Abstract
N-terminal methylation of free α-amino-groups is a post-translational modification of proteins that has been known for 30 years but has been very little studied. In this modification, the initiating M residue is cleaved and the exposed α-amino group is mono- di- or trimethylated by NRMT, a recently identified N-terminal methyltransferase. Currently, all known eukaryotic α-aminomethylated proteins have a unique N-terminal motif, M-X-P-K, where X is A, P, or S. NRMT can also methylate artificial substrates in vitro in which X is G, F, Y, C, M, K, R, N, Q or H. Methylation efficiencies of N-terminal amino acids are variable with respect to the identity of X. Here we use in vitro peptide methylation assays and substrate immunoprecipitations to show that the canonical M-X-P-K methylation motif is not the only one recognized by NRMT. We predict that N-terminal methylation is a widespread post-translational modification, and that there is interplay between N-terminal acetylation and N-terminal methylation. We also use isothermal calorimetry experiments to demonstrate that NRMT can efficiently recognize and bind to its fully methylated products.
Modification of free α-amino group in proteins is a common covalent posttranslational modification. The acetylation of protein N-terminal residues is widely observed, with up to 50% of cytosolic proteins estimated to be acetylated at their N-termini in eukaryotic cells1. In contrast, the alkylation of α-amino groups appears to be less common and has been little studied. To date, approximately two dozen proteins in various organisms have been reported to be N-terminally methylated, mostly in prokaryotes2, 3. The function of N-terminal methylation was completely unknown until recently3, when it was discovered that α-N-methylation of RCC1 (regulator of chromosome condensation 1) is required for its efficient binding to DNA4. However, the functional diversity of proteins reported to be N-terminally methylated suggests this modification likely has additional roles. In bacteria, the known N-terminally methylated proteins are components of large multi-subunit complexes, such as the ribosomal proteins (L11, L16, L33, S11 and the associated IF-3)5, pilins, and the chemotaxis-flagellar apparatus (CheZ protein)5. There is some evidence that these N-termini are located in regions involved in protein-protein interactions5.
Until recently, the enzyme responsible for N-terminal methylation of eukaryotic proteins had not been identified6. Apart from RCC1, eukaryotic proteins shown to be methylated at their α-amino group included histone H2B in Tetrahymena sp. and Drosophila sp., ribosomal protein S25 in Saccaromyces cerevisiae, cytochrome c-557 in Crithidia sp., myosin light chains in rabbit7, and recently SET and Retinoblastoma protein (Rb) in humans6.
The first eukaryotic N-terminal methyltransferase, NRMT, has recently been identified6, 8. The methylation catalyzed by NRMT in eukaryotic cells appears to require a specific consensus on the N-terminus of substrate proteins (M-X-P-K-). The N-terminal Met residue must be removed before N-terminal methylation. X can be one of many different residues but all of the known substrates possess A, S or P6. Based on limited mutagenesis studies, P and K residues in the third and fourth positions also appear to be necessary for efficient N-terminal methylation. Using this consensus several dozen additional proteins are predicted to be α-N-methylated.
Here, we further address the question of substrate specificity of NRMT by expanding its recognition motif, identifying new substrates, and showing that different recognition motifs tend to have variable affinities for NRMT. We also identify for the first time a substrate that can be either N-terminally methylated or acetylated depending on its tissue of origin.
EXPERIMENTAL PROCEDURES
Constructs and antibodies
All mammalian expression constructs were designed as previously described6. Wild-type and the K4Q mutant of ZFP15 and Mina53 vectors were designed as for the RCC1 vectors but using human ZFP15 and Mina53 cDNA3. Triple-GFP fused with the first 10 residues of human RCC1, followed by either a nuclear localization signal (NLS) or a nuclear export signal sequence (NES) was designed on the basis of the pKGFP vector as described previously 9. All His-tagged proteins were purified as previously described3.
Peptide Synthesis
Synthetic peptides were prepared by using standard Fmoc chemistry in an APEX 396 peptide synthesizer (Advanced Automated Peptide Protein Technologies), resuspended at a concentration of 1 mg/ml in aqueous acetic acid 0.1% (v/v) and stored at −35 °C until use. For sequence confirmation, 5 pmol/µl solutions of each peptide were infused into a linear quadrupole ion trap mass spectrometer (Thermo Finnigan LTQ) and analyzed by electrospray ionization (ESI) and collision activated dissociation (CAD)/electron transfer dissociation (ETD). The resulting spectra were interpreted manually. Mixtures of 35–45 synthetic peptides (1 nmol/peptide) were prepared in 0.1% acetic acid, taken to dryness and stored at −35 °C for in vitro methylation assays.
In vitro methylation assays
In vitro methylation reactions were done as described previously6. The peptide library was methylated as follows: The mixtures of 35–45 peptides (0.5 nmol of each peptide) were dissolved in 50 mM Tris, 50mM potassium acetate buffer pH=8.0 followed by addition of 300 µM SAM and 2 µM NRMT to the final volume of 50 µl (final concentration of each peptide = 10 µM). The reaction was run at 30°C for 2 h followed by re-application to Ni-NTA beads to remove NRMT, and microdialysis to remove SAM/SAH (S-adenosylmethionine/S-adenosylhomocysteine). The microdialysis was done using Spectra/Por Biotech Cellulose Ester (CE) Dialysis Membrane (MWCO: 100–500 D) and 50 µl Dialysis Buttons (Hampton Research) filled with methylated sample. The buttons were transferred to NEXTAL hanging Drop Vapor Diffusion Crystallization Plate with reservoirs filled with 2 ml of 50 mM Tris; 50 mM potassium acetate buffer pH 8.0. The dialysis was done overnight at 25°C.
MS Sample Preparation
On-bead samples of proteins immunoprecipitated with anti-FLAG and anti-me3-SPK antibodies were washed 3× with 50 µl aliquots of 100 mM ammonium bicarbonate (pH 8.0), suspended in a 4th aliquot and then reduced, carbamidomethylated, and digested using endoproteinase Glu-C (Roche) at an enzyme:protein ratio of 1:20 as previously described10. The resulting peptides were removed from the beads, acidified with glacial acetic acid (pH 3.5) and stored at −35 °C.
MS Analysis
For all analyses by mass spectrometry, peptides were pressure-loaded onto a capillary precolumn (360 µm o.d., 75 µm i.d.) packed with 5–20 µm C18 resin (YMC). The precolumn was washed with 0.1% acetic acid and connected to an analytical column (360 µm o.d., 50 µm inner i.d.) equipped with an integrated 1 µm emitter tip and packed with 5 µm C18 resin (YMC)11. Peptides were gradient-eluted into the mass spectrometer at 60 nl/min using nanoflow, reverse-phase HPLC and microESI into CAD and front-end ETD-enabled LTQ/Fourier Transform hybrid mass spectrometers (LTQ-FTMS and LTQ-Orbitrap, Thermo Scientific) as previously described12. Mass analyses were conducted using one high resolution (60,000 at 400 m/z) MS scan followed by 5–10 CAD/ETD data-dependent MS2 scans acquired in the linear ion trap. Data resulting from analysis of antime3-SPK Mus musculus immunoprecipitated proteins were searched with OMSSA (version 2.1.1,13) using MS/MS peak lists generated by Bioworks Browser (version 3.3.1 SP1) against the human/rat/mouse RefSeq database (downloaded June 2009). These results were used to guide data analysis. All peptide sequences were confirmed by manual interpretation of the spectra. To determine the relative abundances of each peptide, the areas under the extracted ion chromatogram curves for each peptide in all of its charge states and detected forms were summed. The peak area for each peptide was divided by the total area for all forms of the peptide to obtain an estimate of the percent of each form of modified peptide 14.
In vivo substrate affinity assay
Sequences encoding the following peptides MSPKRIAKRRS, MSPQRIAKRRS, MPPKRIAKRRS were fused to GFP to create a set of competitor constructs. All constructs were based on vectors published previously3. The substrate set was prepared identically but with FLAG-tag on the C-terminus of GFP for efficient immunoprecipitation. Constructs were transfected into 293LT cells using calcium phosphate.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed using an ITC200 microcalorimeter (MicroCal). Recombinant NRMT was dialyzed overnight at 4°C against methyltransferase buffer (50 mM Tris, 50mM potassium acetate buffer pH=8.0). Substrate peptides were purchased from GenScript in lyophilized form, and dissolved in the same buffer. Product peptides were purchased from American Peptide. The measurements were performed at 25°C by titrating 100 µM NMRT with 800–1000 µM peptide solution. Binding curves were analyzed by using the Origin (MicroCal) software package.
RESULTS
NRMT has broad substrate specificity
Our previous RCC1 mutagenesis results suggested that although NRMT can methylate proteins with a variety of residues at the second position in the (M)-X-P-K motif, there is no flexibility allowed at positions 3 and 4. Thus, mutation of K to Q severely lowers the efficiency of methylation. However, the importance of Pro at position 3 had not been thoroughly tested, and we had not investigated any possible supportive role of residue 5. To more rigorously determine the substrate specificity of NRMT, we synthesized a peptide library consisting of the RCC1 N-terminal peptide with varying amino acids in positions 2, 3, 4 and 5, performed in vitro methylation reactions using recombinant NRMT, removed excess SAM, and subjected the purified peptide mixtures to analysis by MS.
The methylation efficiencies of the different substrate peptides from the peptide library are listed in Supplementary Table S1. Remarkably, in addition to the expected methylation substrates with (M)-X-P-K motifs, we identified an unexpectedly large number of other positives: APR, GNK, GPK, GPR, MGKK, MNK, MNR, MPK, MPRR, SAKR, SEKR, SMKR, SNKR, SPRR, SQKK, SQKR, GGKK, GGKR, GSK, and SSKR. This peptide methylation assay reveals that promiscuity of NRMT in terms of position 3 is quite high. Residues on the third position which appear to promote N-terminal methylation in the pool of tested peptides are: P, A, E, M, N, Q, G and S. Arginine, lysine, and residues with aromatic side chains did not result in efficient methylation when on position 3. Similarly, as we have shown previously6, K on position 4 is absolutely crucial for efficient methylation but can be substituted by R in certain situations (Table S1, S2).
Another interesting outcome of the peptide library methylation assays is that different substrates appear to be methylated with different efficiencies in vitro and vary in the total percentage of mono-, di- and tri- (or full) methylation (Table S1). PPK peptides were fully dimethylated with 100% efficiency in contrast to other known NRMT substrates. SPK-RCC1 peptides were mostly mono-, di-, and trimethylated; up to 60%, 3%, 0.8% respectively. These results are in agreement with the ITC experiments, in vivo substrate affinity assays, and in vitro product inhibition assays described below. They suggest that PPK substrates are the best targets for NRMT and that there is a wide range of substrate affinities for NRMT.
On the basis of the above results we were able to predict how abundant each N-terminal methylation motif is in the human genome (See supporting information paragraph) and to divide all potential NRMT substrates into functional groups. A total of 308 potential substrates were identified based on sequence identity to those successfully methylated in the peptide library. The functional analysis of NRMT predicted targets was completed using CDD software15.
Of these targets, 32 (~10%) are defined by the CDD database as Chromatin/DNA binding, 6 (~2%) as ribosomal proteins, there are 27 (~9%) zinc finger containing proteins (not included in the Chromatin/DNA binding group), 12 (~4%) are RNA binding/processing proteins, and 10 (~3%) are myosins.
To test if some of the novel motifs are N-terminally methylated in vivo, we have used mouse spleen lysates as a source of N-terminally methylated proteins. It was shown previously that mouse spleen lysates contain a large number of methylated proteins in comparison to other tissues6. Rabbit anti-trimethylated RCC1 antibody was used to immunoprecipitate (IP) N-terminally methylated proteins6. Three of the co-precipitated proteins were Myosin Light Chain (MLC) isoforms: MLC9, MLC12B, MLCB-like, which begin with an SSKR or SSKK N-terminal sequence. Subsequent MS analysis confirmed that these MLCs are methylated at the N-terminus in vivo (Figure S3 D, E).
The affinity of NRMT for its substrates depends on the substrate consensus sequence
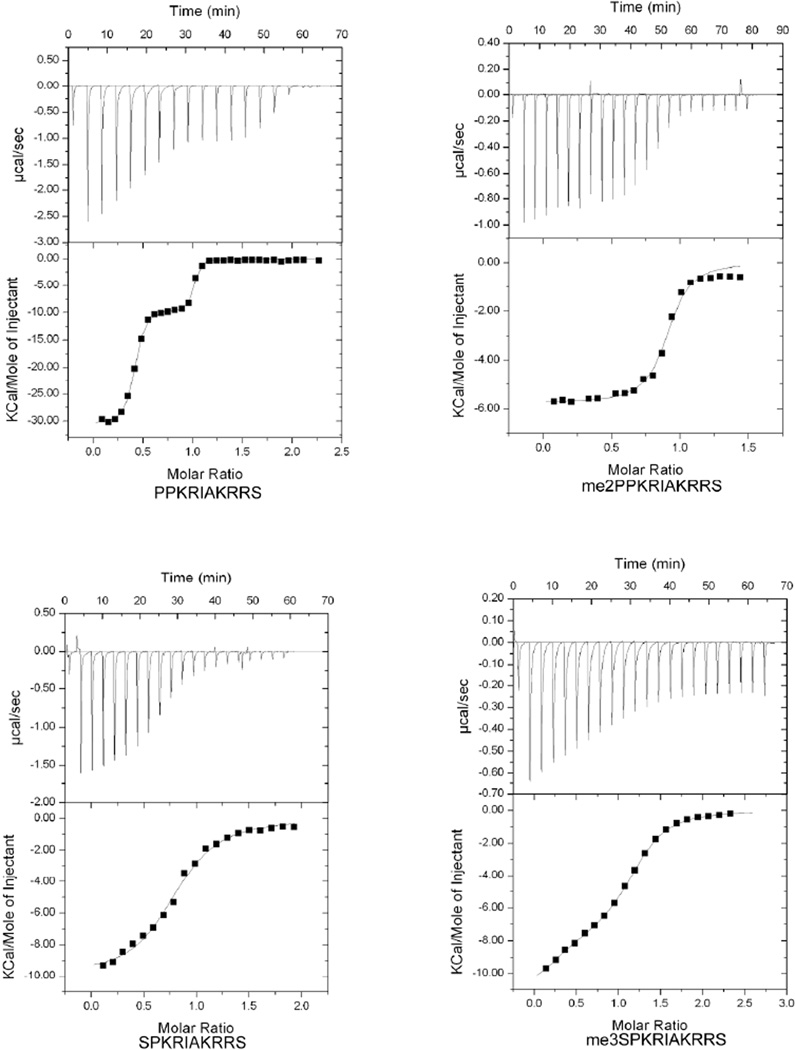
To further characterize NRMT substrate specificity we employed ITC to measure affinity of recombinant NRMT to RCC1 peptides. The following 10 residue RCC1 N-terminal peptides were used for the measurements: SPKRIAKRRS; me3SPKRIAKRRS; PPKRIAKRRS; and me2PPKRIAKRRS. The four peptides that contain Lys at position 4 all bound to recombinant human NRMT with high affinity (Figure 1). The two peptides with Gln at the fourth position (PPQRIAKRRS; SPQRIAKRRS) demonstrated very low affinity binding (in mM range, data not shown) which is in accordance with previous results6.
Figure 1. Substrate and product peptides affinity to NRMT measured using isothermal titration calorimetry.
Wild-type 10-residue SPK-RCC1 N-terminal peptide (bottom left panel) showed efficient binding to NRMT with a Ka 1.0E5±1.10E4 M−1 (Kd 10µM). A PPK-RCC1 N-terminal peptide showed very efficient binding (top left panel) with Ka1 6.60E8±2.0E8 M−1 and Ka2 7.3E6±2.3E6 M−1 (Kd values 1.5nM and 136nM respectively). The 10 residue me3-SPK-RCC1 (Ka 4.8E5±6.75E4 M−1) and me2-PPK-RCC1 (Ka 7.70E5±1.5E5 M−1) N-terminal peptides (left panel) showed efficient binding to NRMT with Kd values of 2.0µM and 1.3µM respectively.
The peptide SPKRIAKRRS represents the wild type N-terminal sequence of human RCC1, a native substrate of NRMT. This peptide demonstrates efficient, binding with Kd = 10 µM (Table 1). The binding of peptide PPKRIAKRRS, representing the N-terminus of mouse RCC1, reproducibly produced a bi-phasic titration curve. Surprisingly, about 40% of the protein binds the peptide with Kd = 1.5nM while the other 60% binds with Kd = 140 nM. We postulate that the PPKRIAKRRS peptide shows this behavior due to very slow isomerization of cis/trans peptide bond in amino acid sequences containing Pro-Pro dipeptide, as has been reported previously16–18. The SPKRIAKRRS peptides (both non-methylated and methylated forms) did not show biphasic binding. Interestingly, binding studies of the methylated me2PPKRIAKRRS peptide also did not yield a bi-phasic curve. To investigate possible explanations for this difference between the methylated and unmethylated PPK peptides, we generated a model of di-methylated tetrapeptide PPAA using ICM-Pro (Internal Coordinate Mechanics Professional)19. Using this model we determined the ten lowest energy conformers, calculated by Marvin Suite ver. 5.9.2, and found that all have the trans conformation of the Pro-Pro peptide bond. There are no energetically favorable conformations for the cis isomer, as calculated by Marvin and ICM-Pro. The search for the lowest energy conformers for the non-methylated tetrapeptide PPAA, (for a subset of ten lowest energy conformers) yielded six of the calculated conformers in the trans conformation and four were found to be cis. We conclude that the addition of a methyl group to the N-terminus of PPKRIAKRRS makes it more likely to adopt a trans conformation (Figure S1A, B) while the cis conformation is not favored due to steric hindrance. As a result me2PPKRIAKRRS no longer can form a stable mixture of cis and trans isomers. Despite the complication of this bi-phasic titration curve, the data clearly indicate that the affinity of NRMT for its substrates is influenced by the N-terminal consensus sequence.
Table 1.
Summary of ITC results
| Peptide | N | Kd | ΔH (cal/mol) | ΔS(cal/mol/deg) |
|---|---|---|---|---|
| SPKRIAKRRS | 0.8 | 10µM | −9943 ±207 | −10.5 |
| PPKRIAKRRS | 0.4/0.6 | 1.5nM/140nM | −3.1E4 ±221/−9428 ±192 | −62.2/−0.2 |
| me3SPKRIAKRRS | 1.2 | 2.0µM | −4333 ±1.2E3 | 11.5 |
| me2PPKRIAKRRS | 0.9 | 1.3µM | −5768 ±93 | 7.6 |
NRMT can also bind its methylated products
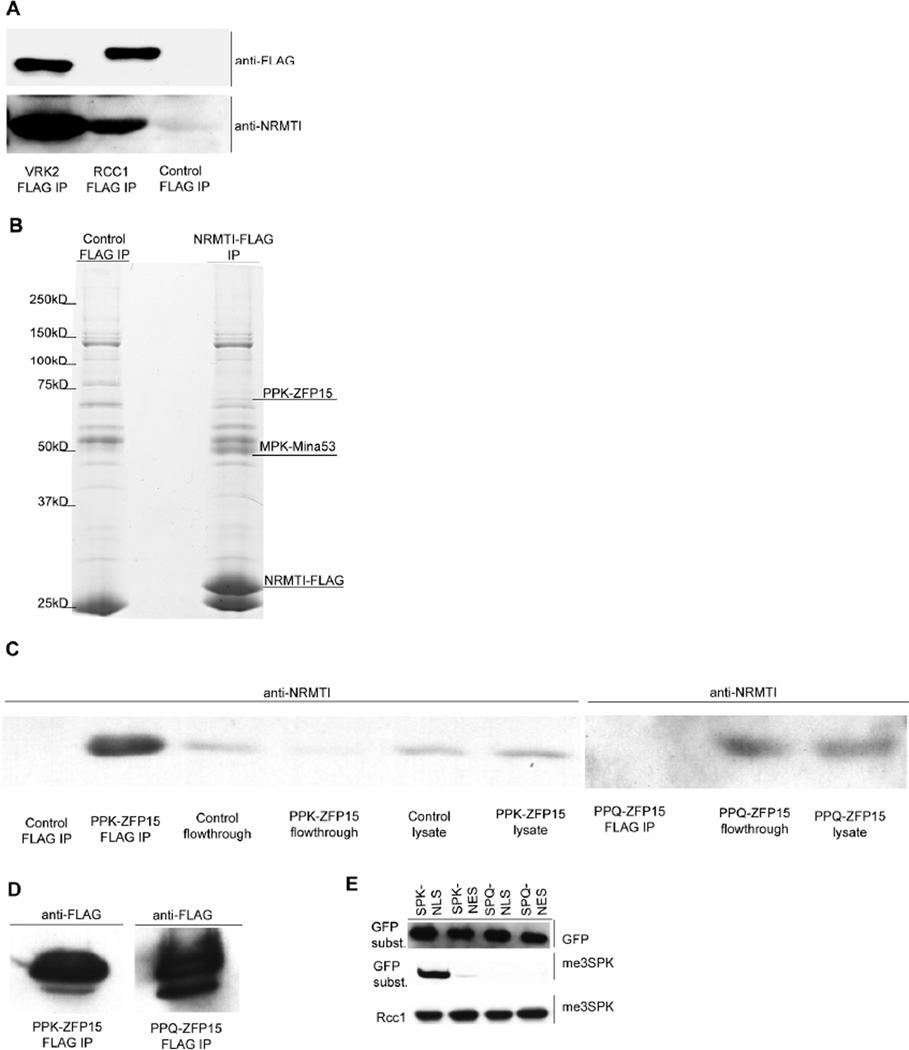
Interestingly, both me3SPKRIAKRRS and me2PPKRIAKRRS peptides, which are products of the enzymatic reaction catalyzed by NRMT, also efficiently bound to recombinant NRMT with Kd values of 2.0 µM and 1.3 µM, respectively (Table 1). This is surprising result, as interactions between an enzyme and its product are usually of very low affinity. It predicts that methylated products would remain bound to NRMT and inhibit further substrate methylation. To test whether such interactions can be detected in vivo, we expressed two target proteins, VRK2 and RCC1, as FLAG-tagged constructs in 293LT cells, and asked if we could immunoprecipitate endogenous NRMT with these proteins. From previous MS studies, we expect that the vast majority of the target proteins will be fully methylated. As shown in Figure 2A, both VRK2-FLAG and RCC1-FLAG efficiently captured endogenous NRMT. Consistent with our in vitro data, the VRK2, which possesses a P-P-K motif, captured more NRMT than the RCC1, which possesses a S-P-K motif.
Figure 2. NRMT can bind to its fully methylated substrate proteins.
A) NRMT is co-precipitated with C-terminally FLAG-tagged substrates (RCC1 and VRK2)
B) C-terminally FLAG-tagged NRMT co-precipitates with endogenous ZFP15 and Mina53. IPs were run on the 10% polyacrylamide gel and stained with Commassie blue.
C) C-terminally FLAG-tagged wild type ZFP15 co-precipitates with endogenous NRMT. The PPQ non-methylatable mutant of ZFP15 does not bind endogenous NRMT.
D) Both PPK- and PPQ-ZFP15 FLAG-tagged proteins are efficiently expressed and immunoprecipitated.
E) N-terminal methylation activity is localized in the nucleus. NLS fused SPK-GFP3 substrates are efficiently methylated, in contrast to NES fused SPK-GFP3.
Next, we took advantage to this unusually strong product binding to help identify new NRMT substrates. To this end, we screened for proteins that were co-precipitated with FLAG-tagged NRMT expressed in 293LT cells. One of the precipitated proteins was identified as ZFP15, which possesses a PPKKQAQ N-methylation motif (Figure 2B). In a reverse precipitation, we also found that FLAG-tagged ZFP15 could efficiently capture endogenous NRMT (Figure 2C, D). We confirmed that majority of precipitated ZFP15 is dimethylated on N-terminal proline residue, as shown by MS (Figure S3A). As expected, a PPQ non-binding/non-methylatable mutant of ZFP15 did not capture endogenous NRMT, suggesting the interaction between NRMT and N-methylated ZFP15 occurs through the methylated N-terminus (Figure 2C, D, Figure S3B).
An additional protein that co-precipitated with FLAG-tagged NRMT, Mina53, possesses a potential N-terminal methylation motif, MPKKAKPT 6. Previously, the MPK N-terminal motif was suggested to be one recognized by NRMT. However, in a reverse precipitation, C-terminally FLAG-tagged Mina53 was unable to capture endogenous NRMT (Figure S2B, C). Subsequent mass spectrometric analysis of both samples demonstrated the initial Met residue was cleaved in both cases with 100% efficiency (data not shown), effectively destroying the N-terminal methylation motif and preventing the methylation and NRMT binding. We hypothesize that a certain fraction of proteins can escape MAP (Methionine Amino Peptidase) activity and retain the initial methionine, which in turn can be subjected to N-terminal methylation.
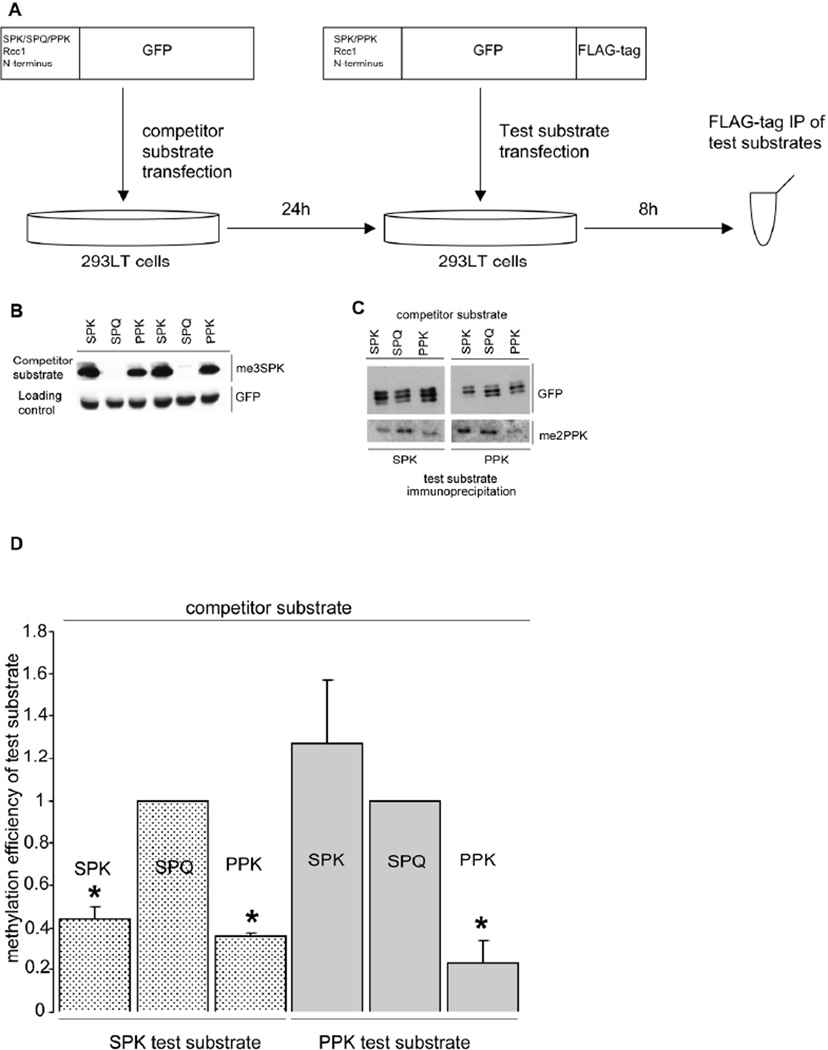
Evidence for NRMT substrate competition in intact cells
To test whether the high affinity substrates/products of NRMT can outcompete other substrates within the intact cell, we designed an experiment in which a test substrate was transiently expressed in cells that had previously been transfected with a potential competitor protein. Eight hours after introduction of the substrate plasmid the cells were lysed and analyzed for methylation of the substrate (Figure 3A). The competitor proteins consisted of SPK-, SPQ- or PPK-RCC1 N-termini fused to GFP. The test substrates consisted of SPK- and PPK-RCC1 N-termini fused to GFP followed by a FLAG tag (Figure 3A). 293LT cells were transfected with the competitors and 24 h later were re-transfected with the substrates. This yielded 6 different combinations of competitors and substrates. The competitor and substrate were expressed together for 8 h. The cells were then lysed, the FLAG-tagged substrates were immunoprecipitated, and immunoprecipitates were blotted for N-methylation and GFP. The results were consistent with the in vitro ITC results. In the presence of either an SPK or PPK competitor, methylation of the SPK substrate was decreased, as compared to the SPQ competitor control (Figure 3 B, C, D). The SPQ competitor is incapable of being methylated (Figure 3B) and does not bind to NRMT, thus it cannot compete with substrate for methylation and is used as a reference control of endogenous NRMT activity.
Figure 3. In vivo substrate affinity assay.
A) Schematic of the in vivo substrate affinity assay
B) Competitor substrate set of SPK-, SPQ- and PPK-GFP is efficiently expressed and N-terminally methylated after 32h of expression following calcium phosphate transfection.
C) The efficiency of N-terminal methylation of immunoprecipitated SPK- and PPK-GFP-FLAG test substrates is variable and depends on the presence of co-transfected competitor substrate.
D) Quantification of the efficiency of N-terminal methylation of SPK- and PPK-GFP-FLAG test substrates in the presence of SPK-, SPQ- or PPK-GFP competitor. Data were normalized and compared to the level of N-terminal methylation of test substrates co-expressed in the presence of unmethylatable SPQ-GFP competitor substrate by two-tailed independent t tests. * indicates P<0.01. n=3 independent repetitions per combination. Error bars: +/− 1 s.d.
When the PPK substrate was co-expressed with the SPK competitor the level of methylation of PPK substrate was comparable to the SPQ competitor control, again consistent with the ITC data showing PPK binds NRMT with a higher affinity than SPK. However, in the presence of the PPK competitor, PPK substrate methylation was substantially reduced (Figure 3 C, D). Together, these data suggest that different substrates and products of NRMT can compete with one another for access to the enzyme, which could result in differential methylation of low affinity versus high affinity substrates.
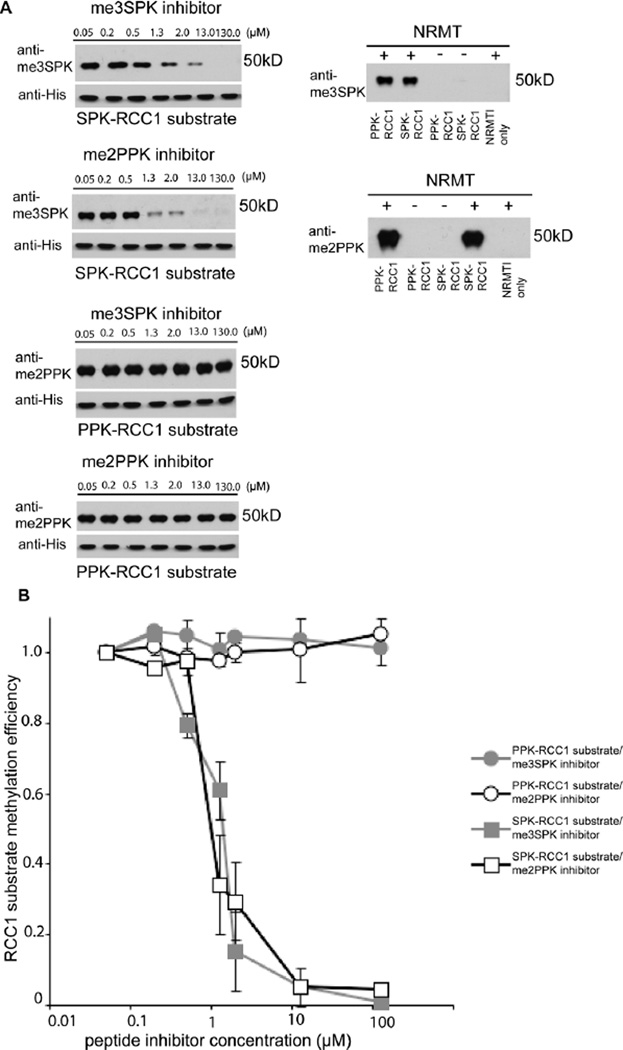
NRMT is inhibited in vitro by me3SPK and by me2PPK product peptides
The experiment described above cannot distinguish whether the methylated or unmethylated competitor was most effectively outcompeting the substrate proteins. Therefore, to test whether the methylated products feedback inhibit NRMT, we monitored the efficiency of N-terminal methylation of SPK- and PPK-RCC1 proteins in vitro in the presence of increasing concentrations of me2PPK or me3SPK product peptides. For the SPK-RCC1 substrate, both me3SPK and me2PPK peptides achieved efficient inhibition at 1.3 µM, which is consistent with the ITC data (Figure 4 A, B). However, as predicted by the ITC studies, inhibition of PPK-RCC1 was not observed (Figure 4 A, B). Together, these data support the concept that methylated NRMT target proteins can efficiently outcompete some types of substrate, thereby limiting their N-methylation. This effect might reduce the number of N-methylated protein in the cell, or enable them to be acetylated instead.
Figure 4.
A) NRMT is inhibited in vitro by me3SPK and by me2PPK product peptides with variable efficiency dependent on the affinity of the enzyme for the substrate.
B) Quantification of inhibition by me3SPK and me2PPK product peptides. Data were normalized and compared to the level of N-terminal methylation of SPK-RCC1 or PPKRCC1 substrates methylated by recombinant NRMT in the presence of 0.05µM me3SPK or me2PPK product peptide by two-tailed independent t tests. P-value is <0.01, n=2 independent repetitions per reaction. Error bars: +/− range.
N-methylation is restricted to proteins with access to the nuclear compartment
A second mechanism that can potentially limit the number of substrates that are N-methylated by NRMT is location. The enzyme is predominantly nuclear, which suggests that cytoplasmic proteins will not be efficiently methylated except during mitosis. Cytoplasmic targets might therefore either be methylated in a cell cycle-dependent manner or not methylated at all. To determine whether substrate location does determine methylation efficiency, we expressed in unsynchronized 293LT cells a triple-GFP fused with the first 10 residues of human RCC1, followed by either a nuclear localization signal (NLS) or a nuclear export signal sequence (NES). After 24 h, only fusion proteins with an NLS sequence were efficiently methylated (Figure 2E). During this period all of the cells are expected to have cycled through at least one mitosis. Therefore, access to the nuclear compartment is essential for N-methylation, and M phase is not sufficient to enable α-N- methylation of cytoplasmically-restricted substrates.
DISCUSSION
The majority of enzymes exhibit low affinities for their products, which promotes efficient catalysis. Strong binding of product peptides and fully methylated proteins (like ZFP15, VRK2 and RCC1) to NRMT is, therefore, a surprising result. The molecular basis for this high affinity is not immediately apparent. However, we noticed that the active site of the NRMT crystal structure contains a conserved triad of aromatic residues (Y19, W20, H141), the spatial arrangement of which is similar to that found in chromo domains, which bind to methylated lysine residues (Figure S2A). Binding specificity results from a cation-π interaction between the tertiary amine group and the aromatic triad 21–23. In NRMT the stoichiometric methylation of the N-terminus puts a stable positive charge on the amino group, making the interaction with the side-chain rings of the aromatic triad very favorable. A product inhibition assay using peptide from histone H1 which contains a tri-methylated lysine residue (K26) (Figure S2D, E) failed to inhibit N-terminal methylation of SPK- or PPK-RCC1. This result suggests that me3 lysine is not recognized by the NRMT aromatic cage.
The high affinity of methylated peptides for NRMT suggests the possibility for feedback inhibition of NRMT by its own products. Indeed, fully methylated product peptides in vitro inhibit N-terminal methylation of SPK-RCC1. This inhibition is achieved at concentrations of inhibitory product peptides consistent with the Kd values obtained from ITC experiments. We have shown that fully methylated FLAG-tagged substrates can bind endogenous NRMT, consistent with the possibility of a product feedback inhibition by methylated substrates as one of the regulatory mechanisms of NRMT. Future experiments will address this possibility.
Diverse substrate and product affinities towards NRMT might be another means by which it is regulation. PPK consensus proteins are much better substrates than others as shown by ITC, in vitro and in vivo methylation assays, and the peptide library screen. PPK peptides are also the only ones that achieve 100% methylation. Additionally, methylation of PPK-RCC1 cannot be inhibited in vitro by fully methylated products. These differences in efficiency of methylation imply some substrate preference of NRMT in vivo.
One surprising result indicates interplay between N-terminal methylation and N-terminal acetylation in different tissues. When we immunoprecipitated MLC12B, MLC B-like, and MLC9 from mouse spleen extract, we found them to be N-terminally methylated. However, when we over-expressed one of the isoforms (MLC9) in 293LT cells, it was not N-terminally methylated but instead was 100% N-terminally acetylated (Figure S3C). When FLAG-tagged on the C-terminus and over-expressed in 293LT cells, N-terminally acetylated MLC9 also cannot IP endogenous NRMT (Figure S3F). This is in agreement with the MS data, but different from the other tested NRMT substrates (Figure 2 A, B). These results suggest that N-terminal methylation is a tissue-specific post-translational modification and can be interchangeable with N-terminal acetylation. Apart from MLC9, which is a novel protein to be N-terminally acetylated, there are other proteins, which could be potentially modified in these two different ways. PMI40 (SNK-N-terminus), MRP23 and MYP2 (SQK-N-terminus), TSNAX protein (SNK-N-terminus), ENSA protein and GBGC (SQK-N-terminus), TTC37 (SSK-N-terminus), protein tyrosine phosphatase PTPCAAX2, PWP1 (MNR-N-terminus), and RP42 (MNK-N-terminus) have all been reported to be N-terminally acetylated in yeast and mammals respectively24,25. In our in vitro peptide library study, peptides with these N-terminal sequences were partially methylated by NRMT. Future studies will address the importance of possible interplay between various N-terminal post-translational modifications.
Subcellular localization is another means by which the number of NRMT targets could be regulated. We have previously shown endogenous NRMT localizes in the nucleus6, and that all N-terminal methylation in the asynchronous 293LT cell culture is achieved in the nucleus. Compartmentalization of N-terminal methyltransferases, and/or exclusion of substrate proteins from the nucleus would, therefore, constrain the number of proteins in the cell which will be efficiently methylated and allow for certain substrates to be acquire competing N-terminal post translational modifications. Abnormal localization of NRMT could then lead to deleterious effects, as it would have access to previously sequestered targets and perhaps alter their normal regulation.
Supplementary Material
Acknowledgements
J.J.P. would like to thank Timothy Errington for valuable discussions and suggestions
Funding
This work was supported by research grants from the National Institutes of Health to I.G.M. (GM50526), D.F.H (GM 037537), C.S.T. (CA158009) and W.M. (GM053163)
Abbreviations
- NRMT
N-terminal RCC1 methyltransferase
- SAM
S-adenosyl methionine
- RCC1
regulator of chromosome condensation 1
- ITC
Isothermal titration calorimetry
Footnotes
Supporting Information Available:
Supplementary Materials and Methods; List of predicted NRMT substrates; Supplementary Table S1 and S2; Supplementary Figures S1, S2, S3. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- Tsunasawa S, Stewart JW, Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J Biol Chem. 1985;260(9):5382–5391. [PubMed] [Google Scholar]
- 2.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220(1):8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 3.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9(5):596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol. 2008;182(5):827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock AM, Stock JB. Purification and characterization of the CheZ protein of bacterial chemotaxis. J Bacteriol. 1987;169(7):3301–3311. doi: 10.1128/jb.169.7.3301-3311.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, Macara IG. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466(7310):1125–1128. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry GD, Winstanley MA, Dalgarno DC, Scott GM, Levine BA, Trayer IP. Characterization of the actin-binding site on the alkali light chain of myosin. Biochim Biophys Acta. 1985;830(3):233–243. doi: 10.1016/0167-4838(85)90279-1. [DOI] [PubMed] [Google Scholar]
- 8.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry. 2010;49(25):5225–5235. doi: 10.1021/bi100428x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol Cell Biol. 2001;21(10):3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76(13):3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 11.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72(18):4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 12.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 14.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1(6):2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gesquiere JC, Diesis E, Cung MT. Tartar, a, Slow Isomerization of Some Proline-Containing Peptides Inducing Peak Splitting during Reversed-Phase High-Performance Liquid-Chromatography. Journal of Chromatography. 1989;478(1):121–129. [Google Scholar]
- 17.Grafl R, Lang K, Wrba A, Schmid FX. Folding mechanism of porcine ribonuclease. J Mol Biol. 1986;191(2):281–293. doi: 10.1016/0022-2836(86)90265-2. [DOI] [PubMed] [Google Scholar]
- 18.Lang K, Schmid FX. Role of two proline-containing turns in the folding of porcine ribonuclease. J Mol Biol. 1990;212(1):185–196. doi: 10.1016/0022-2836(90)90314-C. [DOI] [PubMed] [Google Scholar]
- 19.Abagyan R, Totrov M, Kuznetsov D. Icm - a New Method for Protein Modeling and Design - Applications to Docking and Structure Prediction from the Distorted Native Conformation. Journal of Computational Chemistry. 1994;15(5):488–506. [Google Scholar]
- 20.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416(6876):103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295(5562):2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 23.Blus BJ, Wiggins K, Khorasanizadeh S. Epigenetic virtues of chromodomains. Crit Rev Biochem Mol Biol. 2011;46(6):507–526. doi: 10.3109/10409238.2011.619164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, Mohammed S, Heck AJ. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics. 2010;9(5):928–939. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325(4):595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.