Abstract
Apolipoprotein (apo) E is thought to undergo conformational changes in the N-terminal helix bundle domain upon lipid binding, modulating its receptor binding activity. In this study, site-specific fluorescence labeling of the N-terminal (S94) and C-terminal (W264 or S290) helices in apoE4 by pyrene maleimide or acrylodan was employed to probe the conformational organization and lipid binding behavior of the N- and C-terminal domains. Guanidine denaturation experiments monitored by acrylodan fluorescence demonstrated the less organized, more solvent-exposed structure of the C-terminal helices compared to the N-terminal helix bundle. Pyrene excimer fluorescence together with gel filtration chromatography indicated that there are extensive intermolecular helix-helix contacts through the C-terminal helices of apoE4. Comparison of increases in pyrene fluorescence upon binding of pyrene-labeled apoE4 to egg phosphatidylcholine small unilamellar vesicles suggests a two-step lipid-binding process; apoE4 initially binds to a lipid surface through the C-terminal helices followed by the slower conformational reorganization of the N-terminal helix bundle domain. Consistent with this, fluorescence resonance energy transfer measurements from Trp residues to acrylodan attached at position 94 demonstrated that upon binding to the lipid surface, opening of the N-terminal helix bundle occurs at the same rate as the increase in pyrene fluorescence of the N-terminal domain. Such a two-step mechanism of lipid binding of apoE4 is likely to apply to mostly phospholipids-covered lipoproteins such as VLDL. However, monitoring pyrene fluorescence upon binding to HDL3 suggests that not only apoE-lipid interactions but also protein-protein interactions are important for apoE4 binding to HDL3.
Apolipoprotein (apo) E is a key protein regulating lipid transport and cholesterol homeostasis in the cardiovascular and central nervous systems (1–3). The cholesterol transport functions of apoE are a basis of its anti-atherogenic activity that is achieved through its ability to interact with lipids and cell surface molecules such as the low-density lipoprotein (LDL) receptor family and glycosaminoglycans (4, 5). In the brain, apoE shuttles cholesterol from astrocytes to neurons via lipoprotein complexes in which cholesterol required for neuronal maintenance is supplied from astrocytes by apoE-mediated transport, and excess cholesterol is effluxed to apoE-containing lipoproteins (6, 7).
Human apoE is a 299-residue polypeptide that folds into two tertiary structure domains; a 22-kDa N-terminal domain (residues 1–191) and a 10-kDa C-terminal domain (residues 216–299) linked by a hinge region (8–10). The N-terminal domain is folded into a four-helix bundle of amphipathic α-helices (11, 12) and contains the region (residues 136–150) that binds to the LDL receptor superfamily (13). The C-terminal domain contains amphipathic α-helices that are involved in binding to lipoproteins with high affinity (14–16) and mediate ATP-binding cassette transporter A1-dependent cholesterol efflux (17). Lipid-free apoE exists as oligomer in solution (15, 16, 18) and the C-terminal domain is responsible for this self-association through the intermolecular coiled-coil formation (19, 20). These helix-helix contacts are converted to helix-lipid interaction upon binding of apoE to lipoprotein surfaces, with the N-terminal four-helix bundle being in either an open or a closed conformation depending upon the surface area availability (21, 22). Since the conformational reorganization of the N-terminal domain upon lipid interaction is associated with enhanced receptor binding activity (13, 23), the balance between such two lipid-bound states of apoE on lipoprotein particles is assumed to determine the extent to which apoE can mediate the interaction with the LDL receptor family (3, 24).
Based on the two lipid-bound states of apoE on spherical lipid particles, we (25) and others (26) have proposed that there is a two-step mechanism for lipid binding of apoE like apoA-I (27); after an initial interaction through the C-terminal domain, the second slower step involves opening of the N-terminal helix bundle domain. Indeed, recent surface plasmon resonance analysis for the binding of apoE to lipoprotein particles such as very low-density lipoprotein (VLDL) and high-density lipoprotein (HDL) indicated that the interaction of apoE with lipid is a two-step process (25). In addition, recent crystal structures of a C-terminal truncated apoA-I (28) and the apoA-IV core domain (29), and NMR structure of full-length apoE3 (26) suggested that domain swapping might be a common mechanism for exchangeable apolipoproteins to open the helix bundle upon lipid binding. However, the direct experimental evidence for the conformational opening of the N-terminal helix bundle upon binding to lipoprotein particles is still lacking.
In the present study, we examined the lipid-bound conformations of apoE on spherical lipid particles by employing site-directed fluorescence labeling of apoE, as we have done successfully for apoA-I bound to spherical phospholipid vesicles (30). The results demonstrate that upon binding to lipid vesicles or VLDL where apoE-lipid interaction is dominant, opening of the N-terminal helix bundle indeed occurs on the lipid surface. Upon binding to HDL3, however, interactions of apoE with resident proteins are also involved in the binding.
EXPERIMENTAL PROCEDURES
Materials
Human apoE4 variants were expressed in E. coli as thioredoxin fusion proteins and isolated and purified as described previously (22, 31). The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was employed to introduce the S94C, W264C, or S290C point mutations into apoE4. The apoE preparations were at least 95% pure as assessed by SDS−PAGE. In all experiments, the apoE sample was freshly dialyzed at 4°C from a 6 M guanidine hydrochloride (GdnHCl) and 1% β-mercaptoethanol solution into Tris buffered saline (TBS; 10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.02 % NaN3, pH 7.4) before use. HDL3 and VLDL were purified by sequential ultracentrifugation from a pool of normolipidemic human plasma as described previously (16, 25). Egg yolk phosphatidylcholine (PC) was obtained from Kewpie (Tokyo, Japan). N-(1-pyrene)maleimide and 6-acryloyl-2-dimethylaminonaphthalene (acrylodan, Ac) were purchased from Invitrogen (Eugene, OR).
Fluorescence Labeling
Cysteine-containing apoE4 variants were incubated with 10-fold molar excess of tris(2-carboxyethyl)phosphine hydrochloride (Thermo Scientific, Rockford, IL) for 1 h to reduce the sulfhydryl group. A 10 mM stock solution of N-(1-pyrene)maleimide (in dimethylsulfoxide) or acrylodan (in dimethylformamide) was added so that a final molar ratio of probe to protein was 10:1. The reaction mixtures were then incubated at room temperature for 3 h in the dark, and unreacted probe was removed by extensive dialysis at 4 °C in TBS. The degree of labeling was determined using the extinction coefficients of 38,200 M−1 cm−1 at 338 nm for pyrene and 19,200 M−1 cm−1 at 391 nm for acrylodan, respectively.
Preparation of Small Unilamellar Vesicles (SUVs
SUVs were prepared as described (32, 33). Briefly, a film of egg PC on the wall of a glass tube was dried under vacuum overnight. The lipid was then hydrated in TBS and sonicated on ice under nitrogen. After removing titanium debris, the samples were centrifuged in a Beckman 70.1Ti rotor for 1.5 h at 4 °C at 40,000 rpm to separate any remaining large vesicles. The PC concentration of SUV was determined using an enzymatic assay kit from Wako Pure Chemicals (Osaka, Japan).
Fluorescence Measurements
Fluorescence measurements were carried out with a Hitachi F-4500 fluorescence spectrophotometer at 25 °C in Tris buffer (pH 7.4). Acrylodan emission fluorescence was collected from 380 to 600 nm using a 360 nm excitation wavelength. Steady-state fluorescence anisotropy of acrylodan was measured with excitation at 360 nm and emission at 485 nm, as described (34). The generalized polarization (GP) is given by GP = (IB − IR)/(IB + IR), where IB and IR are the emission intensities at the blue (480 nm) and red (530 nm) edges of the emission spectrum, respectively (35). For monitoring chemical denaturation, proteins at a concentration of 25 µg/ml were incubated overnight at 4 °C with GdnHCl at various concentrations. The equilibrium constant of denaturation, KD, at a given GdnHCl concentration was calculated from the GP values and, the free energy of denaturation, ΔGD°, the midpoint of denaturation, D1/2, and m value which reflects the cooperativity of denaturation in the transition region, were determined by the linear equation, ΔGD = ΔGD° − m[GdnHCl], where ΔGD = − RT ln KD(33, 35). For fluorescence resonance energy transfer (FRET) experiments, the emission spectra of acrylodan-labeled and unlabeled apoE4 variants were measured from 300 to 600 nm with excitation of 290 nm. FRET efficiency (E) was calculated according to E = 1 − FDA/FD, where FDA is the fluorescence intensity of the donor with acrylodan attached and FD is the fluorescence intensity of the donor lacking acrylodan. The FRET distance (R) was calculated according to E = R06 /(R 06 + R6), where R0 is the Förster radius for energy transfer from Trp to acylodan in a protein (2.7 nm) (35).
Pyrene emission fluorescence was recorded from 360 to 500 nm using a 342 nm excitation wavelength. Excimer to monomer ratios were calculated by dividing the area under the excimer emission peak from 450 to 490 nm by that for the monomer emission peak from 370 to 410 nm. In binding experiments to SUV, HDL3 or VLDL, the increase in emission fluorescence of pyrene-labeled apoE4 variants was assessed from the area under the emission from 370 to 410 nm using a 4 × 4 mm cuvette to reduce the effect of light scattering caused by lipid particles.
Binding parameters of pyrene-apoE4 to SUV were derived as described (30). Briefly, the fraction of bound apoE4, θ was calculated according to θ = Pb/PT = (F – F0/Fmax–F0), where Pb and PT are bound and total apoE4 concentrations, respectively, and F and F0 are integrated fluorescence intensities for pyrene-apoE4 in the presence and absence of SUV, respectively, and Fmax represents the fluorescence intensity when pyrene-apoE4 completely binds to SUV. Assuming that binding of apoE4 to lipid particles is described by a one-site binding model, Pb/[PC] = BmaxPf/(Kd + Pf), where Pf is unbound apoE4 concentration, [PC] is PC concentration of SUV, and Kd and Bmax are the dissociation constant and the maximal binding capacity, respectively, binding data were analyzed by linear regression based on the Hanes-Woolf equation, [PC]Pf/Pb = Kd/Bmax + Pf/Bmax.
RESULTS
Acrylodan Fluorescence
To introduce fluorescence probes into the nonpolar face of a putative amphipathic helix located at either the N- or C-terminal domains of apoE4, S94 in the N-terminal helix bundle domain, W264 or S290 in the C-terminal domain were replaced with a cysteine residue. The mutations of S94C and W264C, both of which are located in buried, helical structure in the lipid-bound state, were previously shown to retain the structural properties of apoE4 (23, 36, 37). S290 is likely to be in the loop structure in the lipid-free state (26), but this position is expected to be located at the hydrophobic face to interact with lipids (38). Thus, fluorescence signals from probes attached at these sites can be used to monitor the conformational change or lipid-binding behavior of apoE4.
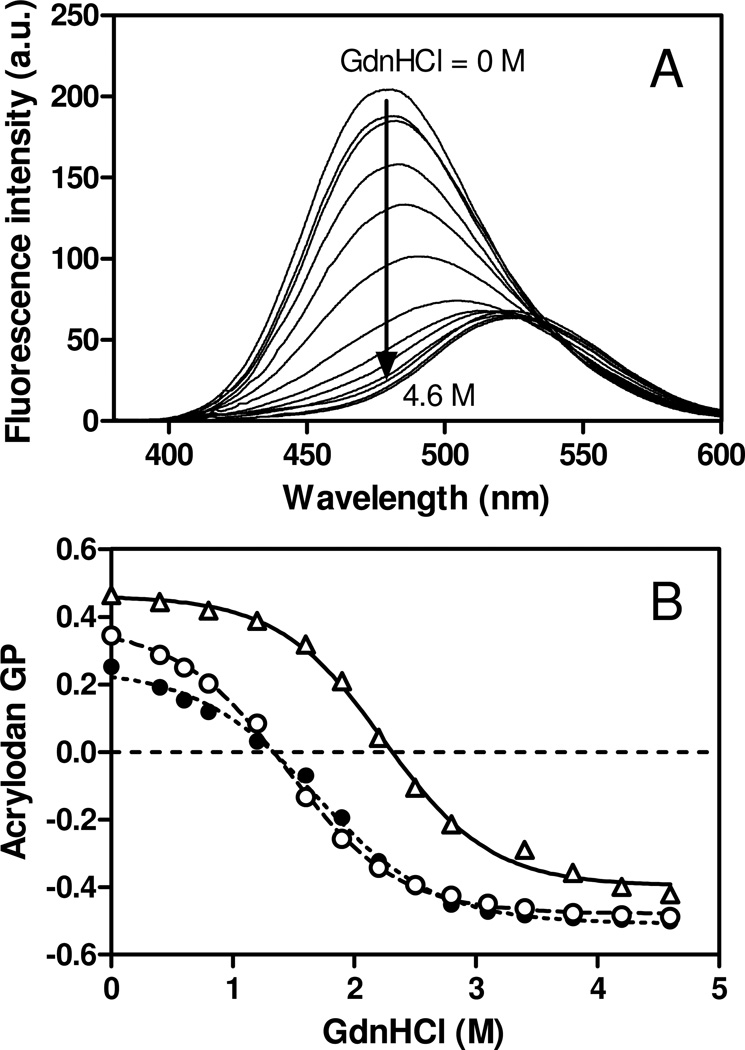
We first employed acrylodan labeling of apoE4 cysteine variants to probe site specific conformation in the proteins (35, 39, 40). Fig. 1A shows the change in acrylodan fluorescence emission spectra of apoE4 S94C-Ac when incubated at different concentrations of GdnHCl. Significant decreases in fluorescence intensity and red shifts in wavelength of maximum fluorescence (WMF) of acrylodan were observed with increasing concentrations of GdnHCl, indicating transfer of acrylodan molecule from the hydrophobic interior in a folded protein into an aqueous environment. Since the GP value signifies the change in the environment polarity of acrylodan, the denaturation curves for apoE4 cysteine variants monitored by acrylodan GP (Fig. 1B) reflect the conformational stability of the N- or C-terminal helices in apoE4 against GdnHCl denaturation. The fact that thermodynamic parameters of denaturation for apoE4 S94C-Ac are similar to those for apoE4 22-kDa monitored by Trp fluorescence (Table 1) indicates that the change in GP value indeed reflects the conformational change of the protein. Comparison of thermodynamic parameters of denaturation for apoE4 cysteine variants (Table 1) demonstrated that S94 is more stable than both W264 and S290 which have similar stability, consistent with the notion that the N-terminal helix bundle domain is much more stable than the C-terminal domain in apoE4 (41, 42). Table 1 also shows WMF and fluorescence anisotropy of acrylodan for apoE4 cysteine variants. Compared to apoE4 W264C-Ac, the S290C-Ac exhibited higher WMF and lower anisotropy value, indicating that S290 is located in less motionally restricted, more exposed structure compared to W264 in the C-terminal domain (26).
Fig. 1.
(A) Acrylodan fluorescence emission spectra of apoE4 S94C-acrylodan at different concentrations of GdnHCl (from 0 to 4.6 M). (B) The change in acrylodan GP for apoE4 S94C-acrylodan (Δ), W264C-acrylodan (○), and S290C-acrylodan (●) as a function of GdnHCl concentration. Protein concentration was 25 µg/ml.
Table 1.
WMF, Fluorescence Anisotropy, and GdnHCl Denaturation Parameters of Acrylodan Fluorescence of ApoE4 Variants
| GdnHCl denaturation | |||||
|---|---|---|---|---|---|
| apoE4 variant | WMFa | fluorescence anisotropy |
|||
| ΔGD° | m | D1/2 | |||
| nm | kcal/mol | M | |||
| apoE4 S94C-acrylodan | 479 | 0.253 ± 0.003 | 2.8 ± 0.1 | 1.3 ± 0.1 | 2.2 ± 0.1 |
| apoE4 W264C-acrylodan | 479 | 0.230 ± 0.005 | 2.0 ± 0.1 | 1.3 ± 0.1 | 1.5 ± 0.1 |
| apoE4 S290C-acrylodan | 488 | 0.194 ± 0.004 | 1.9 ± 0.1 | 1.2 ± 0.1 | 1.6 ± 0.1 |
| apoE4 22-kDa (Trp) | n.d.b | n.d.b | 3.1 ± 0.2 | 1.5 ± 0.1 | 2.1 ± 0.1 |
Estimated error is within ±2 nm.
not determined
Pyrene Excimer Fluorescence
We next performed pyrene labeling of apoE4 cysteine variants to monitor the self-association and lipid binding behaviors of apoE4. Pyrene fluorescence emission spectra display unique features that give information on the spatial proximity and polarity of the environment where the probe is located. Although pyrene typically exhibits fluorescence emission maxima at 375 and 395 nm, attributed to a monomeric state, an additional broad and red-shifted emission peak appears generally at around 470 nm when two pyrene rings are within 10 Å of each other, attributed to formation of an excited state dimer (43). Taking advantage of this unique property, we evaluated intermolecular spatial proximity of pyrene-labeled apoE4 variants because apoE is known to exist as a tetramer in solution (15, 16, 18).
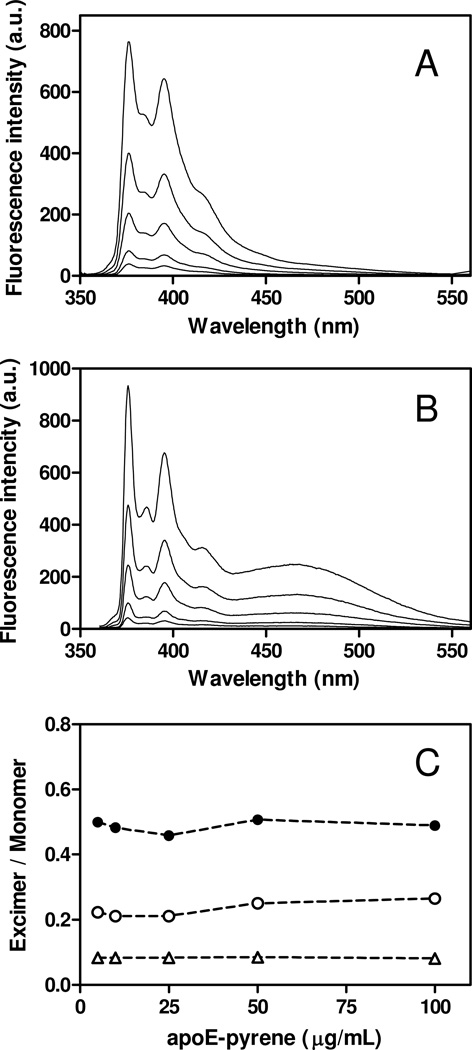
Fig. 2 shows pyrene fluorescence emission spectra and excimer/monomer ratios for pyrene-labeled apoE4 variants at protein concentrations ranging from 5 to 100 µg/ml, in which apoE4 is thought to be predominantly in tetramer form (8, 18). In fact, gel filtration experiments demonstrated that all apoE4 variants used exist predominantly as tetramer (Figs. S2 and S3 of the Supporting Information). ApoE4 S94C-pyrene exhibited virtually no excimer peak (Fig. 2A) whereas an intense excimer peak was observed for apoE4 S290C-pyrene (Fig. 2B), consistent with the notion that the C-terminal domain is responsible for the self-association of apoE through the intermolecular coiled-coil formation (19, 20). Interestingly, apoE4 W264C-pyrene displayed relatively lower excimer/monomer ratio compared to the S290C-pyrene, suggesting large reduction of the tetramer formation in the case of apoE4 W264C-pyrene. Indeed, comparison of gel filtration chromatography of apoE4 W264C without or with pyrene labeling revealed that addition of pyrene to the position 264 strongly inhibits apoE4 self-association (Figs. S3C and D of the Supporting Information). These results are consistent with prior findings that W264 is one of the key residues critical for the oligomerization of the C-terminal domain of apoE (44, 45).
Fig. 2.
Pyrene fluorescence emission spectra of apoE4 S94C-pyrene (A) and S290C-pyrene (B) at different concentrations of proteins (5–100 µg/ml). (C) The pyrene excimer (at 470 nm)/monomer (at 375 nm) intensity ratio for apoE4 S94C-pyrene (Δ), W264C-pyrene (○), and S290C-pyrene (●) as a function of apoE4 concentration. The excitation wavelength was 342 nm.
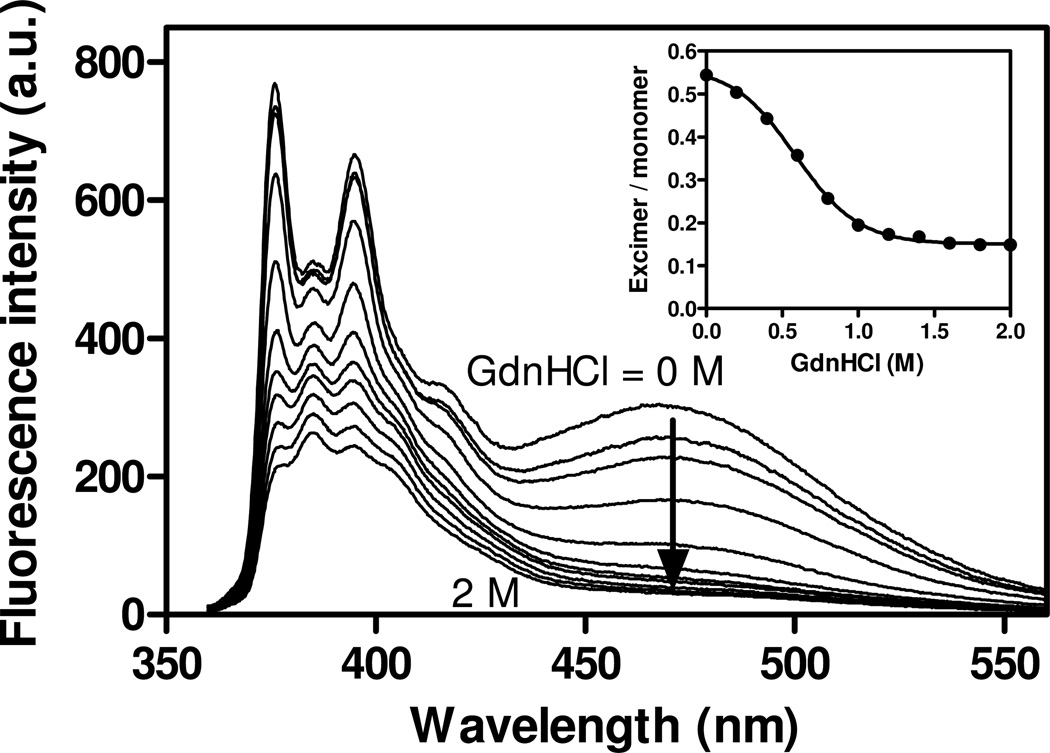
We also evaluated GdnHCl-induced dissociation of tetramer to monomer by pyrene excimer fluorescence. Fig. 3 shows the change in pyrene fluorescence emission spectra of apoE4 S290C-pyrene at different concentrations of GdnHCl. The excimer/monomer ratio gradually decreases with increasing concentration of GdnHCl, with the midpoint being around 0.6 M GdnHCl (Fig. 3, inset). This indicates that the dissociation to monomer of apoE4 S290C-pyrene occurs at a much lower concentration of GdnHCl than that for denaturation of apoE4 S290C-Ac (1.6 M GdnHCl in Table 1).
Fig. 3.
Pyrene fluorescence emission spectra of apoE4 S290C-pyrene (25 µg/ml) at different concentrations of GdnHCl (from 0 to 2 M). The inset shows the change in the excimer/monomer ratio as a function of GdnHCl concentration. The excitation wavelength was 342 nm.
SUV Binding of ApoE4 Monitored by Pyrene Fluorescence
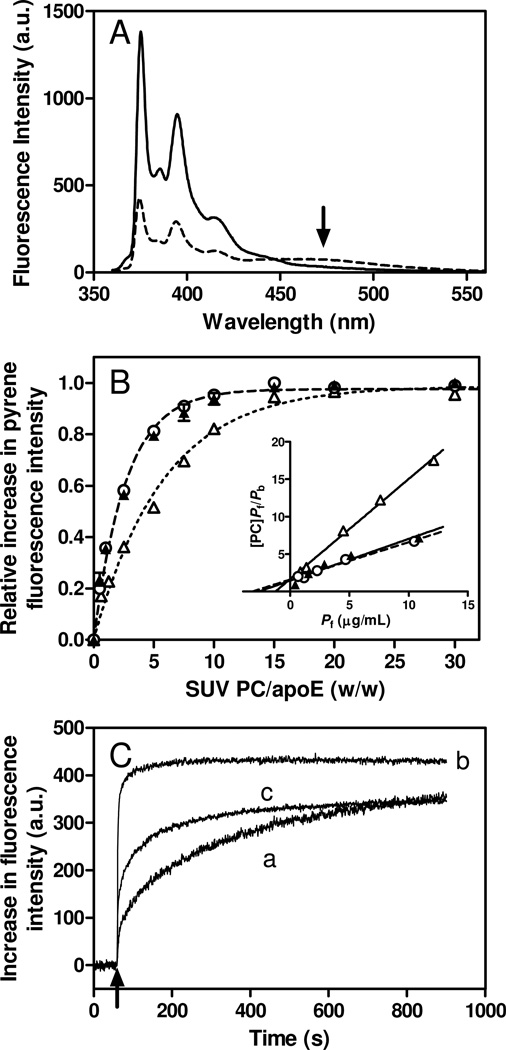
Binding behavior of pyrene-labeled apoE4 to SUV was estimated by the increase in pyrene fluorescence intensity upon lipid binding (30, 46). Fig. 4A shows fluorescence emission spectra of apoE4 S290C-pyrene in the absence or presence of SUV. The addition of SUV caused a large increase in fluorescence intensity of apoE4 S290C-pyrene, suggesting that the pyrene moiety attached to the protein is embedded in a hydrophobic environment of lipid membrane. The arrow at around 470 nm is indicative of an oligomer formation of apoE4 S290C-pyrene in the lipid-free state but not in the lipid-bound state (47).
Fig. 4.
(A) Pyrene fluorescence emission spectra of apoE S290C-pyrene in the lipid-free (dashed line) or bound to egg PC SUV (solid line). The arrow draws attention to the excimer peak at around 470 nm. Protein and PC concentrations were 25 µg/ml and 1.5 mg/ml, respectively. (B) Increases in fluorescence intensity of apoE4 S94C-pyrene (Δ), W264C-pyrene (○), and S290C-pyrene (▲) upon binding to egg PC SUV as a function of the weight ratio of PC to apoE4. Protein concentration was 25 µg/ml. The inset shows the linearized plots according to Hanes-Woolf equation (see Experimental Procedures). (C) Time courses of increases in fluorescence intensity upon binding to egg PC SUV for apoE4 S94C-pyrene (trace a), W264C-pyrene (trace b), and S290C-pyrene (trace c). SUV was added to apoE4 variants at final concentrations of 10 µg/ml protein and 0.4 mg/ml PC. Pyrene fluorescence was monitored at 385 nm with an excitation of 342 nm.
Fig. 4B shows the increases in fluorescence intensity of the pyrene-labeled apoE4 variants with increasing weight ratio of PC to protein (binding titration curves). Although all variants became saturated at high PC-to-protein ratio, both apoE4 W264C-pyrene and S290C-pyrene exhibited a greater tendency to reach saturation at a lower PC-to-protein ratio compared to the S94C-pyrene variant, indicating that the pyrene molecule attached at the N- or C-terminal helices in apoE4 binds differently to the SUV surface. From the linear regression lines based on the Hanes-Woolf equation shown in Fig. 4B, inset, Kd and Bmax values for pyrene-apoE4 variants were obtained (Table 2). The Kd values for all apoE4 variants are in submicromolar range and consistent with prior result (22). The Bmax values for the C-terminal pyrene-labeled apoE4 variants (W264C-pyrene and S290C-pyrene) were much higher than that of the N-terminal variant (S94C-pyrene). Since the increase in fluorescence intensity of a C-terminal pyrene-labeled apoE variant monitors the total amount of apoE bound while that for an N-terminal labeled apoE monitors only those molecules that are bound with the helix bundle open conformation, the observed variations in Bmax values indicate that apoE adopts multiple lipid-bound conformations at a lipid surface with the N-terminal helix bundle being closed and out of contact with lipid at near maximal binding (22, 48).
Table 2.
Parameters of Binding of ApoE4 Variants to Egg PC SUV
| apoE4 variant | Kd | Bmax |
|---|---|---|
| µg/ml | amino acids/mol PC | |
| apoE4 S94C-pyrene | 1.3 ± 0.3 | 0.75 ± 0.17 |
| apoE4 W264C-pyrene | 3.2 ± 0.6 | 2.0 ± 0.40 |
| apoE4 S290C-pyrene | 2.7 ± 1.2 | 1.8 ± 0.26 |
| apoE4 S94C-acrylodan | 2.3 ± 0.3 | 0.62 ± 0.09 |
We also monitored the time course of the increase in fluorescence upon binding of the pyrene-apoE4 variants to SUV (Fig. 4C). There was a rapid increase in pyrene fluorescence after the addition of SUV for the W264C-pyrene whereas the S94C-pyrene exhibited slow kinetics, suggesting the two-step mechanism for lipid binding of apoE4 like apoA-I (27, 30): initial binding occurs rapidly through the C-terminal α-helices followed by relatively slow conformational reorganization of the N-terminal helix bundle. The averaged half times for the increase in pyrene fluorescence obtained by fitting to the bi-exponential kinetic equation were 229, 27, and 112 s for apoE4 S94C-pyrene, W264C-pyrene, and S290C-pyrene, respectively. Based on the fact that apoE4 S290C-pyrene predominantly exists as tetramer whereas does apoE4 W264C-pyrene exists as monomer (Fig. S3 of the Supporting Information), the relatively slower increase in pyrene fluorescence of apoE4 S290C-pyrene compared to the apoE4 W264C-pyrene is likely due to the required dissociation of apoE4 S290C-pyrene tetramers to monomers prior to lipid binding. In fact, the rate constant for the slow phase of apoE4 S290C-pyrene (5.8 × 10−3 s−1) is in the same order to that for the tetramer dissociation of apoE4 (47).
FRET from Trp Residues to Acrylodan in ApoE4
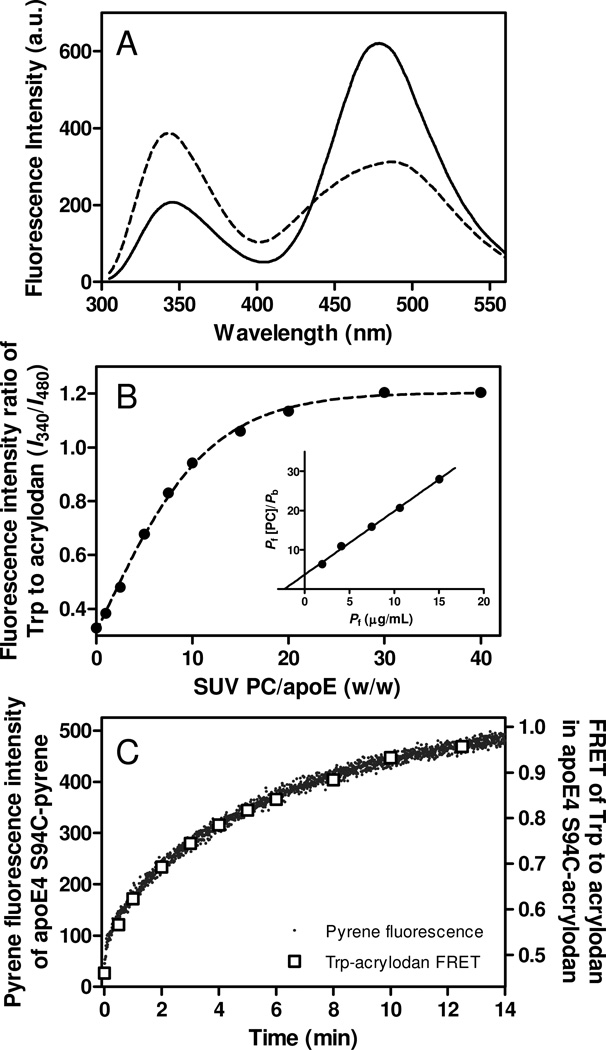
To further explore the conformational reorganization of the N-terminal helix bundle in apoE4 upon SUV binding, we measured FRET from intrinsic Trp residues (four in the N-terminal domain and three in the C-terminal domain) to acrylodan attached at S94C in apoE4 (47, 49). The occurrence of FRET in apoE4 S94C-Ac was confirmed by comparing Trp emission fluorescence spectra of unlabeled and acrylodan-labeled apoE4 S94C protein (Fig. S5 of the Supporting Information). Fig. 5A shows fluorescence emission spectra of apoE4 S94C-Ac in the absence or presence of SUV. A large increase in Trp emission fluorescence at around 340 nm and the concomitant decrease in acrylodan fluorescence at around 480 nm upon SUV binding indicate a significant reduction in the efficiency of FRET, consistent with the notion that the conformational rearrangement of the N-terminal helix bundle occurs upon lipid binding (22, 49, 50). Since almost no excimer peak of apoE4 S94C-pyrene was detected (Fig. 2), the observed FRET comes predominantly from intramolecular (interdomain or intradomain) interactions in apoE4. In addition, the similar dependency of FRET efficiency (E) in apoE4 S94C-Ac and increase in pyrene fluorescence of apoE4 S94C-pyrene on the SUV interaction (Fig. S5C of the Supporting Information) indicates that the change in FRET mainly reflects the conformational change of the N-terminal domain upon lipid binding, that is, the helix bundle opening of this domain. Taking the fluorescence intensity ratio of Trp residues at 340 nm to acrylodan at 480 nm as an indicator of FRET efficiency, binding titration curves of apoE4 S94C-Ac to SUV were derived (Fig. 5B). The resultant Kd and Bmax values for apoE4 S94C-Ac obtained from the linear regression lines in Fig. 5B, inset were similar to those for apoE4 S94C-pyrene (Table 2), indicating that the increased pyrene fluorescence of apoE4 S94C-pyrene upon SUV binding reflects the helix bundle opening of the N-terminal domain in apoE4. Consistent with this, the change in FRET from Trp residues to acrylodan in apoE4 S94C-Ac upon SUV binding occurs at the same rate as the increase in pyrene fluorescence of apoE4 S94C-pyrene (Fig. 5C).
Fig. 5.
(A) FRET between Trp residues and acrylodan in apoE4 S94C-acrylodan. Fluorescence emission spectra excited at 290 nm were recorded in the lipid-free (solid line) or SUV-bound (dashed line) states. Protein and PC concentrations were 25 µg/ml and 1.0 mg/ml, respectively. (B) Increases in fluorescence intensity ratio of Trp residues at 340 nm to acrylodan at 480 nm as a function of the weight ratio of PC to apoE4. Protein concentration was 25 µg/ml. The inset shows the linearized plots according to Hanes-Woolf equation. (C) Comparison of time courses of increase in pyrene fluorescence intensity and fluorescence intensity ratio of Trp to acrylodan for apoE4 S94C-acrylodan upon binding to egg PC SUV.
Lipoprotein Binding of ApoE4 Monitored by Pyrene Fluorescence
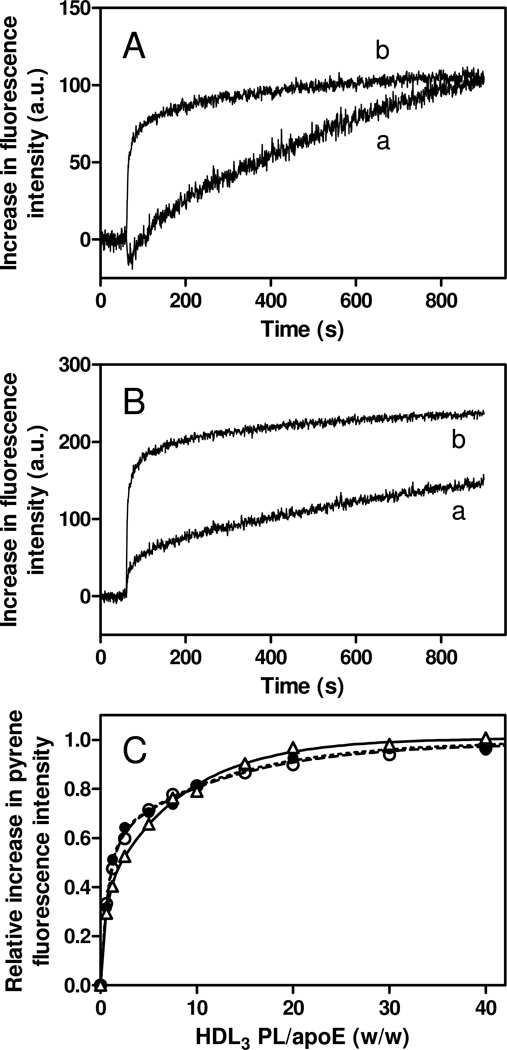
We also explored the binding of pyrene-labeled apoE4 variants to VLDL and HDL3 particles by monitoring pyrene fluorescence. The strong light scattering from VLDL particles made it difficult to obtain reliable binding titration curves of pyrene-labeled apoE4 variants to VLDL. Nevertheless, the measurements of time courses of binding of apoE4 N-terminal helix bundle domain (S94C-pyrene) and the C-terminal domain (W264C-pyrene) to VLDL (Fig. 6A) revealed that the kinetics of increase in pyrene fluorescence upon binding to VLDL for apoE4 S94C-pyrene is much slower than that for apoE4 W264C-pyrene (averaged half times are 631 and 182 s, respectively), similarly to the case of SUV (Fig. 4C). Interestingly, the kinetics of interaction of the N-terminal domain (S94C-pyrene) with HDL3 was markedly different from VLDL, whereas that of the C-terminal domain (W264C-pyrene) was similar for both lipoproteins (Fig. 6B). In addition, the binding titration curves shown in Fig. 6C demonstrate that the difference in binding behavior of the N- and C-terminal domains of apoE4 variants to the lipid surface as observed on SUV (Fig. 4B) is almost eliminated with HDL3. These results indicate that protein-lipid interactions are dominant for binding of the N-terminal helix bundle domain of apoE4 to the VLDL surface, whereas not only protein-lipid interactions but also protein-resident protein interactions modulate the binding process to the HDL3 surface (25).
Fig. 6.
Time courses of increases in fluorescence intensity upon binding to VLDL (A) and HDL3 (B) for apoE4 S94C-pyrene (trace a) and W264C-pyrene (trace b). VLDL or HDL3 was added to apoE4 variants at final concentrations of 10 µg/ml apoE4 and 0.1–0.4 mg/ml PL. Pyrene fluorescence was monitored at 385 nm with an excitation of 342 nm. (C) Increases in fluorescence intensity of apoE4 S94C-pyrene (Δ), W264C-pyrene (○), and S290C-pyrene (●) upon binding to HDL3 as a function of the weight ratio of PL to apoE4. Protein concentration was 25 µg/ml.
DISCUSSION
Given that apoE can exist in two lipid-bound states on spherical lipid particles with the N-terminal helix bundle in either open or closed conformations (21, 22, 48), it is likely that there is a two-step mechanism for binding of apoE to a lipid surface. Recently, surface plasmon resonance analysis to monitor in real time the reversible binding of apoE to VLDL and HDL3 particles indicated that apoE/lipoprotein interaction is indeed a two-step process: after an initial interaction the second slower step appears to involve opening of the N-terminal helix bundle domain of the apoE molecule (25). In this study, we applied fluorescence spectroscopy measurements with pyrene or acrylodan-labeled apoE4 to monitor such a conformational change of the protein on spherical lipid particles, as we and others have done successfully for apolipophorin III (51) and apoE4 (52) in discoidal complexes and apoA-I on SUV (30).
The fluorescence analysis of acrylodan- or pyrene-labeled apoE4 variants in the lipid-free state allowed us to examine the site-specific structure and stability of the protein. The results of GdnHCl denaturation for the acrylodan-labeled apoE4 variants (Fig. 1 and Table 1) demonstrated that the C-terminal domain, especially the most C-terminal region around S290 has a less organized and more exposed conformation than the N-terminal domain (41, 42), consistent with the high-resolution NMR structure of apoE3 where both S94 and W264 are located in helical structure whereas S290 is in a loop region (26). The comparison of pyrene excimer/monomer fluorescence of pyrene-labeled apoE4 variants (Fig. 2) indicated that the C-terminal helices are responsible for self-association of apoE4 in solution (15, 20, 42). The fact that apoE4 W264C-pyrene exhibited much less excimer fluorescence than S290C-pyrene (Fig. 2C), together with the gel filtration results demonstrating that addition of pyrene to the cysteine residue at position 264 strongly inhibits apoE4 self-association (Figs. S3C and D of the Supporting Information), indicates that W264 is of the key residues for the self-association. This finding agrees with prior reports revealing that five bulky hydrophobic residues in the C-terminal region (F257, W264, V269, L279, and V287) participate in stabilizing interactions of the tetramer (44, 45). The fact that the GdnHCl-induced decrease in pyrene excimer fluorescence of apoE4 S290C-pyrene (Fig. 3) occurs at a much lower concentration than that for the decrease in acrylodan fluorescence of apoE4 S290C-Ac (Fig. 1) indicates that a loss in quaternary self-association precedes the denaturation of secondary/tertiary structure of apoE4 (20).
Taking advantage of the fact that there is a significant increase in fluorescence intensity when a pyrene-labeled helix makes contact with a lipid surface (30, 46), we monitored the behaviors of the N- and C-terminal helices upon binding of apoE4 to spherical lipid particles (Fig. 4). The increase in fluorescence intensity observed with a C-terminal pyrene-labeled apoE molecule monitors the total amount of apoE bound because all C-terminal helices are in contact with the surface of the lipid particle (22, 27). In contrast, the increase in fluorescence intensity with an N-terminal pyrene-labeled apoE molecule monitors only those molecules that are bound with the helix bundle open because, when the helix bundle is closed, the pyrene label is not in contact with the lipid surface (30). Thus, the significant difference in fluorescence intensity increase observed with N-terminal labeled apoE4 (S94C-pyrene) and C-terminal labeled apoE4 (W264C-pyrene and S290C-pyrene) (Fig. 4B) implies that there are two lipid-bound conformations of apoE4 in which the N-terminal helix bundle adopts either open or closed conformations. The existence of two such lipid-bound states underlies the two-step kinetics of lipid binding of apoE4 in which the rate of increase in pyrene fluorescence of the N-terminal labeled apoE4 reflects the rate of helix bundle opening that is much slower than that observed with the C-terminal labeled apoE4 (Fig. 4C). Interestingly, the relatively slower kinetics of lipid binding of apoE4 S290C-pyrene compared to apoE4 W264C-pyrene (Fig. 4C) coupled with the finding that tetramer formation is essentially eliminated with the W264C-pyrene variant but not with the S290C-pyrene variant (Fig. S3 of the Supporting Information) indicates that tetramer dissociation is the rate-determining step for interaction of the C-terminal domain of apoE4 with a lipid surface (18).
FRET results with the N-terminal acrylodan-labeled apoE4 provide more direct evidence for the conformational reorganization of the N-terminal helix bundle domain of apoE4 upon binding to lipid particles. Significant reductions of the FRET efficiency from Trp residues to acrylodan in apoE4 S94C-Ac with increasing PC-to-protein ratio (Fig. 5B) directly indicate a conformational change of the N-terminal helix bundle domain upon SUV binding, consistent with the concept that the opening of the N-terminal four-helix bundle occurs upon lipid interaction (8, 22, 49). The fact that the change in FRET of apoE4 S94C-Ac upon SUV binding occurs at the same rate as the increase in pyrene fluorescence of apoE4 S94C-pyrene (Fig. 5C) confirms that the increase in pyrene fluorescence of the N-terminal pyrene-labeled apoE4 indeed reflects the conformational reorganization of the N-terminal helix bundle upon lipid binding.
In contrast to binding to lipid particles like SUV, it is thought that not only apoE-lipid interactions but also apoE-resident protein interactions are involved in the binding to lipoprotein particles (25). Similar kinetics of fluorescence changes from the N- or C-terminal pyrene-labeled apoE4 variants upon binding to VLDL (Fig. 6A) compared to SUV (Fig. 4C) implies that lipid interactions dominate binding of apoE4 to a VLDL particle (25). In contrast, the markedly different kinetics of the N-terminal pyrene-labeled apoE4 variant (Fig. 6B) together with the similar binding behaviors between the N- and C-terminal domains of apoE4 variants (Fig. 6C) observed upon binding to HDL3 indicate that lipid interactions play a lesser role in this case. Based on the estimation that ~80% of the HDL3 particle surface is protein-covered whereas ~60% of the VLDL particle surface is covered by phospholipid (53), it is likely that protein-protein interactions between apoE4 N-terminal helix bundle domain and apolipoproteins resident on the HDL3 surface are involved in the binding to HDL3 (25, 53). It should be noted that there was no apoE4-induced displacement of HDL3 proteins in our experimental conditions (Fig. S6 of the Supporting Information). Since the LDL receptor binding site in apoE is not recognized when the N-terminal helix bundle is in the closed conformation (8, 13), such different interactions of the N-and C-terminal domains of apoE4 with the surfaces of VLDL and HDL3 particles would play a critical role in lipoprotein metabolism. Consistent with this idea, an electron paramagnetic resonance spectroscopy study indicated that apoE4 associated with VLDL can have a more extended conformation than when associated with either LDL or HDL (24).
In summary, application of site-directed fluorescence labeling of apoE4 has provided insights into the binding mechanisms of the N- and C-terminal domains of apoE4 to spherical lipid particles and lipoproteins. ApoE4 binds to the lipid surface by a two-step mechanism in which initial binding occurs rapidly through the C-terminal domain followed by relatively slower conformational reorganization of the N-terminal helix bundle domain. The differences in the nature of the surfaces of VLDL and HDL3 in which the former is largely phospholipid-covered whereas the latter is mostly covered by the resident apolipoproteins appear to determine whether apoE-lipid or apoE-protein interactions dominate binding of apoE4 to the lipoprotein surface. Such differences in binding behavior of apoE4 to VLDL and HDL3 seem to underlie its binding selectivity to these lipoprotein particles (25, 54).
Supplementary Material
Abbreviations
- Ac
acrylodan
- apoE
apolipoprotein E
- FRET
fluorescence resonance energytransfer
- GdnHCl
guanidine hydrochloride
- GP
generalized polarization
- HDL
high-densitylipoprotein
- LDL
low-density lipoprotein
- PC
phosphatidylcholine
- PL
phospholipid
- SUV
small unilamellar vesicle
- TBS
Tris-buffered saline
- VLDL
very low-density lipoprotein
- WMF
wavelength of maximum fluorescence
Footnotes
This work was supported by NIH grant HL56083 and Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (No. 22590046).
Supporting Information Available
Far-UV CD spectra, GdnHCl denaturation curves of apoE4 variants monitored by CD (Figure S1), gel filtration profiles of apoE4 WT and monomeric variant (Figure S2), apoE4 cysteine variants without and with pyrene labeling (Figure S3), fluorescence emission spectra of acrylodan-labeled apoE4 variants (Figure S4), effect of SUV binding on the FRET between Trp residues and acrylodan in apoE4 S94C-Ac (Figure S5), gel filtration of 14C-labeled HDL (Figure S6) are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J. Lipid Res. 2009;50(Suppl):S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Getz GS, Reardon CA. Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall. J. Lipid Res. 2009;50(Suppl):S156–S161. doi: 10.1194/jlr.R800058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser PS, Narayanaswami V, Ryan RO. Apolipoprotein E: from lipid transport to neurobiology. Prog Lipid Res. 2011;50:62–74. doi: 10.1016/j.plipres.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 5.Mahley RW, Huang Y. Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. J Clin Invest. 2007;117:94–98. doi: 10.1172/JCI30889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raffai RL, Weisgraber KH. Cholesterol: from heart attacks to Alzheimer's disease. J. Lipid Res. 2003;44:1423–1430. doi: 10.1194/jlr.R300007-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 9.Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 12.Sivashanmugam A, Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J. Biol. Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaiou M, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH, Segall ML, Phillips MC, Lund-Katz S. Apolipoprotein E-low density lipoprotein receptor interaction: Influences of basic residue and amphipathic alpha-helix organization in the ligand. J. Lipid Res. 2000;41:1087–1095. [PubMed] [Google Scholar]
- 14.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 15.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 16.Sakamoto T, Tanaka M, Vedhachalam C, Nickel M, Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Contributions of the carboxylterminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms. Biochemistry. 2008;47:2968–2977. doi: 10.1021/bi701923h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vedhachalam C, Duong PT, Nickel M, Nguyen D, Dhanasekaran P, Saito H, Rothblat GH, Lund-Katz S, Phillips MC. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 2007;282:25123–25130. doi: 10.1074/jbc.M704590200. [DOI] [PubMed] [Google Scholar]
- 18.Garai K, Frieden C. The association-dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry. 2010;49:9533–9541. doi: 10.1021/bi101407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choy N, Raussens V, Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 20.Patel AB, Khumsupan P, Narayanaswami V. Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 21.Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 22.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Lund-Katz S, Phillips MC. Lipid binding-induced conformational change in human apolipoprotein E. Evidence for two lipid-bound states on spherical particles. J. Biol. Chem. 2001;276:40949–40954. doi: 10.1074/jbc.M106337200. [DOI] [PubMed] [Google Scholar]
- 23.Hatters DM, Voss JC, Budamagunta MS, Newhouse YN, Weisgraber KH. Insight on the molecular envelope of lipid-bound apolipoprotein E from electron paramagnetic resonance spectroscopy. J. Mol. Biol. 2009;386:261–271. doi: 10.1016/j.jmb.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tetali SD, Budamagunta MS, Simion C, den Hartigh LJ, Kalai T, Hideg K, Hatters DM, Weisgraber KH, Voss JC, Rutledge JC. VLDL lipolysis products increase VLDL fluidity and convert apolipoprotein E4 into a more expanded conformation. J. Lipid Res. 2010;51:1273–1283. doi: 10.1194/jlr.M000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry. 2009;48:3025–3032. doi: 10.1021/bi9000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad.Sci. USA. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 28.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 2011;286:38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng X, Morris J, Dressmen J, Tubb MR, Tso P, Jerome WG, Davidson WS, Thompson TB. The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association. Structure. 2012;20:767–779. doi: 10.1016/j.str.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kono M, Okumura Y, Tanaka M, Nguyen D, Dhanasekaran P, Lund-Katz S, Phillips MC, Saito H. Conformational flexibility of the N-terminal domain of apolipoprotein a-I bound to spherical lipid particles. Biochemistry. 2008;47:11340–11347. doi: 10.1021/bi801503r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrow JA, Arnold KS, Weisgraber KH. Functional characterization of apolipoprotein E isoforms overexpressed in Escherichia coli. Protein Expr. Purif. 1999;16:224–230. doi: 10.1006/prep.1999.1069. [DOI] [PubMed] [Google Scholar]
- 32.Saito H, Dhanasekaran P, Nguyen D, Deridder E, Holvoet P, Lund-Katz S, Phillips MC. α-Helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 2004;279:20974–20981. doi: 10.1074/jbc.M402043200. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M, Dhanasekaran P, Nguyen D, Ohta S, Lund-Katz S, Phillips MC, Saito H. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 2006;45:10351–10358. doi: 10.1021/bi060726t. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Minamida T, Arimoto I, Handa T, Miyajima K. Physical states of surface and core lipids in lipid emulsions and apolipoprotein binding to the emulsion surface. J. Biol. Chem. 1996;271:15515–15520. doi: 10.1074/jbc.271.26.15515. [DOI] [PubMed] [Google Scholar]
- 35.Koyama M, Tanaka M, Dhanasekaran P, Lund-Katz S, Phillips MC, Saito H. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 2009;48:2529–2537. doi: 10.1021/bi802317v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of apolipoprotein E structure by domain interaction: differences in lipidbound and lipid-free forms. J. Biol. Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- 37.Tetali SD, Budamagunta MS, Voss JC, Rutledge JC. C-terminal interactions of apolipoprotein E4 respond to the postprandial state. J. Lipid Res. 2006;47:1358–1365. doi: 10.1194/jlr.M500559-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, Bielicki JK. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46:2583–2593. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]
- 39.Tricerri MA, Behling Agree AK, Sanchez SA, Jonas A. Characterization of apolipoprotein A-I structure using a cysteine-specific fluorescence probe. Biochemistry. 2000;39:14682–14691. doi: 10.1021/bi0014251. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Breydo L, Makarava N, Yang Q, Bocharova OV, Baskakov IV. Site-specific conformational studies of prion protein (PrP) amyloid fibrils revealed two cooperative folding domains within amyloid structure. J. Biol. Chem. 2007;282:9090–9097. doi: 10.1074/jbc.M608623200. [DOI] [PubMed] [Google Scholar]
- 41.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 2006;45:4240–4247. doi: 10.1021/bi060023b. [DOI] [PubMed] [Google Scholar]
- 43.Bains G, Patel AB, Narayanaswami V. Pyrene: a probe to study protein conformation and conformational changes. Molecules. 2011;16:7909–7935. doi: 10.3390/molecules16097909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan D, Li Q, Korando L, Jerome WG, Wang J. A monomeric human apolipoprotein E carboxyl-terminal domain. Biochemistry. 2004;43:5055–5064. doi: 10.1021/bi035958w. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 46.Tamamizu-Kato S, Kosaraju MG, Kato H, Raussens V, Ruysschaert JM, Narayanaswami V. Calcium-triggered membrane interaction of the alphasynuclein acidic tail. Biochemistry. 2006;45:10947–10956. doi: 10.1021/bi060939i. [DOI] [PubMed] [Google Scholar]
- 47.Garai K, Baban B, Frieden C. Dissociation of apolipoprotein E oligomers to monomer is required for high-affinity binding to phospholipid vesicles. Biochemistry. 2011;50:2550–2558. doi: 10.1021/bi1020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC, Lund-Katz S. Effects of polymorphism on the lipid interaction of human apolipoprotein E. J. Biol. Chem. 2003;278:40723–40729. doi: 10.1074/jbc.M304814200. [DOI] [PubMed] [Google Scholar]
- 49.Fisher CA, Ryan RO. Lipid binding-induced conformational changes in the N-terminal domain of human apolipoprotein E. J. Lipid Res. 1999;40:93–99. [PubMed] [Google Scholar]
- 50.Fisher CA, Narayanaswami V, Ryan RO. The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E. J. Biol. Chem. 2000;275:33601–33606. doi: 10.1074/jbc.M002643200. [DOI] [PubMed] [Google Scholar]
- 51.Sahoo D, Narayanaswami V, Kay CM, Ryan RO. Pyrene excimer fluorescence: a spatially sensitive probe to monitor lipid-induced helical rearrangement of apolipophorin III. Biochemistry. 2000;39:6594–6601. doi: 10.1021/bi992609m. [DOI] [PubMed] [Google Scholar]
- 52.Drury J, Narayanaswami V. Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence. J. Biol. Chem. 2005;280:14605–14610. doi: 10.1074/jbc.M414019200. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen D, Dhanasekaran P, Nickel M, Nakatani R, Saito H, Phillips MC, Lund-Katz S. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 2010;49:10881–10889. doi: 10.1021/bi1017655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisgraber KH. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990;31:1503–1511. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.