragment containing the Gpam promoter (−489 to +79) was purified, which was followed by treatment with SssI (CpG methylase) as described previously (17). Methylation was confirmed by digestion with a methylation-sensitive restriction enzyme, HpaII. The luciferase assay was performed as described in (19,24).
Chromatin immunoprecipitation analysis.
Chromatin immunoprecipitation (ChIP) was performed using an assay kit (Upstate, Temecula, CA) (25). The lysate was incubated with DynaBeads protein G-conjugated (Life Technologies, Carlsbad, CA) anti–SREBP-1c (sc-8984X; Santa Cruz Biotechnology), anti-Dnmt1 (sc-20701; Santa Cruz Biotechnology), anti-Dnmt3a (ab2850; Abcam), anti-Dnmt3b (ab2851; Abcam), anti-trimethyl-histone H3 (Lys4) (#07-473; Upstate), anti-acetyl-histone H3 (Lys9) (#06-911; Millipore, Temecula, CA), anti-dimethyl-histone H3 (Lys9) (#9753; Cell Signaling Technology, Danvers, MA) antibody, or rabbit normal IgG (sc-2027; Santa Cruz Biotechnology). The ChIP-enriched DNA samples were analyzed by quantitative PCR. PCR primers were designed to locate SREs of the Gpam or Scd1 promoter (Supplementary Table 3).
Liver TG analysis.
The liver TG levels were measured by enzymatic colorimetry as described (19).
Statistical analysis.
Statistical analysis was performed using Student t test and ANOVA followed by Scheffe test. Data were expressed as the mean ± SE. A P value of <0.05 was considered statistically significant.
RESULTS
Lipogenic gene and SREBP-1c mRNA expression in the neonatal and adult livers.
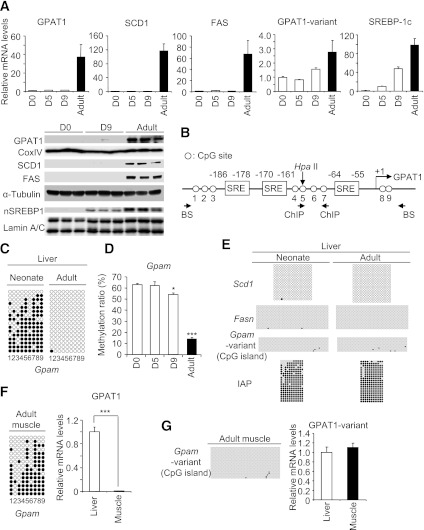
We examined lipogenic gene expression in the neonatal and adult livers before and after weaning. The GPAT1 mRNA was only slightly expressed in the neonatal liver (days 0, 5, and 9), but markedly increased (∼40-fold) in the adult liver (10 weeks of age). Similar to GPAT1, both SCD1 and FAS mRNA levels, as well as protein levels of GPAT1, SCD1, and FAS, and GPAT-mediated TG/diacylglycerol (DG) biosynthesis were increased in the adult liver relative to the neonatal liver (Fig. 1A and Supplementary Fig. 2). There is a GPAT1 splicing-variant (GPAT1-variant) with an alternative first noncoding exon (GenBank accession number: NM_008149) using an alternative promoter that is ∼30 kb downstream from the Gpam promoter containing SREs, thus producing the identical coding GPAT1 protein. In this study, its mRNA expression showed a relatively small increase (approximately threefold) from neonates (day 0) to adults (Fig. 1A).
FIG. 1.
Lipogenic gene expression and DNA methylation in the neonatal and adult mice livers. A: Relative lipogenic gene and SREBP-1c mRNA levels in the neonatal (days after birth: 0, 5, and 9) and adult livers (after weaning, 10 weeks of age), examined by quantitative real-time PCR. Values for day 0 were set at 1. Western blot analysis of GPAT1, SCD1, FAS, and nuclear form of SREBP-1 (nSREBP1) in neonatal (days 0 and 9) and adult livers (10 weeks of age). For GPAT1, mitochondrial fraction was used. For FAS/SCD1 and SREBP-1, cytoplasmic and nuclear fractions were used, respectively. α-Tubulin and Lamin a/c are cytoplasmic and nuclear markers, respectively, used for loading controls. B: Schematic representation of the Gpam promoter region. The Gpam promoter contains three SREs (−186 to −178, −170 to −161, and −64 to −55, transcription start site counted as +1). Each circle denotes a CpG site, numbered 1–9. The HpaII site is indicated (arrow). Position of PCR primers for bisulfite PCR analysis (BS) and ChIP analysis are indicated. C: Bisulfite analysis of the Gpam promoter. PCR primers used in bisulfite analysis for the Gpam promoter are shown in B. Filled circles are methylated and open circles are unmethylated CpGs. The numbers beneath are as shown in B. The left panel is a neonatal liver sample (day 0), and the right panel is an adult liver sample (10 weeks of age). Representative results of three independent animals of each group with similar results are shown. D: DNA methylation of the Gpam promoter, estimated by HpaII digestion followed by quantitative real-time PCR. Genomic DNA treated with BamHI alone was prepared as an uncut control (i.e., a fully methylated control [100%]) and diluted as a standard dilution series. *P < 0.05, ***P < 0.001 compared with D0 sample; n = 3. E: Bisulfite analysis of the Scd1 and Fasn promoters and the alternative Gpam promoter, containing CpG island, and IAP in the neonatal and adult liver. Representative results of three independent animals of each group with similar results are shown. F: Bisulfite analysis and quantitative real-time PCR mRNA analysis of the Gpam promoter in the adult skeletal muscle (10 weeks of age). Values for the liver were set at 1. n = 3. For bisulfite analysis, a typical result from three independent experiments with similar results is shown. ***P < 0.001. G: Bisulfite analysis of the alternative Gpam promoter in the adult skeletal muscle and relative GPAT1-variant mRNA levels in the adult liver and skeletal muscle. n = 3. CoxIV, cytochrome oxidase IV, a mitochondrial marker.
SREBP-1c mRNA was weakly expressed in the neonatal liver (day 0) and increased markedly (∼100-fold) in the adult liver (10 weeks of age). At day 9, approximately half the level seen in the adults was observed (Fig. 1A). SREBP-1c protein level was very low on day 0 and modestly and markedly increased on day 9 and in adults, respectively (Fig. 1A). Namely, on day 9, SREBP-1c was expressed but its target genes were not; they showed delayed expression than SREBP-1c, suggesting that the activity of SREBP-1c is regulated posttranslationally.
DNA methylation of lipogenic gene promoters in neonatal and adult livers.
The Gpam promoter region contains three SREs (Fig. 1B) (6). Bisulfite analysis revealed high DNA methylation levels of the Gpam promoter containing SREs in the neonatal liver (day 0) (Fig. 1C). By contrast, less DNA methylation levels were observed in the adult liver (Fig. 1C). We also confirmed the differential DNA methylation of the Gpam promoter in the neonatal and adult livers, based upon digestion with a methylation-sensitive enzyme, HpaII, for which the recognition site locates between the first and second SREs (Fig. 1B), and a subsequent quantitative real-time PCR analysis. Consistent with the data of bisulfite analysis, DNA methylation levels were high in the neonatal liver (day 0 and 5; ∼60%, day 9; ∼55%) relative to the adult liver (∼15%) (Fig. 1D). By contrast, appreciable DNA methylation of the Scd1 and Fasn promoters, containing SREs, was not observed in both the neonatal (day 0) and adult (10 weeks of age) livers (Fig. 1E).
In a CpG island of the alternative promoter of the Gpam, there was also no significant DNA methylation in the neonatal (day 0) and adult (10 weeks of age) livers (Fig. 1E). In contrast, DNA methylation levels were high in the repetitive element of intracisternal A particle (IAP) (26), one of the markers of global DNA methylation, in both the neonatal and adult livers (Fig. 1E). It is therefore likely that DNA methylation differs in certain regions of the genome between neonatal and adult livers.
DNA methylation of the Gpam promoters in the adult skeletal muscle.
In contrast to the liver, high DNA methylation levels in the Gpam promoter were observed in the skeletal muscle of adult mice, where its mRNA expression was very low (Fig. 1F). DNA methylation levels of the alternative Gpam promoter were low in the skeletal muscle and liver, and GPAT1-variant expression levels were similar between the skeletal muscle and liver (Fig. 1G). These observations, taken together, suggest that DNA methylation of the Gpam promoter, containing SREs, and its mRNA expression are inversely correlated in the skeletal muscle.
DNA methylation of the Gpam promoter in primary culture of neonatal mouse hepatocytes.
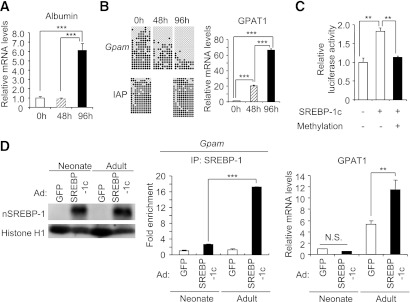
We next examined DNA methylation of the Gpam promoter in primary culture of neonatal mouse hepatocytes. In this study, the gene expression of albumin, a marker of mature hepatocytes, increased during the course of culture, suggesting the maturation of primary hepatocytes (Fig. 2A). The DNA methylation levels of the Gpam promoter was high at 0 h but gradually decreased at 48 and 96 h, and GPAT1 mRNA levels were low at 0 h and increased at 48 and 96 h (Fig. 2B). By contrast, DNA methylation levels of IAP remained high at both 0 and 96 h (Fig. 2B), suggesting that decreased methylation is not likely due to total demethylation of the whole genome. These observations also suggest an inverse correlation between DNA methylation of the Gpam promoter and its mRNA expression in primary culture of neonatal mouse hepatocytes.
FIG. 2.
In vitro analysis of DNA methylation and SREBP-1 recruitment of the Gpam promoter. mRNA expression levels of albumin (A) and DNA methylation and mRNA expression levels (B) of the Gpam in neonatal primary hepatocytes, cultured at 0, 48, and 96 h. The upper panels are the results of the Gpam promoter and lower panels are those of IAP. ***P < 0.001. C: In vitro methylation reporter assay. Relative luciferase activities are shown. Values without DNA methylation and in the absence of SREBP-1c expression vector are set at 1. **P < 0.01; n = 4. D: Left: Western blot analysis of primary hepatocytes from day 0 (neonate) and 10 weeks (adult) of age, overexpressing SREBP-1c and control GFP. Middle: ChIP analysis; recruitment of SREBP-1c to the Gpam promoter in those cells. Right: Quantitative real-time PCR analysis of GPAT1 mRNA. Values for the neonatal hepatocytes with GFP were set at 1. **P < 0.01, ***P < 0.001; n = 3.
Reporter activity from the Gpam promoter with in vitro DNA methylation.
In an in vitro reporter assay, we examined whether DNA methylation of the Gpam promoter affects the SREBP-1c–induced transcriptional activity. Cotransfection of the Gpam promoter-Luc without DNA methylation and SREBP-1c expression plasmid in HEK293 cells caused increased reporter activity from the Gpam promoter (Fig. 2C). By contrast, upon methylation of the Gpam promoter, Luc activity was not increased above the basal level even in the presence of the SREBP-1c expression plasmid (Fig. 2C).
Adenovirus-mediated SREBP-1c overexpression and ChIP analysis of the Gpam promoter in neonatal and adult primary hepatocytes.
To address whether DNA methylation of the Gpam promoter is critical for SREBP-1c–dependent GPAT1 expression, we overexpressed SREBP-1c in neonatal (high DNA methylation) and adult (low DNA methylation levels in the Gpam promoter) primary hepatocytes through the adenoviral technique. We observed similarly high expression of nuclear form of SREBP-1c protein in neonatal and adult primary hepatocytes (Fig. 2D). In this experiment, we observed less expression of GPAT1 with lower recruitment of SREBP-1 to the Gpam promoter in the neonatal primary hepatocytes relative to adult primary hepatocytes (Fig. 2D). The data indicated that GPAT1 expression is low even in the presence of considerable SREBP-1c expression, suggesting that DNA methylation of the Gpam promoter is critical for SREBP-1c–dependent GPAT1 expression.
ChIP analysis of the Gpam promoter in neonatal and adult liver.
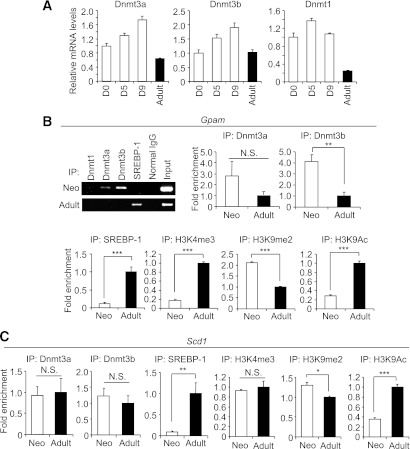
Dnmt1, -3a, and -3b mRNA expression was detected in the neonatal and adult livers, although the hepatic Dnmt1 mRNA levels were lower in the adult liver than in the neonatal liver (Fig. 3A). Therefore, we performed ChIP analysis to compare the recruitment of SREBP-1 and Dnmts to the Gpam promoter between the neonatal and adult livers. In this study, SREBP-1 was not recruited to the Gpam promoter in the neonatal liver (day 0 after birth), although it was clearly recruited in the adult liver (Fig. 3B). Importantly, Dnmt3b was strongly recruited to the Gpam promoter in the neonatal liver, but not in the adult liver (Fig. 3B). Dnmt3a was also recruited, although weakly, to the Gpam promoter in the neonatal liver, but not in the adult liver (Fig. 3B). We also found that levels of histone H3 lysine-4-trimethylation (H3K4me3) and lysine-9-acetylation (H3K9Ac), two transcriptionally active histone codes, are increased and those of the repressive histone H3 lysine-9-di-methylation (H3K9me2) are decreased at the Gpam promoter in the adult liver relative to the neonatal liver (Fig. 3B). The recruitment of Dnmt3a and Dnmt3b to the Scd1 promoter did not differ significantly between the neonatal and adult livers (Fig. 3C). In this study, we observed that the recruitment of SREBP-1 and level of H3K9Ac are increased and that of H3K9me2 is decreased at the Scd1 promoter in the adult liver relative to the neonatal liver (Fig. 3C).
FIG. 3.
Recruitment of SREBP-1c and Dnmts to the Gpam and Scd1 promoters containing SREs, with changes in histone modification. A: Relative Dnmts mRNA levels in the neonatal (days after birth: 0, 5, and 9) and adult livers (10 weeks of age) examined by quantitative real-time PCR. Values for day 0 were set at 1. n = 3 to 4. B: ChIP analysis. Neonatal (day 0) and adult (10 weeks) livers were used for ChIP analysis with the indicated antibodies. Amplified PCR primers are as shown in Fig. 1B. The top left panel is a representative gel electrophoresis photo. Input is a PCR product from the aliquot of liver lysate before immunoprecipitation (IP). The graphs demonstrate quantitative PCR analysis. The graph of Dnmt1 is not shown because signals from the neonatal and adult livers are lower than the negative control IgG signals. C: ChIP analysis in the Scd1 promoter in the samples used in B. Values of adult sample are set at 1. *P < 0.05, **P < 0.01, ***P < 0.001, n = 3 to 4.
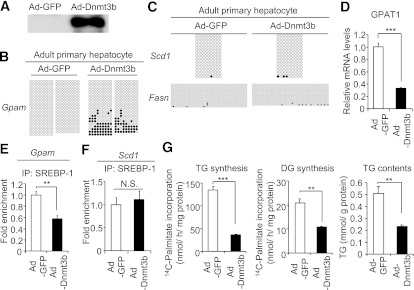
DNA methylation of the Gpam promoter and GPAT-mediated TG/DG synthesis in primary hepatocytes with Dnmt3b overexpression.
With adenoviral transduction of Dnmt3b, which was strongly recruited to the Gpam promoter in the neonatal liver (Fig. 3B), we next examined DNA methylation of the Gpam promoter, its mRNA expression, and TG/DG synthesis in primary hepatocytes, which were obtained from adult mice with low DNA methylation levels of the Gpam promoter. In this study, we confirmed Dnmt3b overexpression causes significant Dnmt3b protein expression (Fig. 4A). We found increased DNA methylation of the Gpam promoter (Fig. 4B), but not Scd1 and Fasn promoters (Fig. 4C), in Dnmt3b-expressing hepatocytes relative to control GFP-expressing hepatocytes. The GPAT1 mRNA levels were markedly lower in hepatocytes with Dnmt3b overexpression than those with control GFP (Fig. 4D). In the Gpam promoter (Fig. 4E), but not the Scd1 promoter (Fig. 4F), SREBP-1 was less recruited in Dnmt3b-expressing hepatocytes than in control GFP-expressing hepatocytes. In addition, TG/DG synthesis and TG contents were lower in Dnmt3b-expressing hepatocytes than in GFP-expressing hepatocytes (Fig. 4G). These data suggest that increased DNA methylation suppresses SREBP-1 recruitment in the Gpam promoter, decreased GPAT1 expression, and TG/DG synthesis.
FIG. 4.
DNA methylation level of the Gpam promoter and GPAT-mediated TG/DG synthesis in the adult primary hepatocytes overexpressing Dnmt3b. A: Protein expression levels of Dnmt3b in adult primary hepatocytes overexpressing Dnmt3b and control GFP. Endogenous Dnmt3b protein expression is known to be high in early embryo and embryonic stem cells, but very low in adult tissues (33) and was not detected in this experiment using adult primary hepatocyte samples. B: DNA methylation level of the Gpam promoter in adult primary hepatocytes overexpressing Dnmt3b and control GFP. Results of two independent dishes from each group are shown. C: DNA methylation level of the Scd1 and Fasn promoters. D: Expression of GPAT1 mRNA levels and recruitment of SREBP-1 to the Gpam (E) and Scd1 (F) promoters. Values of GFP cells are set at 1. G: TG/DG synthesis levels and TG contents. **P < 0.01, ***P < 0.001. n = 3.
DNA methylation of the Gpam promoter in the neonatal offspring liver of the HF/HS diet–fed dams.
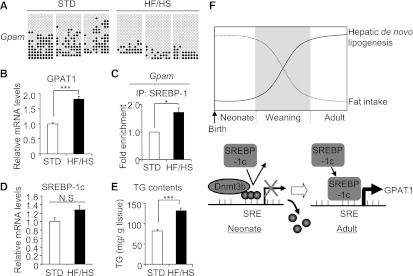
We next examined whether the observed DNA methylation of the Gpam promoter is affected by environmental factors in vivo. A high-fat or high-calorie diet fed to female animals during gestation and lactation has been reported to increase lipogenic gene expression and TG levels in the liver of offspring (27). In this study, we fed a high-calorie, lipogenic HF/HS diet to dams and examined DNA methylation of the Gpam promoter in the offspring. In pups of the HF/HS diet–fed dams, DNA methylation levels of the Gpam promoter were lower, and GPAT1 mRNA levels and SREBP-1 recruitment, but not SREBP-1c mRNA levels, were higher than those in the pups of standard diet–fed dams (Fig. 5A–D). In this study, hepatic TG levels increased in the pups of the HF/HS diet–fed dams relative to those of the standard diet–fed dams (Fig. 5E).
FIG. 5.
DNA methylation of the Gpam promoter and its mRNA expression in pups of dams fed an HF/HS diet during gestation and lactation. A: DNA methylation of the Gpam promoter in pups (day 5) of the HF/HS or standard (STD) diet–fed dams. Representative bisulfite sequencing data from three animals in each group. Relative GPAT1 mRNA levels (B), recruitment of SREBP-1 to the Gpam promoter (C), and relative SREBP-1c mRNA levels (D) in pups of the HF/HS and STD diet–fed dams. E: Liver TG levels of neonatal liver (day 5). Open bar, STD-fed dams; filled bar, HF/HS diet-fed dams. *P < 0.05, ***P < 0.001; n = 7 (open bars), n = 4 (filled bars). F: Schematic model of the DNA methylation-mediated regulation of Gpam expression in the neonatal and adult livers. The graph shows the time course of the dietary fat content and rate of hepatic de novo lipogenesis before and after weaning. In the neonatal liver, the Gpam promoter may be highly methylated at least in part by Dnmt3b, when SREBP-1c cannot make access to the promoter. In the adult liver, the Gpam promoter is less methylated, with SREBP-1c being recruited to the promoter, thereby activating GPAT1 mRNA expression. Environmental factors, such as nutritional state, may change the DNA methylation status. Closed circle denotes methyl group.
DISCUSSION
The DNA methylation status has been considered to be relatively stable except during embryogenesis and carcinogenesis. However, recent studies showed that DNA methylation can be modulated in normal tissues even after birth (28). In this study, we show that expression of Gpam, which encodes a rate-limiting enzyme for TG biosynthesis, increases in the liver during weaning in response to the physiologic demand of TG biosynthesis. This may be related to the dynamic change in the DNA methylation status of the Gpam promoter during liver maturation. In the neonatal liver, the Gpam promoter region, containing three SREs, shows high DNA methylation levels with low Gpam expression, whereas in the adult liver, it shows low DNA methylation levels with high Gpam expression. Importantly, SREBP-1c is recruited to the promoter in the adult liver but not in the neonatal liver. In this study, in vitro analysis revealed that DNA methylation of the Gpam promoter can suppress the SREBP-1c–mediated transcriptional activation. Less SREBP-1 recruitment in the Gpam promoter in Dnmt3b-overexpressing hepatocytes suggests that DNA methylation of the Gpam promoter inhibited SREBP-1c recruitment. These observations, taken together, suggest the role of DNA methylation in the suppressed Gpam expression in the neonatal liver even in the presence of SREBP-1c. This above discussion is consistent with a previous observation that the extent of hepatic de novo lipogenesis in the neonatal liver is lower than that in the adult liver (1). Consistent with the decreased DNA methylation, transcriptionally active H3K4me3 and H3K9Ac are increased, whereas repressive H3K9me2 is decreased at the Gpam promoter in the adult liver. It is conceivable that in the neonatal liver, DNA methylation of the Gpam promoter plays roles in the formation and/or maintenance of transcriptionally repressive chromatin conformation, thereby inhibiting the recruitment of SREBP-1c. In contrast, in the adult liver, decreased DNA methylation and increased active histone modifications may lead to the recruitment of SREBP-1c to the Gpam promoter.
The mechanism underlying the altered DNA methylation of the genome has not been thoroughly investigated. In this study, we found that Dnmt3b, which is implicated in de novo DNA methylation, is strongly recruited to the Gpam promoter in the neonatal liver but not in the adult liver. Dnmt3b overexpression markedly increases DNA methylation levels of the Gpam promoter in primary adult hepatocytes. Dnmt3b plays important roles in embryogenesis (10). It has been recently reported to be involved in colon cancer (29) and hormonal gene regulation in renal tubular cells (30). Therefore, it is likely that the decreased recruitment of Dnmt3b plays a role in the decreased DNA methylation of the Gpam promoter, and additional mechanism(s) for DNA demethylation may be involved in the process.
The H3K4 methylation level is reported to be reciprocal with the DNA methylation level, and H3K4me2/3 and DNA methylation occur mutually exclusively (11,31). In the region of H3K4me2/3, the DNA methylation levels are generally low. Alternatively, Dnmt3 may preferentially bind to genomic DNA without H3K4 methylation. In this study, the H3K4me3 level in the Gpam promoter was low in the neonatal liver and high in adult liver, whereas the H3K4me3 level in the Scd1 promoter was similar in neonatal and adult livers. Thus, it is possible that histone modification is involved in the regulation of specificity of DNA methylation in Gpam and Scd1 promoters. In addition, because GPAT1 contributes to the transfer of fatty acid to glycerol, whereas SCD1 and FAS are involved in fatty acid biosynthesis (3), the differential positioning of GPAT1, SCD1, and FAS in the TG biosynthesis pathway may explain the different mechanisms of their gene expression. Further studies are required to elucidate how Scd1 and Fasn transcription is regulated in the neonatal liver.
In this study, we also found that GPAT1 mRNA levels are much lower in the adult skeletal muscle than those in the adult liver, although SREBP-1c is highly expressed in both tissues (19). This is consistent with a previous report that the GPAT1 enzymatic activity in the skeletal muscle is much lower than that in the liver (32). The low expression of Gpam in the adult skeletal muscle may be because the high DNA methylation of the Gpam promoter suppresses the SREBP-1c–mediated transcriptional activation. Therefore, it is conceivable that DNA methylation of the Gpam promoter is involved in the SREBP-1c–dependent tissue-specific regulation of Gpam expression. In contrast, the expression of the GPAT1-variant transcript from the alternative promoter without SRE in the neonatal liver is roughly comparable to that in the adult liver. This is consistent with our observation that the alternative promoter is unmethylated in both the neonatal and adult livers, thereby suggesting the role for DNA methylation in the promoter-specific regulation of Gpam in the liver.
The change in hepatic gene expression before and after weaning could be regulated in response to nutritional demand. Indeed, we demonstrated that the lipogenic HF/HS diet fed to female mice during pregnancy and lactation results in decreased DNA methylation of the Gpam promoter in the liver of offspring, suggesting that the DNA methylation status can be modulated, at least in part, by the nutritional factors. Thus, the change in DNA methylation of the Gpam promoter is likely to be affected by the fetal and neonatal environments, such as nutrition, and/or be a programmed process of liver maturation.
Whether DNA methylation would affect hepatic de novo lipogenesis later in life is an important issue to be addressed. Previous reports showed that maternal overnutrition in animals contributes to the development of nonalcoholic fatty liver disease in adult offspring (27), suggesting that hepatic lipid metabolism is nutritionally affected early in life. In this study, maternal overnutrition caused decreased Gpam promoter methylation with increased GPAT1 expression and TG level in the liver of the offspring; however, the possibility cannot be excluded that increased lipid flux from dam, as well as increased de novo TG biosynthesis, might have affected the liver TG content of offspring. Further studies are required to understand whether DNA methylation can affect hepatic de novo lipogenesis and susceptibility to fatty liver-related diseases in later life.
In conclusion, this study is the first demonstration of reciprocal change in DNA methylation of the Gpam promoter and its mRNA expression in the mouse liver before and after weaning (Fig. 5F). Our data suggest that in the neonatal liver, DNA methylation of the Gpam promoter containing SREs, which is likely to be induced by Dnmt3b, inhibits the recruitment of SREBP-1c, whereas in the adult liver, the decreased DNA methylation may result in active chromatin conformational change, thereby allowing the recruitment of SREBP-1c. This is the first detailed analysis of the DNA methylation-dependent regulation of TG biosynthesis gene in the liver, thereby leading to the better understanding of the molecular mechanism underlying the epigenetic regulation of metabolic genes and thus metabolic diseases.
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Ministry of Health, Labour and Welfare of Japan and research grants from “To promote basic research by research personnel in private-sector business (Japan Science and Technology Agency),” Mitsukoshi Health and Welfare Foundation, The Danone Institute of Japan Foundation, Nestle Nutrition Council, Novo Nordisk Study Award for Growth and Development, Ono Medical Research Foundation, and The Morinaga Foundation for Health and Nutrition. No other potential conflicts of interest relevant to this article were reported.
T.E., Y.K., and Y.O. designed the research. T.E., Y.K., M.Tak., X.Y., S.K., E.T., M.Tan., T.Y., and S.M. performed research. T.S. and M.O. analyzed data. O.E. and M.O. contributed new reagents and analytic tools. Y.K. and Y.O. wrote the manuscript. All authors contributed to the discussion. Y.O. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Dr. Hitoshi Shimano (Tsukuba University, Japan) for the kind gift of Ad-SREBP-1c and Drs. Jun Inoue and Ryuichiro Sato (University of Tokyo, Japan) for advice on SREBP-1c Western blotting. The authors also thank Dr. Shigeomi Shimizu (Tokyo Medical and Dental University, Japan) for advice on mitochondrial preparation, Drs. Eriko Misawa and Miyuki Tanaka (Morinaga Milk Industry Co. Ltd., Kanagawa, Japan) for assistance in TG synthesis measurement, Kozue Ochi (Tokyo Medical and Dental University, Tokyo Japan) for technical assistance, and Hiroko Takahashi (Tokyo Medical and Dental University) for secretarial assistance.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1834/-/DC1.
REFERENCES
- 1.Perez-Castillo A, Schwartz HL, Oppenheimer JH. Rat hepatic mRNA-S14 and lipogenic enzymes during weaning: role of S14 in lipogenesis. Am J Physiol 1987;253:E536–E542 [DOI] [PubMed] [Google Scholar]
- 2.Decaux JF, Ferré P, Robin D, Robin P, Girard J. Decreased hepatic fatty acid oxidation at weaning in the rat is not linked to a variation of malonyl-CoA concentration. J Biol Chem 1988;263:3284–3289 [PubMed] [Google Scholar]
- 3.Wendel AA, Lewin TM, Coleman RA. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim Biophys Acta 2009;1791:501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindén D, William-Olsson L, Ahnmark A, et al. Liver-directed overexpression of mitochondrial glycerol-3-phosphate acyltransferase results in hepatic steatosis, increased triacylglycerol secretion and reduced fatty acid oxidation. FASEB J 2006;20:434–443 [DOI] [PubMed] [Google Scholar]
- 5.Hammond LE, Gallagher PA, Wang S, et al. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol Cell Biol 2002;22:8204–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ericsson J, Jackson SM, Kim JB, Spiegelman BM, Edwards PA. Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1- and sterol regulatory element-binding protein-responsive gene. J Biol Chem 1997;272:7298–7305 [DOI] [PubMed] [Google Scholar]
- 7.Tabor DE, Kim JB, Spiegelman BM, Edwards PA. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem 1999;274:20603–20610 [DOI] [PubMed] [Google Scholar]
- 8.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell 2009;136:1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefan N, Kantartzis K, Häring HU. Causes and metabolic consequences of Fatty liver. Endocr Rev 2008;29:939–960 [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257 [DOI] [PubMed] [Google Scholar]
- 11.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007;39:457–466 [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen TS, Xu Z, Zhang X, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010;143:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466:253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007;450:908–912 [DOI] [PubMed] [Google Scholar]
- 15.Barrès R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab 2009;10:189–198 [DOI] [PubMed] [Google Scholar]
- 16.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 2009;5:401–408 [DOI] [PubMed] [Google Scholar]
- 17.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol 2009;7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamei Y, Suganami T, Ehara T, et al. Increased expression of DNA methyltransferase 3a in obese adipose tissue: studies with transgenic mice. Obesity (Silver Spring) 2010;18:314–321 [DOI] [PubMed] [Google Scholar]
- 19.Kamei Y, Miura S, Suganami T, et al. Regulation of SREBP1c gene expression in skeletal muscle: role of retinoid X receptor/liver X receptor and forkhead-O1 transcription factor. Endocrinology 2008;149:2293–2305 [DOI] [PubMed] [Google Scholar]
- 20.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 2008;36(Web Server issue):W170–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuso A, Scarpa S, Grandoni F, Strom R, Lucarelli M. A reassessment of semiquantitative analytical procedures for DNA methylation: comparison of bisulfite- and HpaII polymerase-chain-reaction-based methods. Anal Biochem 2006;350:24–31 [DOI] [PubMed] [Google Scholar]
- 22.Honkakoski P, Moore R, Gynther J, Negishi M. Characterization of phenobarbital-inducible mouse Cyp2b10 gene transcription in primary hepatocytes. J Biol Chem 1996;271:9746–9753 [DOI] [PubMed] [Google Scholar]
- 23.Hayhurst GP, Strick-Marchand H, Mulet C, et al. Morphogenetic competence of HNF4 α-deficient mouse hepatic cells. J Hepatol 2008;49:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumadaki S, Matsuzaka T, Kato T, et al. Mouse Elovl-6 promoter is an SREBP target. Biochem Biophys Res Commun 2008;368:261–266 [DOI] [PubMed] [Google Scholar]
- 25.Suganami T, Yuan X, Shimoda Y, et al. Activating transcription factor 3 constitutes a negative feedback mechanism that attenuates saturated Fatty acid/toll-like receptor 4 signaling and macrophage activation in obese adipose tissue. Circ Res 2009;105:25–32 [DOI] [PubMed] [Google Scholar]
- 26.Kato Y, Kaneda M, Hata K, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 2007;16:2272–2280 [DOI] [PubMed] [Google Scholar]
- 27.Bruce KD, Cagampang FR, Argenton M, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology 2009;50:1796–1808 [DOI] [PubMed] [Google Scholar]
- 28.Waterland RA, Kellermayer R, Rached MT, et al. Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum Mol Genet 2009;18:3026–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steine EJ, Ehrich M, Bell GW, et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J Clin Invest 2011;121:1748–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature 2009;461:1007–1012 [DOI] [PubMed] [Google Scholar]
- 31.Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007;448:714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewin TM, Granger DA, Kim JH, Coleman RA. Regulation of mitochondrial sn-glycerol-3-phosphate acyltransferase activity: response to feeding status is unique in various rat tissues and is discordant with protein expression. Arch Biochem Biophys 2001;396:119–127 [DOI] [PubMed] [Google Scholar]
- 33.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 2002;277:38746–38754 [DOI] [PubMed] [Google Scholar]