Abstract
U2AF65 is essential for pre-mRNA splicing in most eukaryotes. Two consecutive RNA recognition motifs (RRM) of U2AF65 recognize a polypyrimidine tract at the 3′ splice site. Here, we use small-angle X-ray scattering to demonstrate that the tandem U2AF65 RRMs exhibit a broad range of conformations in the solution ensemble. The majority of U2AF65 conformations exhibit few contacts between the RRMs, such as observed in the crystal structure. A subpopulation adopts tight inter-RRM contacts, such as independently reported based on paramagnetic relaxation enhancements. These complementary structural methods demonstrate that diverse splice sites have the opportunity to select compact or extended inter-RRM proximities from the U2AF65 conformational pool.
Pre-mRNA splicing is a critical step of eukaryotic gene expression that regulates most human transcripts1. The pre-mRNA splice sites are marked by consensus sequences, including a branch point sequence (BPS) and a nearby polypyrimidine (Py) tract at the 3′ splice site. The essential splicing factor U2AF65 recognizes the Py tract during the early stages of pre-mRNA splicing2, and stabilizes association of the U2 small nuclear ribonucleoprotein (RNP) particle with the upstream BPS. Two central RNA recognition motifs (RRM1 and RRM2) of U2AF65, tethered by a 30-residue linker, are responsible for targeting the Py tract3 (Figure 1A, Table S1).
Figure 1.
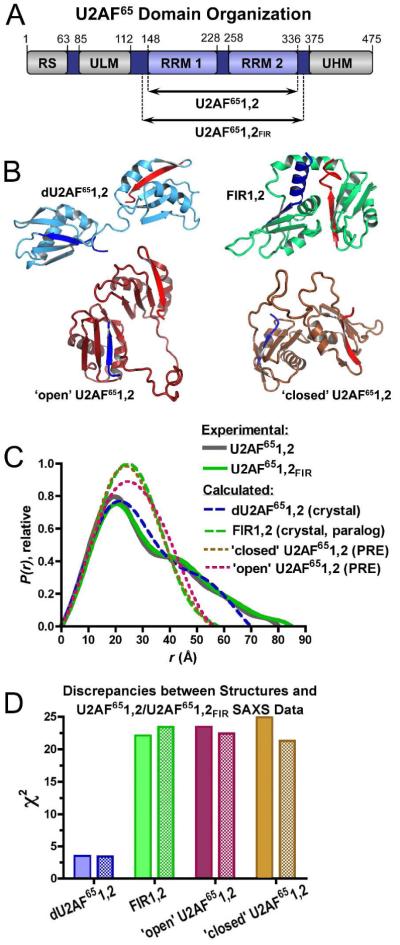
Comparison of U2AF structures and SAXS data. (A) Human U2AF65 domain organization and constructs used in this study. (B) Comparison of dU2AF651,2 (blue, PDB ID 2G4B) or FIR1,2 (green, PDB ID 2QFJ) crystal structures and the ‘open’ (maroon, PDB ID 2YH1) or ‘closed’ (brown, PDB ID 2YH0) NMR models of U2AF651,2. RNA ligands are omitted for clarity. The FIR1,2 structure in the absence of ligand is nearly identical (PDB ID 3UWT). N-and C-terminal secondary structures are colored dark blue or red, respectively. (C) Experimental P(r) functions of U2AF651,2 (gray, solid line) and U2AF651,2FIR (green, solid line) compared with calculated P(r) functions of the structures (dashed lines, colored as in A). Plots were scaled by the integrated area of the curves. (D) The χ2 values between the experimental U2AF651,2 (filled) U2AF651,2FIR (hatched) data and each of the structures shown in A, calculated as described in Supplementary Methods.
A structural understanding for how U2AF65 recognizes the Py tract is still emerging (Figure 1B). The crystal structure of the tandem U2AF65 RRMs connected by a shortened interdomain linker (dU2AF651,2) shows that conserved ribonucleoprotein (RNP) motifs of each RRM recognize the Py tract4. The two RRMs of the dU2AF65 polypeptide appear to act independently and lack substantial contacts between the RRMs. An average ab initio model determined by small-angle X-ray scattering (SAXS) confirms that the wild-type U2AF65 RRMs (U2AF651,2) exhibit a bilobal shape in solution5. Nevertheless, a ‘closed’ arrangement of the tandem U2AF65 RRM1 and RRM2 domains (U2AF651,2) was proposed recently based on paramagnetic relaxation enhancement (PRE) data6. In the ‘closed’ model, the RRM2 ‘backside’, or α-helical face occludes the RNP face of RRM1 (Figure 1B). A distinct ‘open’ form, in which the RRMs remain in contact but are oriented for RNA binding, was reported to increase prevalence following titration with a minimal Py tract6.
We individually compared each of the three available structures of the linked U2AF65 RRMs with the U2AF651,2 SAXS data (Figure 1C-D, Table S2, Figure S1). As noted previously5, the skewed, bimodal curves of the U2AF651,2 paired-distance distribution [P(r)] plots are consistent with an ellipsoidal overall shape and two independent RRM domains such as observed for the dU2AF651,2 crystal structure (Figure 1C). A peak at ~18 Å distances corresponds to the doubly weighted intra-RRM vectors (from both RRM1 and RRM2), whereas the shoulder at ~45 Å corresponds to vectors between RRM1 and RRM2. The lower maximum dimension (Dmax) of the crystal structure is expected given a 20-residue deletion in the inter-RRM linker, and is consistent with the experimentally determined Dmax of the identical construct5 (70 and 65±5 Å respectively). The dU2AF651,2 crystal structure (polypeptide coordinates) produces a reasonable fit with the U2AF651,2 SAXS data considering a 20-residue deletion within the inter-RRM linker (χ2 3.5) (Figure 1D, Table S2).
Conversely, large discrepancies between the NMR-based structures and the U2AF651,2 SAXS data were observed (Figure 1C-D, Table S2). Rather than bimodal curves, the P(r) functions calculated for the ‘closed’ or ‘open’ NMR-based U2AF651,2 structures exhibited the inverted parabolic curves of compact spherical shapes. Although PRE data portrayed a dominant ‘closed’ conformation in the absence of RNA, the discrepancy between the ‘closed’ structure and the U2AF651,2 SAXS data was severe (χ2 23.4). The discrepancy between the ‘open’ NMR structure and the U2AF651,2 SAXS data also was high (χ2 24.9).
Considering that the PRE data is limited to distances within the approximate diameter of an RRM (20 Å), we investigated whether a mixture of the three available structures in the U2AF65 solution ensemble could account for the apparent discrepancy (Figure S2). We input the available NMR and crystal structures as a starting pool for a minimal ensemble search (MES)7. Although not a rigorous thermodynamic algorithm, the MES algorithm selected the dU2AF651,2 crystal structure as contributing 90.1% of the solution conformations in an ensemble to best fit the U2AF651,2 scattering data, and the ‘open’ and ‘closed’ NMR structures as contributing 6.6% and 3.3% of the conformational ensemble, respectively.
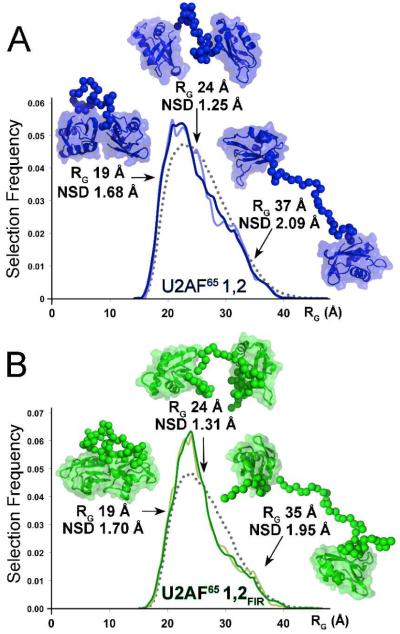
Next, an unbiased pool of randomized structures composed of U2AF65 RRM1 and RRM2 connected in a variety of orientations and proximities by ab initio linkers was tested as the starting pool to fit the SAXS data with a conformational ensemble. Ensembles of one, two, three, four, five, twenty, or fifty conformations were selected for the U2AF651,2 SAXS data using the EOM algorithm8 (Figure S3). Changing the selection from a single conformation, to a pool of two conformations with otherwise identical input parameters improved the discrepancies dramatically (respectively, from χ2 2.3 to 0.9). Further increases in ensemble size slightly improved the fits (Figure 2, Figure S3A). The improved fit of ‘ensembles’ composed of two over a single conformation indicated that U2AF651,2 populates at least two major classes of conformations in solution (Figure SB,C). One of the two selected structure classes is compact (RG ~20 Å), consistent with either ‘open’ or ‘closed’ NMR structures (RG 19.5-20.5 Å, Table S2), since the details of the RRM1-RRM2 interactions are obscure in the SAXS analyses (which is comparable to ~20 Å resolution). The second class lacks direct contacts between RRM1 and RRM2 (RG ~29 Å), as observed for the dU2AF651,2 crystal structure (RG 23.6 Å, Table S2).
Figure 2.
The 20-PDB (light color) or 50-PDB (dark color) ensemble fits of (A) U2AF651,2 (blue) and (B) U2AF651,2FIR (green) SAXS data.The radii of gyration (RG) are plotted on the x-axis, and the frequency of a structure with a given RG on the y-axis. Gray dashed lines plot the randomized starting pool; Solid lines the selected pool. The most typical or divergent selected structures are inset.
Selections of larger 20- or 50-PDB ensembles from the randomized starting pool further indicate flexible RRM1-RRM2 proximities (Figure 2A). A broad distribution of selected RRM1-RRM2 distances (normalized spatial discrepancy, NSD 1.48 ± 0.20 Å for the selected 20-PDB pool) resembles the inter-RRM distances of the randomized starting pool. The most representative structures (NSD 1.25 Å) lack direct contacts between the RRMs, but suggest some structural organization effectively shortens the RRM1-RRM2 linker (RG 23.7 Å). The most divergent structures (respective NSD 1.68 or 2.09 Å) either tightly pack (RG 18.5 Å) or fully extend (RG 37.4 Å) U2AF65 RRM1 and RRM2, consistent with a low selection of compact conformations in the minimal ensemble search.
We further considered whether slight differences in the constructs used for these distinct structural studies could contribute to discrepancies between the techniques (Table S1). The NMR structures include six additional residues at the C-terminus compared with the U2AF651,2 boundaries of the SAXS experiment. It was unlikely that a 6-residue size difference could directly account for the large discrepancy values; For comparison, the U2AF651,2 and dU2AF651,2 constructs differed by 20-residues in length, yet produce SAXS data in reasonable agreement5. Nevertheless, it remained possible that the six residues indirectly influenced the U2AF651,2 conformational pool. By analogy, the U2AF65 paralogue FIR (also called Puf60) contains tandem RRM domains (FIR1,2) packed in a qualitatively similar manner as the ‘closed’ NMR-based conformation of U2AF651,2 (9, Joint Center for Structural Genomics PDB ID 3UWT). One distinction is that FIR1,2 RRM1 surface is available for RNA binding, in contrast with RRM2 of ‘closed’ U2AF651,2 (Figure 1B). Residues flanking the core FIR RRMs are integrated within and appear to stabilize the ‘closed’ conformation. Based on comparison with FIR, we characterized a longer construct (U2AF651,2FIR). The U2AF651,2FIR protein included 12- and 11-residue extensions at the N- and C-termini respectively compared with the U2AF651,2 constructs used for X-ray studies (Figure 1A, Table S1). The U2AF651,2FIR boundaries correspond to those of the FIR1,2 structure and extend a few residues beyond the U2AF651,2 NMR construct.
The U2AF651,2FIR SAXS samples were monodisperse and data were collected in the 0.011–0.32 Å−1 q range (Figure S1). Size exclusion chromatography, dynamic light scattering, Porod volumes and concentration-independent Guinier RG or ensemble fits (Figure S1, Figure S4, Supplementary Methods) verified that the scattering data were not influenced by dimerization or other aggregates. The U2AF651,2FIR and U2AF651,2 P(r) plots were similar in overall dimensions and bimodal shapes consistent with independent action of the RRM1 and RRM2 domains (Figure 1C). The U2AF65 1,2FIR SAXS data remained a significantly better fit with the crystal structure (χ2 3.4) compared with either ‘closed’ or ‘open’ NMR structures (χ2 22.4 or 21.3, respectively) (Figure 1D).
Ensemble fits of U2AF651,2FIR scattering data decreased the χ2 from 2.2 for an ‘ensemble’ of a single structure to 1.0 for two co-existing structures in solution (Figure S2). Like U2AF651,2, one type of selected conformation in the 2-PDB ensemble exhibited direct contacts between the RRMs consistent with NMR structures, whereas the other lacked inter-RRM contacts as observed in the crystal structure. Ensembles of 20- or 50-PDBs further improved the fit (χ2 0.9). The structural variability within the 20- and 50-PDB ensembles remained broad in the presence of the additional U2AF651,2FIR residues (average NSD 1.48 ± 0.20 Å) (Figure 2B). As for U2AF651,2, the most populated U2AF651,2FIR structures (NSD 1.31 Å) lacked direct contacts between the RRMs (Figure 2B). We concluded that N- and C-terminal residues bordering the tandem U2AF65 RRMs did not detectably promote compact conformations such as the FIR1,2 or PRE-derived U2AF651,2 structures.
The ensemble fits potentially reconcile the apparent discrepancies between the SAXS data and NMR models. Indeed, the PRE data already indicated the presence of ‘open’ as well as ‘closed’ U2AF651,2 conformations in the absence of RNA. The SAXS method is sensitive to conformations that lack contacts between RRM1 and RRM2, which would have little effect on PRE signals. Whereas both SAXS and PRE-derived data are consistent with ensembles of U2AF651,2 conformations, the SAXS data unambiguously demonstrate that U2AF65 conformations with loosely associated RRMs substantially contribute to the solution ensemble. Altogether, these results emphasize the importance of crystallography, SAXS, and NMR as complementary methods to fully portray macromolecular structures.
The SAXS analyses presented here has important implications for pre-mRNA splice site recognition. The U2AF65 RRMs are not locked in the ‘closed’ state, rather are available to independently seek compatible binding sites in diverse pre-mRNAs. Future structures of U2AF65 bound to distinct splice sites are needed to reveal how the conformations can locally and globally adapt to distinct RNA sequences.
Supplementary Material
ACKNOWLEDGMENT
We thank an anonymous reviewer, S. D. Kennedy, M. R. Green, G. Hura and J. E. Wedekind for insightful comments.
Funding Sources
This work was supported by National Institutes of Health Grant R01 GM070503.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information. Tables S1-S2, Figures S1-S3, and experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes. SAXS data for U2AF651,2 and U2AF651,2FIR have been deposited in BIOISIS and are available with accession codes 1U2FRP and 2U2FRP respectively.
REFERENCES
- (1).Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zamore PD, Green MR. Proc Natl Acad Sci U S A. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zamore PD, Patton JG, Green MR. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- (4).Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jenkins JL, Shen H, Green MR, Kielkopf CL. J Biol Chem. 2008;283:33641–33649. doi: 10.1074/jbc.M806297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- (7).Pelikan M, Hura GL, Hammel M. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- (9).Crichlow GV, Zhou H, Hsiao HH, Frederick KB, Debrosse M, Yang Y, Folta-Stogniew EJ, Chung HJ, Fan C, De la Cruz EM, Levens D, Lolis E, Braddock D. EMBO J. 2008;27:277–289. doi: 10.1038/sj.emboj.7601936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.