Abstract
The excision of uracil bases from DNA is accomplished by the enzyme uracil DNA glycosylase (UNG). Recognition of uracil bases in free DNA is facilitated by uracil base pair dynamics, but it is not known whether this same mechanistic feature is relevant for detection and excision of uracil residues embedded in nucleosomes. Here we investigate this question using nucleosome core particles (NCPs) generated from X. laevis histones and the high-affinity “Widom 601” positioning sequence. The reactivity of uracil residues in NCPs under steady-state multiple turnover conditions was generally decreased as compared to free 601 DNA, mostly due to anticipated steric effects of histones. However, some sites in NCPs had equal or even greater reactivity than free DNA, and the observed reactivities were not readily explained by simple steric considerations, or by global DNA unwrapping models for nucleosome invasion. In particular, some reactive uracils were found in occluded positions, while some unreactive uracils were found in exposed positions. One feature of many exposed reactive sites is a wide DNA minor groove, which allows penetration of a key active site loop of the enzyme. In single-turnover kinetic measurements, multi-phasic reaction kinetics were observed for several uracil sites, where each kinetic transient was independent of the UNG concentration. These kinetic measurements, and supporting structural analyses, support a mechanism where some uracils are transiently exposed to UNG by local, rate-limiting nucleosome conformational dynamics, followed by rapid trapping of the exposed state by the enzyme. We present structural models and plausible reaction mechanisms for the reaction of UNG at three distinct uracil sites in the NCP.
The recognition and repair of damaged DNA bases is largely the task of the base excision repair pathway. This pathway is initiated by a variety of DNA glycosylases, each with a different specificity for DNA damage. A common mechanistic problem encountered by these enzymes is the structural obstacle imposed by duplex DNA, which obscures the damaged base within the DNA duplex. Thus by necessity, these diverse glycosylases have evolved a common strategy to extrude damaged bases from the confines of the DNA duplex and then dock the base in their active sites for catalysis to ensue.1 This process of “base flipping” requires substantial binding interactions with the DNA backbone, ultimately resulting in substantial DNA bending. An intriguing mechanistic question is how do these enzymes operate when a damaged base is embedded in a large protein complex such as a nucleosome, rather than in free duplex DNA?
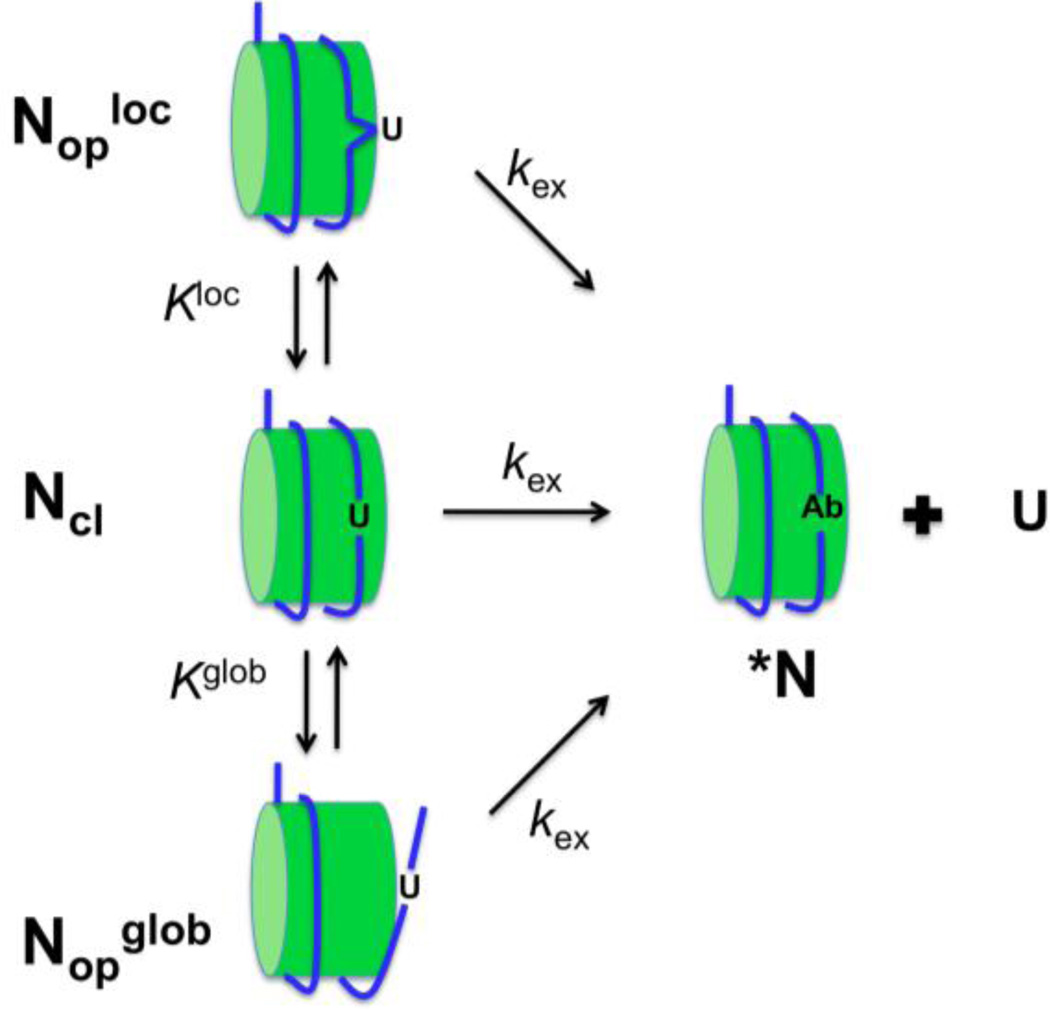
The enzyme uracil DNA glycosylase (UNG) is the most catalytically robust of DNA glycosylases2and shows a remarkable plasticity to locate and excise uracils in duplex or single stranded DNA contexts, and remarkably, mononucleosomes3, 4, 5, 6 The enzyme utilizes the favorable opening dynamics of uracil base pairs in free DNA to initiate the process of base flipping7, 8, suggesting that nucleosome-induced changes in individual base pair dynamics could have a profound effect on the activity of UNG. In this regard, several distinct models can be envisioned to explain the reaction of uracil bases embedded in a nucleosome core particle (NCP) (Figure 1). The simplest model involves direct excision of a uracil without a prerequisite conformational transition in the NCP that exposes the site (kex for Ncl, Fig. 1). Such a mechanism might hold for sites that are exposed on the surface of the NCP, but steric conflicts between the enzyme and the NCP, or reduced base pair dynamics arising from structural aspects of the site could still hinder the reaction as compared to free DNA. Another model, that has already been established for access of restriction enzymes to their sites,9, 10 is complete unwrapping of the DNA from the histone octamers (Kglobal, Fig. 1). With unwrapping as the primary means of UDG accessing nucleosomal DNA, one would expect DNA near the entry/exit sites to react more quickly than internal sites located near the nucleosome dyad axis.11, 12 Indeed, the probability for unwrapping internal sites may be 10,000-fold smaller than end sites13, leading to extremely low rates for these sites. A third route for UDG action could involve local NCP dynamics, rather than global unwrapping (Klocal, Fig. 1). The timescale for such dynamics would be determined by local structural features of the site, which could be quite varied for DNA bound to histones. A unique feature of local dynamics is that rapid exposure of sites could occur for sites distant from the DNA ends.
Figure 1.
Three possible models for uracil excision in the context of a NCP. The NCP is viewed as existing in a closed form where the DNA is wrapped tightly around the nucleosome (Ncl), and two open forms that differ with respect to whether conformational transitions involving global unwrapping (Nopglob) or local release (Noploc) of the DNA have occurred. Model 1 (middle pathway, Ncl). A uracil site is exposed even in the context of the closed NCP, and the uracil is excised as rapidly as in free DNA (kex). Model 2 (bottom pathway, Nopglob). The site is inaccessible to UNG and the lowest energy pathway to exposing the site is global unwrapping of the DNA from the NCP. This may be described by an unfavorable equilibrium (Kglob). Once exposed, the site may react with the same excision rate as the free DNA. Model 3 (top pathway, Noploc). The site is inaccessible to UNG and the lowest energy pathway to exposing the site is local release of the DNA from the NCP. This may be described by an unfavorable equilibrium (Kloc). Once exposed, the site may react with the same excision rate as the free DNA.
In this report, we have assembled NCPs containing X. laevis histones and the 147 bp high-ffinity “Widom 601” positioning sequence. Our constructs are identical to a recently published X-ray crystal structure (except for single T/A to U/A substitutions at specific locations),14 and therefore allow direct interpretation of our kinetic and dynamic measurements using structural parameters. We find that although simple steric considerations and burial of uracils can affect their reactivity with UNG, some uracil sites are reactive even when the crystal structure would indicate a lack of accessibility, and many exposed sites are unreactive. We now propose mechanistic explanations for the reactivities of individual uracil sites in NCPs based on the DNA and histone structural features obtained from the crystal model, as well as small molecule structural probes (KMnO4 and hydroxyl radical) and single-turnover kinetic experiments. In addition, molecular docking of UNG to a highly reactive site in the NCP provides a detailed structural basis for the mechanism of uracil recognition.
Materials and Methods
DNA Sequences and Nomenclature
The NCPs were assembled using a minor variant of the 147 bp Widom SELEX 601 DNA sequence employed by Makde et al in their crystallographic work14, 15:
601-147b (strand one): 5'-ATCGGATGTATATATCTGACACGTGCCTGGAGACTAGGGAGTAATCCCCTTGGCGGTTAAAACGCGGGGGACAGCGCGTACGTGCGTTTAAGCGGTGCTAGAGCTGTCTACGACCAATTGAGCGGCCTCGGCACCGGGATTCTCGAT-3'
601-147b (strand two): 5'-ATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGAT-3'
The two complementary strands of the Widom 601 sequence (strand one and strand two, given above) are called N1 and N2, respectively, and for the thymine of interest replaced with uracil, the number of nucleotides from the dyad is indicated with a superscript. Following the convention of Richmond and coworkers,16 the superscript position is either positive or negative, depending on whether the nucleotide of interest is 3′ or 5′ relative to the nucleosome dyad, respectively. Strands one and two of the Widom 601 sequence correspond to chains j and i, respectively, in the crystal structure of the 601 nucleosome reported by Tan and coworkers (PDB 3MVD)14. In that structure, the dyad is located at nucleotide 74 on each strand. Therefore, the numbering convention used here can be converted to the nucleotide numbering of the crystal structure by addition of +74. For example, U+22N1 corresponds to nucleotide 96 on chain j.
Preparation of 601 DNA Containing Randomly Incorporated Uracil Residues
147-nucleotide nucleosomal DNA strands were ordered from Integrated DNA Technologies (IDT) and were purified by PAGE using a 6 % sequencing gel. Sequenase 2.0 (USB/Affymetrix) was used to randomly incorporate uracil into each strand of DNA at less than 1 uracil/molecule. Polymerase reactions were carried out by using either strand one or strand two as a template (400 nM) and 400 nM of a 5’-Fam labeled 25mer primer complementary to the first 25 bases at the 3’ end of each template. After heating to 95 °C for 5 min, strands were annealed by slow cooling in a buffer consisting of 20 mM Tris-HCl pH 7.5, 10 mM MgCl2 and 25 mM NaCl. Polymerization reactions contained 10 µM DTT, 300 µM of each standard dNTPs, 30 µM dUTP, and 1 unit Sequenase per nmole duplex. Sequencing ladders were constructed using the same conditions except that dUTP was omitted and 30 µM ddATP or ddTTP was added. The reaction was carried out at 37 °C for 45 minutes and the extended duplex was isolated using a PCR cleanup kit (Fermentas).
Formation of Nucleosomes
Recombinant Xenopus laevis histones were expressed individually in bacteria and the octamer was reconstituted as previously described17. NCPs were prepared by adding 400 nM duplex DNA in 2 M NaCl, 10 mM Tris pH 7.5, 1 mM DTT and 10 mM EDTA to an equal volume of ~440 nM reconstituted histone octamer in 2 M NaCl, 10 mM Tris pH 7.5, 1 mM DTT and 0.25 mM EDTA. The mixture was incubated on ice for 30 minutes and dialyzed into 10 mM Tris pH 7.5, 1 mM EDTA, 1 mM DTT and decreasing amounts of NaCl (1.2 M, 1.0 M and 0.6 M NaCl dialysis steps were performed for 1 hour each). The final step of dialysis into a zero NaCl solution was performed for 3 hours. Complete nucleosomal formation was confirmed using electrophoresis on a native 6% polyacrylamide (60:1 acrylamide:bisacrylamide) gel in 0.25 × TBE running buffer, run at 4 °C and 100V (Fig. S1). Only preparations showing a single nucleosome species and where > 95% of the input DNA was in the bound form were used for experiments.
Global Measurements of UNG Activity on Uracilated DNA
The catalytic domain of human uracil DNA glycosylase (UNG) was expressed in bacteria and purified as previously described18. Activity was measured on the randomly uracilated free DNA or in complex with histones under the following conditions: 50 nM nucleosomes (or free 601 DNA), 0.5 nM UNG, 20 mM Hepes pH 7.5, 10 mM NaCl, 1 mM DTT, 2.5 mM MgCl2, 0.002% Brij 35 and 0.1 mg/ml BSA. Reactions were performed at room temperature and were quenched at various times with 1% SDS. At this time a 5′-FAM labeled duplex DNA internal standard was added to each quenched reaction to allow correction for loading differences during gel electrophoresis. For the strand one and strand two templated experiments, 95 and 72 base pair standards were added, respectively. These standards were designed to migrate at positions in the gel where no uracil excision fragments would be found. The reaction mixtures were extracted with equal volumes of phenol:chloroform:isoamyl alcohol (PCA, 25:24:1, v/v). The aqueous phases were separated and treated with 1/10th volume of 1 M NaOH at 95 °C for 10 min. To the resulting solution were added 1/10th volume of 3M NaOAc pH 5.2, 2.5 µL 20 mg/mL glycogen and 3 volumes of cold ethanol. The samples were frozen in −80 °C for 1 hr and spun down at 15000 g for 15 min. The precipitates were dried, dissolved in 5 µL formamide DNA loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue) and loaded on a 6% denaturing polyacrylamide gel. The sequencing gel was run at constant 70 W until the bromophenol blue reached the bottom. The FAM-labeled DNA was detected on the gel with a Typhoon (GE Healthcare) imager with a 520 nm filter and quantified with ImageQuant software (GE Healthcare). After normalization to the internal standard on the gel, the fractional reaction of all uracil cleavage bands in the nucleosome and free DNA were calculated using the endpoint of the free DNA reactions as 100%. The resulting fractional reactions were plotted against time and initial rates were determined using linear regression analysis.
KMnO4 Footprinting
75 fmol freshly-prepared nucleosomes were dissolved in 25 µL buffer (10 mM Tis-HCl pH7.5, 1mM EDTA, 1mM DTT) to a final concentration of 3 nM. The resulting solution was treated with 2 µL of 100 mM KMnO4 (final concentration 8 mM) for 2 min at room temperature, and then quenched by 25 µL 1.5 M NaOAc (pH = 6.2) containing 7% BME. DNA was precipitated using 2 µL 20 mg/mL glycogen and 150 µL cold EtOH at 80°C. The supernatant was carefully removed and the pellet was dried. Subsequently, 50 µL of 1 M piperidine was added to dissolve the pellet and the solution was incubated at 90°C for 30 min, followed by ethanol precipitation as described above. The pellet was then washed with 75% cold ethanol, and then precipated again at 80°C as described above. The supernatant was carefully removed and the pellet was dried by gentle air flow. The pellet was dissolved in 7 µL formamide DNA loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue). A 3 µL portion was loaded on a 6% polyacrylamide gel containing 7M urea and electrophoresis was performed at 75 W with 1X TBE running buffer until bromophenol blue reached the bottom. The gel was imaged with a Typhoon 8600 (GE Healthcare) imager with a 520 nm filter and quantified with Image J (http://rsbweb.nih.gov/ij/).
Steady-State Kinetic Measurements
NCPs containing a single uracil at sites U+44, U+22, U−32 were dissolved in 20 µL of buffer containing 20 mM Tris pH 7.5, 3 mM EDTA, 1 mM DTT and 0.002% Brij 35, giving final NCP concentrations of 60 nM, 40 nM, 30 nM and 15 nM. hUNG was diluted to 0.6 nM in the same buffer and reactions were initiated by mixing equal volumes of UNG and NCPs. Reactions were quenched with a 2X volume of 0.5 M HCl at various reaction times. Specific reaction times were selected such that less than 30% of the substrate was reacted and each reaction was performed in triplicate for error estimates. Quenched reaction mixtures were extracted with PCA, and 1/10th volume of 10 M piperidine was added, followed by vortexing and centrifugation to separate the aqueous and organic phases. The aqueous layers were heated at 95°C for 20 min. The resulting solutions were dried in vacuo and re-suspended in 150 µL water followed by the addition of 1/10th volume of 3M NaOAc (pH 5.2), 5 µL glycogen and 3 vol cold EtOH. The samples were frozen at −80°C for >1 hr and centrifuged for 10 min at 15000g at 4°C. The precipitates were dried, dissolved in 5 µL 1X formamide load buffer and loaded on a 6% denaturing polyacrylamide gel. Electrophoreses was performed at 20 W with 1X TBE running buffer until bromophenol blue reached the bottom. The gel was imaged with a Typhoon 8600 (GE Healthcare) imager and quantified with Image J software. Initial velocities with standard errors were calculated by dividing the amount of product at the time of quench by the elapsed time. Observed steady-state rate constants (kobsss) were obtained by dividing the initial velocities by the UNG concentration. Because saturation was not achievable with NCP substrates, the second-order rate constant for the steady-state reaction (k´ss) was obtained from the linear slope of a plot of kobsss against substrate concentration using linear regression methods (see Supplemental Materials).
Single-turnover Kinetic Measurements
Measurements were performed by manual mixing or by using a Kintek RQ-3 chemical quench-flow instrument depending on the magnitude of the reaction rates. NCPs or free 601 DNA containing a single uracil at sites U+44, U+22 and U−32 were dissolved in a buffer containing 20 mM Tris (pH 7.5), 3 mM EDTA, 1 mM DTT and 0.002% Brij 35, giving a final concentrations of DNA or NCPs of 20 nM. UNG was diluted to final concentrations of 100 nM, 200 nM or 400 nM in the same buffer. Reactions were performed using pseudo-first order reaction conditions by mixing equal volumes of hUNG with NCPs (or free 601 DNA). Reactions were quenched with a 2X volume of 0.5 M HCl at various reaction times. The quenched reactions were extracted with 1:1:1 PCA, and 1/10th volume of 10 M piperidine was added, followed by vortexing and centrifugation to separate the aqueous and organic phases. The aqueous layers were heated at 95°C for 20 min. The resulting solutions were dried in vacuo and re-suspended in 150 µL water followed by the addition of 1/10th volume of 3M NaOAc (pH 5.2), 5 µL 20 mg/mL glycogen and 3 vol cold EtOH. The samples were frozen at −80°C for >1 hr and centrifuged for 10 min at 15000g at 4°C. The precipitates were dried, dissolved in 5 µL 1X formamide load buffer and loaded on a 6% denaturing polyacrylamide gel. Electrophoresis was performed at 20 W with 1X TBE running buffer until the bromophenol blue dye front reached the bottom of the gel. The gel was imaged with a Typhoon 8600 (GE Healthcare) imager and band volumes were quantified using Image J19. The fractional extents of reaction were plotted against reaction times and fitted to a single exponential expression, or the sum of two or three exponentials as required. For the DNA substrate U+22 and its NCP version U+22N1, the single-turnover observed rate constants (kobssto) were plotted against the concentration of UNG and fitted to eq 1, where kexsto is the maximal uracil excision rate, and K0.5 is the enzyme concentration that gives the half-maximal rate. The ratio kexsto/K0.5 = k′sto is the second-order rate constant for single-turnover uracil excision.
| (1) |
Molecular Docking
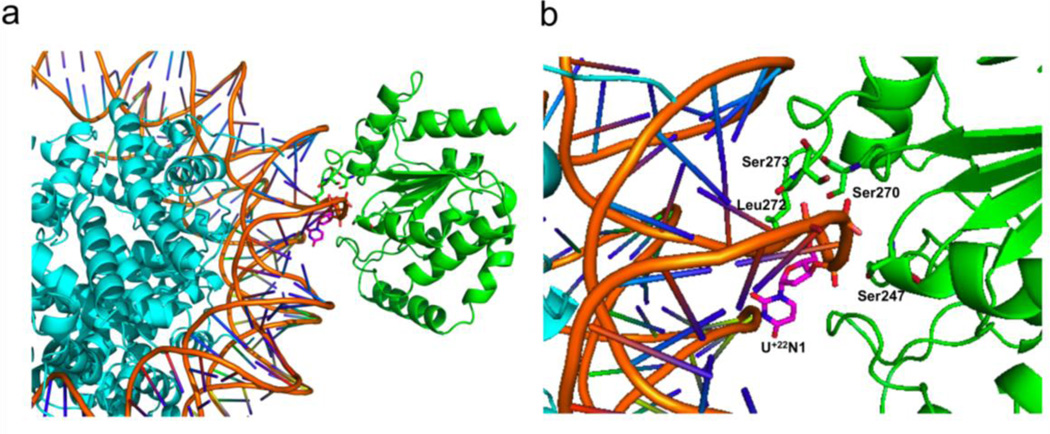
To create a model of enzyme-nucleosome binding at U+22N1, the high-resolution part of the RosettaDock protocol was used for all-atom optimization of both backbone rigid-body orientation and side-chain conformation20, 21. Using the visualization software PyMOL (http://www.pymol.org), the starting structure of docking partners was prepared by superimposing the DNA segment of the UNG-DNA complex (PDB 2OXM) onto the nucleosome DNA (PDB 3MVD). In the superposition, the extrahelical base B5 in the enzyme-DNA complex was aligned to match the position of the probed base U+22N1 in the nucleosome DNA. Based on the UNG-DNA binding mode, base U+22N1 must flip out of the DNA helix to allow the residue Leu272 in the UNG active site to enter into the minor groove. To satisfy this precondition, the backbone atom coordinates of nucleotides +21N1 to +25N1 in the nucleosome DNA (PDB 3MVD) were replaced by the B4–B8 backbone coordinates in the superimposed DNA segment (PDB 2OXM), without changing the original DNA sequence. Using PyRosetta22, the high-resolution docking simulations were then conducted with the structure of the superimposed enzyme and the modified nucleosome applying Rosetta’s DNA scoring function23. No artificial constraint potentials were employed to bias the binding. The simulation generated 20 candidate models. The model with lowest interface energy was selected and shown in Figure 7.
Figure 7.
Docking based model for recognition of uracil at the highly reactive site U+22N1. (a) View of docking complex (b) Expanded view of the docking complex. U+22N1, Ser247, Ser247, Ser273 and Leu272 are labeled and shown in sticks.
Results and Discussion
Three substantial advantages of studying the high affinity 601 DNA sequence as opposed to other lower affinity histone-DNA binding sequences are (i) the DNA is held in a single register in the NCP, diminishing the contribution of alternative binding modes, (ii) a high resolution crystal structure has recently been solved of the 601 DNA in complex with Xenopus laevis histones (PDB 3MVD),14 and (iii) restriction enzyme reactivities have been mapped along the sequence, providing a valuable point of comparison12. We utilize these favorable attributes in the analyses of the UNG reactivity data.
Global Assessment of UNG Activity at U/A Pairs in Free and Nucleosome Bound 601 DNA
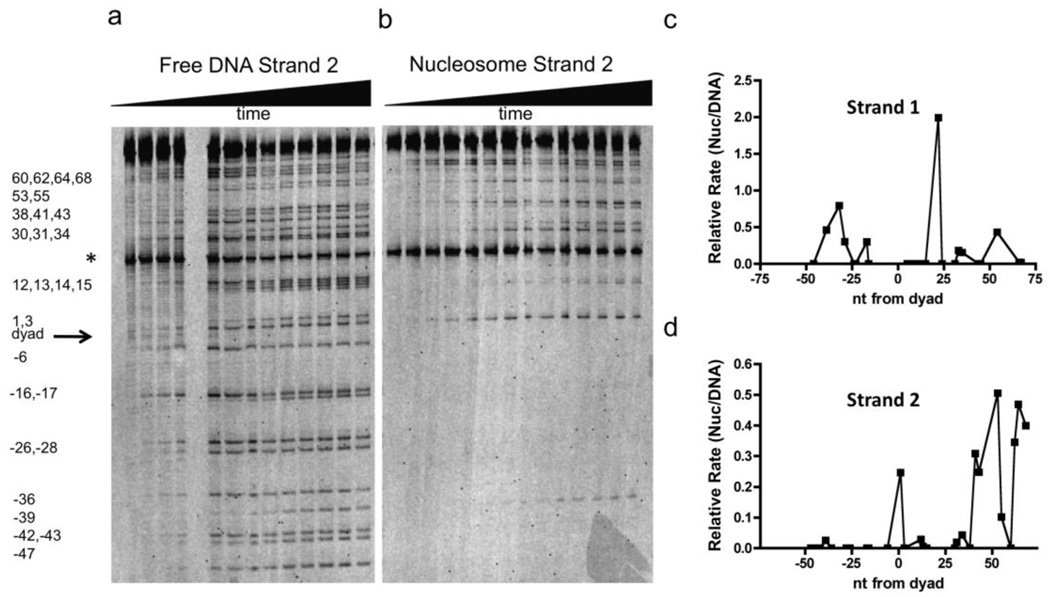
Using an appropriate dUTP/TTP ratio, 601 DNA strands containing on average one U/A pair per strand were synthesized by DNA polymerase primer extension, thereby generating a population of DNA molecules where each thymine was substituted with uracil. Such constructs allow the global assessment of the steady-state initial rates for hUNG reaction at each uracil site along the DNA for both free and nucleosome-bound DNA (Fig. 2a, b and Fig. S2). Due to the modest flanking sequence preferences of hUNG for free 601 DNA (Fig. 2a), different U/A base pairs showed reactivities that differed by as much as 5-fold (Fig. S3a)24. Preferred cleavage sequences were AUA, TUA and CUA, and less reactive sequences were TUC, AUT and GUC, which is similar to the flanking base trends previously observed with Escherichia coli UNG25. In contrast to free DNA, the cleavage rates in the context of the NCP vary by over two-orders of magnitude (Fig. 2b and S3b), with no obvious sequence trends, suggesting that DNA sequence plays a minor role, and that other determinants dominate.
Figure 2.
Reaction of UNG with free and histone-bound 147 bp 601 DNA containing global T→U substitutions. (a) Denaturing polyacrylamide gel separation of the DNA fragments resulting from base-catalyzed cleavage of the abasic sites generated from UNG excision of uracils. The DNA strand contained a 5′Fam-label and was reacted with hUNG for 0 s, 5 s, 10 s, 20 s, 30 s, 1 min, 2.5 min, 5 min, 10 min, 15 min, 30 min, 60 min and 95 min (lanes 1–13, left to right). The asterisk (*) indicates the migration position of a 5′ Fam-labeled DNA internal standard that was added post-reaction to normalize for lane loading differences (see Methods). (b) Same analysis as (a) except the DNA is in the context of the NCP. Reaction times are 0 s, 10 s, 32 s, 1 min, 2 min, 5 min, 10.9 min, 20 min, 30 min, 45 min, 60 min, 90 min, 120 min and 190 min (lanes 1–14, left to right). (c), (d) Relative activity of UNG for uracil sites in strand one and two of free and histone bound 601 DNA. The relative activity is defined as the initial rate of a uracil site in the NCP divided by that of the same site measured in the free DNA.
Unlike restriction enzymes, which show a ~10−4 decrease in the efficiency of cleaving their sites as the site is moved from near the DNA entry/exit sites towards the NCP dyad axis12, the distribution of UNG reactive and unreactive uracil sites across the entire 601 sequence are not clearly related to position relative to the DNA dyad (Fig. 2c, 2d). Thirty-six of the total fifty-two NCP uracil sites showed zero to 10% of the activity compared to the same site in free DNA, fourteen showed 10 to 60% of the activity, and two sites showed roughly equal, or even two-fold greater activity than free 601 DNA (sites U−32N1 and U+22N1, respectively) (Fig. S4). These reactivity trends, at the very least, indicate that additional mechanisms above and beyond DNA unwrapping must be operative. Thus, local dynamic events or the unique conformations of individual sites must also be important in determining UNG reactivity.
Correlations Between Uracil Site Reactivity and Small Molecule Probes of Duplex Structure
It is reasonable to expect that UNG-reactive uracil sites might be correlated with their rotational exposure (or lack of steric hindrance) in the crystal structure of the 601 NCP (PDB 3MVD). The simplest expectation would be that rotationally exposed uracils (i.e. oriented away from the histone surface as observed in the structure PDB 3MVD), would be the most reactive, and buried uracils (oriented against the histone surface) would be the least reactive, as has been previously suggested5, 6. However, it is also possible that otherwise rotationally exposed residues may also be hindered to the action of UNG due to changes in duplex structural properties or interfering histone side chains.
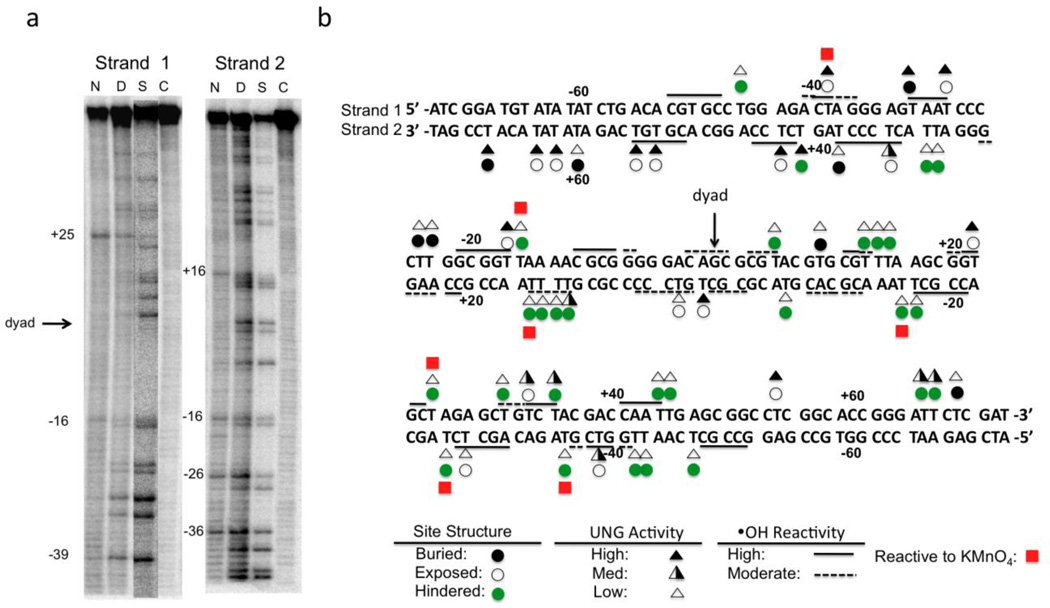
We looked for possible correlations of the site reactivities with the sensitivity of the 601 DNA sequence to both potassium permanganate, which probes accessibility of the 5, 6 double bond of thymidine residues (a surrogate for uracil in the uracilated 601 substrate, Fig. 3a), and hydroxyl radicals, which probe the width of the DNA minor groove26, 27. Only seven thymidine sites on both DNA strands in the context of NCPs were found to be especially permanganate sensitive (red squares, Fig. 3a, 3b). These sites were also reactive in the free 601 DNA, suggesting that the DNA sequence context of these thymidines plays a substantial role in determining their sensitivity to permanganate in both the free DNA and in the NCP. Importantly, all seven of the permanganate sensitive sites were rotationally exposed in the structure, but only one (U−39N1) was reactive to UNG. Comparing UNG reactivity results with previously reported NCP hydroxyl radical sensitivity data28, we found that only 15/26 sensitive sites were also UNG reactive (Fig. 3b, solid and dashed lines). Thus we find no strong relationship between the reactivity of UNG and small molecule structural probes, even though the reactivity of the small molecules corresponds well with rotational exposure of the site. We conclude that the reactivity of UNG must have higher-order requirements and that these requirements may involve duplex structural properties, the presence of interfering histone side chains, or the flexibility of the site in addition to its exposure (see below).
Figure 3.
Comparison of UNG reactivity with structural properties of the uracil sites in the NCP. (a) KMnO4 footprinting of each strand of the 147mer DNA in the context of the NCP (see Methods). Abbreviations: N, NCP; D, free DNA; S, single strand T ladder; C, control (free DNA, no KMnO4). (b) Comparison of the reactivity of hUNG with site exposure in the crystal structure, and KMnO4 and hydroxyl radical footprinting data28. Key legend: Triangles represent the ratio (R) between the initial rates for uracil excision in the nucleosome site as compared to the same site in free DNA, with filled, half-filled and unfilled triangles corresponding to R > 0.2, 0.2 > R > 0.02, and R < 0.02, respectively. High and moderate sensitivities to hydroxyl radical are indicated by solid and dashed lines. Red squares indicate high sensitivity to KMnO4 oxidation. Open, black and green circles represent rotationally exposed, buried, and rotationally exposed (but otherwise sterically hindered to UNG) uracil sites, respectively.
Correlations Between Uracil Reactivity in the NCP and Site Structure
The expectation that rotational exposure of a uracil site may correlate with its reactivity is largely substantiated because 14/20 of the rotationally exposed uracils are moderately or highly reactive, while only 2/28 of the buried (or sterically hindered) uracils show similar reactivity (Fig. 3). However, we also found that nine uracil sites on strand one (U+13,+14,+15N1, U+25N1, U+31N1, U+44,+45N1, and U+66,+67N1), and five on strand two (U−6N2, U−16, −17N2, U−26N2, and U−47N2) were unreactive even though they had a rotational position on the nucleosome that appears to expose the site. Closer inspection of these rotationally exposed sites reveals structural features that would likely lead to poor reactivity. The specific features of these sites include one or more of the following: (i) a very narrow minor groove that would impede interactions with an important active site loop of hUNG (see further Results below), (ii) direct hydrogen bonds to the uracil base or phosphate atoms from histone side-chains (as inferred from the thymidines in the structure), and (iii) the presence of one or more histone side-chains positioned in the minor groove. In addition, site exposure alone fails to explain the absence of reactivity of six highly exposed uracil sites on strand two where no obvious structural impediments exist (U+3N2, U+12,+13N2, U+34N2, U−28N2, U−39N2), nor does it explain the reactivity of two buried sites (U−32N1 and U+68N2). We conclude that more complicated mechanisms–possibly involving NCP dynamics–must also contribute to the reactivity of these individual sites.
NCP DNA Structural Parameters and Site Reactivity
We used the DNA structural analysis program Curves and the input file PDB 3MVD to investigate whether perturbed 601 base pair structural parameters correlate with the observed uracil reactivities (Fig. S5)29. The first question we explored was whether the subset of exposed sites that were highly reactive had perturbed base pair parameters that might destabilize base stacking and hydrogen bonding interactions at these sites, thereby leading to their enhanced reactivity. For this analysis, a reactive uracil site was defined as an exposed site with >20% of the reactivity of the same site in free DNA. The most highly reactive site in strand one (U+22N1), showed large perturbations in twist, roll, buckle, tip and inclination that were amongst the largest observed across the entire NCP (Fig. S5). Thus, these perturbations may increase the dynamic flexibility of this site and lead to its 2-fold greater reaction rate as compared to the same site in free DNA. The two other exposed sites in strand one (U+54N1 and U−39N1) were ~five-times less reactive than U+22N1 and displayed normal B-DNA values for the duplex parameters, except for tip. For the five reactive and exposed sites in strand two (U+1N2, U+53N2, U+55N2, U+62N2 and U+64N2), only one site showed substantial deviations in several duplex parameters (U+1N2), and the other sites were not remarkable in this regard. Moreover, we also observed that a number of unreactive exposed sites had perturbed base pair parameters (U+3N2, U+34N2, U−28N2, U−39N2)(Fig. S5). We conclude from the above analyses that extreme base pair structural parameters can make a given exposed site hyperreactive (i.e. U+22N1), but that these considerations alone are in general poorly predictive of the reactivity of exposed sites. One possible explanation for the poor reactivity of some exposed sites is that strong interactions between the histones and DNA at these sites hinders the conformational flexibility required to extrude a uracil base from the duplex. Such energetic effects may arise from interactions at a distance from the site, and are therefore impossible to discern from inspection of the crystal structure.
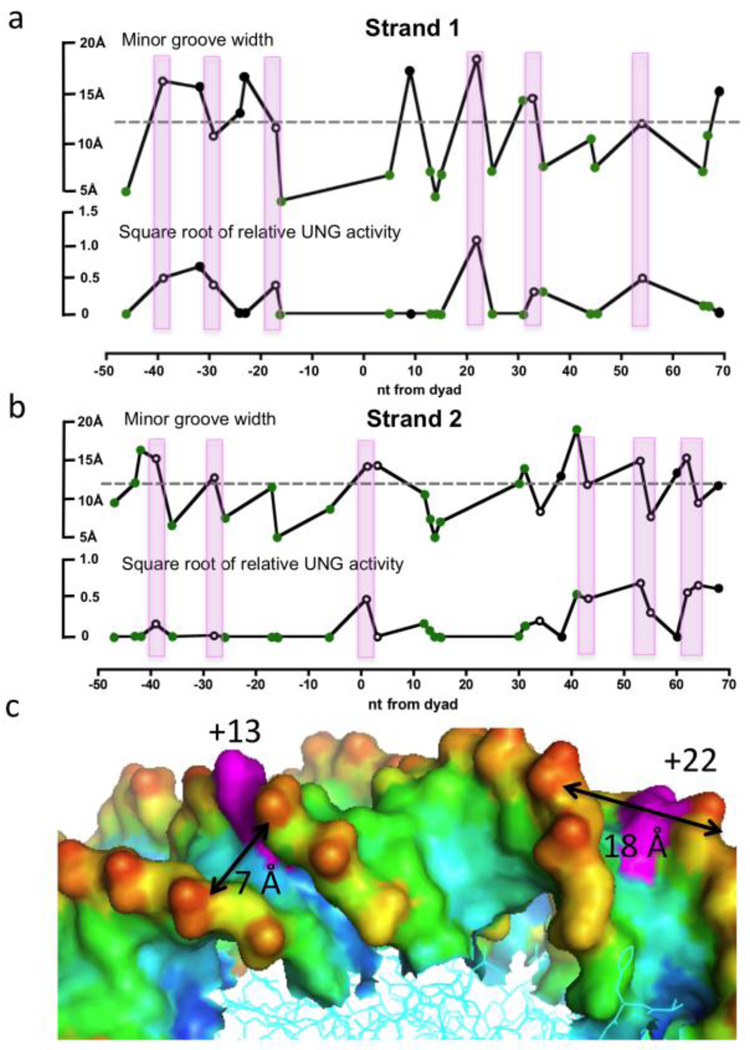
One additional feature of several exposed and reactive uracil sites was an exceptionally wide minor groove, and conversely, exposed sites with narrow minor grooves were often unreactive (Fig. 4a, 4b). This is best illustrated in the structural comparison shown in Figure 4c, where the dramatic differences in the minor groove widths for the highly reactive U+22N1 (~18 Å) and the unreactive U+13N1 (~7 Å) sites can be discerned. In terms of the UNG reaction mechanism, a narrow minor groove would place a severe steric impediment to the insertion of the active site loop containing the “wedge” residue Leu27218, 30, 31, 32, 33. Insertion of this conserved side chain serves to prevent the retrograde motion of the uracil base back into the base stack during the early base flipping step of the reaction33, 34. It is notable that the most reactive +22 site is located in a region that has been previously suggested to be dynamic. Structurally, this region has been shown to have the capability of “absorbing” one extra base pair, or alternatively, “stretching” if one base pair is deleted14. In addition, HIV and intercalating small molecules preferentially insert into this region, which has been attributed to the wide minor groove35, 36. We surmise that the unusual flexibility or dynamics of this region could very well explain the reactivity of this region to hUNG and to a variety of modifications.
Figure 4.
Correlations between DNA minor groove width in the NCP and the relative activity of UNG. (a) Strand one. (b) Strand two. The minor groove widths were obtained by analysis of the X-ray structure using the program Curves. The relative UNG activities (vNCP/vfree DNA) are based on our experimental data and are displayed as (relative activity)−1/2 to facilitate presentation of small and large values. In these panels, rotationally exposed and buried residues are indicated by open and closed circles, respectively. The green filled circles represent residues that are rotationally exposed but show other structural features that might inhibit reactivity (histone side chain interactions or narrow minor grooves). The vertical colored bars indicate those sites that have the following properties: rotational exposure, accessibility to UNG and average or wider minor groove widths. (c) Structural features of two rotationally exposed residues (magenta) corresponding to U+22N1 (reactive) and U+13N1 (unreactive), which are located in sites with dramatically different minor groove widths. The other colors in this panel reflect atom depth as calculated using the program SADIC using a probe radius corresponding to a water molecule (1.4 Å)46. Red and blue colors represent the most solvent exposed and buried atoms, respectively.
Steady-state and Single-turnover Kinetic Reactivities of Individual Uracil Sites Reveals NCP Conformational Transitions
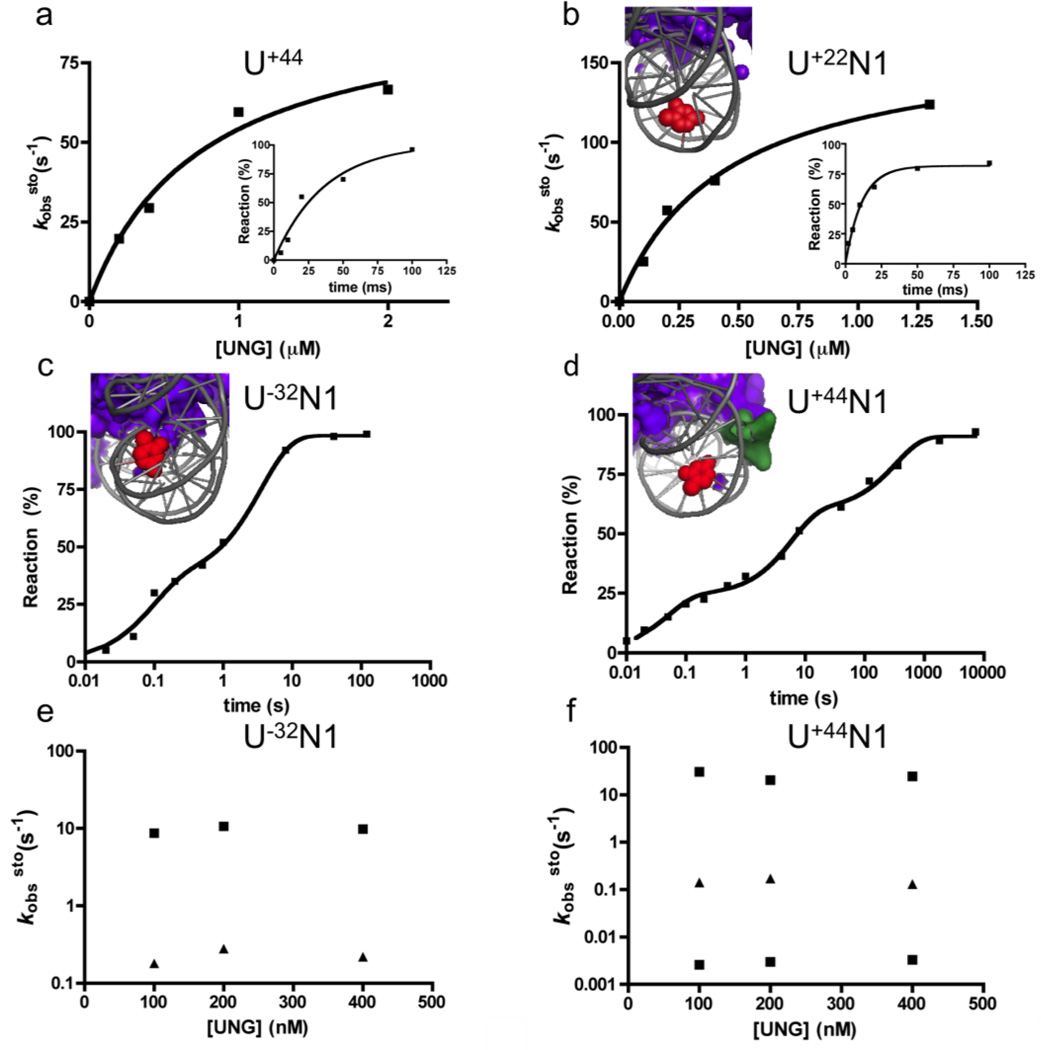
To further understand the reactivities of uracils embedded in distinct positions of the nucleosome we turned our attention to three distinct sites with intriguing properties. These sites were investigated individually using both steady-state and single-turnover kinetic methods (Table 1). Site U+22N1 is exposed and reacts at a diffusion controlled rate of k′ss = 4 × 108 M−1 s−1 in steady-state measurements of second-order reactions rates (Fig. S6). Site U−32N1 is buried yet still reacts with a substantial rate of k′ss = 0.5 × 108 M−1 s−1 (~40% of the average uracil reactivity in free DNA). Finally, site U+44N1 is rotationally exposed, but reacts almost 50 times slower than the exposed site U+22N1 in steady-state reactions (k′ss = 8 × 106 M−1 s−1). The poor reactivity of U+44N1 is likely due to interactions of the flexible N-terminal tail of histone H2A with the minor groove at this site (Ala12, Lys13, Ala14, Lys15), which is discussed further below.
Table1.
Steady-state and single turnover kinetic parameters for excision of uracil sites in free 601 DNA and in NCPsa
|
k′ss (M−1s−1) × 108 |
kexsto (s−1) |
K0.5 (µM) |
k′stob (M−1s−1) × 108 |
|
|---|---|---|---|---|
| U+44 | 1.33 ± 0.03 | 96 ± 13 | 0.76 ± 0.24d | 1.3 ± 0.7d |
| U−32N1 | 0.51 ± 0.01 | 10.6 ± 0.2 (36%)c | ||
| 0.29 ± 0.01 (64%) | ||||
| U+22N1 | 3.9 ± 0.1 | 166 ± 12 (100%) | 0.45 ± 0.08d | 3.7 ± 0.7d |
| U+44N1 | 0.08 ± 0.01 | 24.0 ± 0.1 (23%) | ||
| 0.17 ± 0.01 (41%) | ||||
| 0.003 ± 0.001 (36%) |
Errors for the steady-state kinetic measurements were obtained from three replicate experiments performed on two independently prepared nucleosome preparations (two experiments were performed on the first preparation and the third on the second). The errors shown for the single-turnover kinetic measurements are the standard deviations of the data from the fitted curve for reactions containing 20 nM NCP and 200 nM UNG. Each time point from the rapid-quench experiment was obtained in duplicate and averaged for plotting. The experiment for U−32N1 and U+44N1 were repeated at three UNG concentrations (100, 200 and 400 nM) with identical results (Fig. 5e, f). These single-turnover data were also collected using two independently prepared nucleosome preparations (the experiments with 100 and 200 nM UNG were performed on one preparation and the experiment with 400 nM UNG was performed on the second preparation).
Obtained from the ratio kexsto/K0.5.
Relative amplitude of each kinetic phase.
Each kinetic phase was independent of UNG concentration (see Fig. 5e, f).
To further understand the distinct reactivity of these three sites, single-turnover kinetic measurements were performed (Fig. 5, Fig. S7). For comparison, we first measured the single-turnover uracil excision rate for a site in free 601 DNA. Such measurements have been previously made for both E. coli and human UNG37, 38, 39, 40, but these measurements were repeated here by placing a single U/A base pair at site U+44 in the free 601 DNA (Fig. 5a). Similar to previous kinetic findings, the maximal rate of uracil excision at site U+44 in free 601 DNA was kexsto = 96 s−1 (22 °C)(Table 1), and the observed second-order rate constant was k´sto = 1.3 × 108 M−1 s−1, which was identical to the diffusion-controlled second-order rate (k′ss) observed in the steady-state reaction with this same site in free 601 DNA (Fig. S6).
Figure 5.
Single-turnover kinetic experiments on free DNA (U+44) and NCPs containing T→U substitutions at three single sites (U+22N1, U−32N1 and U+44N1). (a) Observed rate constants (kobssto) for reaction of 20 nM free 601 DNA containing a uracil at site U+44 with various concentrations of UNG. The insert shows the kinetic time course with 0.4 µM UNG. (b) Observed rate constants (kobssto) for reaction of 20 nM NCP containing a uracil at site U+22N1 with various concentrations of UNG. The insert shows the kinetic time course with 1.3 µM UNG. (c) Time course (log scale) for reaction of 20 nM NCP containing uracil at site U−32N1 with 0.2 µM UNG. (d) Time course (log scale) for reaction of 20 nM NCP containing uracil at site U+44N1 with 0.2 µM UNG. In panels b-c the graphic inserts display the structural properties of each NCP site. The uracils are in red, the histones are in purple and the DNA is in grey. (e, f) Absence of hUNG concentration dependence for the observed rate constants for single-turnover excision of uracil from nucleosome sites U+44N1 and U−32N1.
The three uracil sites embedded in NCPs showed very different kinetic behavior as compared to the free control DNA. Exposed site U+22N1 reacted with a single-exponential appearance of product and a hyperbolic dependence on UNG concentration (Fig. 5b). The excision rates for U+22N1 at each UNG concentration exceeded that of the free DNA by 1.5 to 2-fold, indicating enhanced second-order (k´sto) and maximal excision rates (kexsto) for this site (Table 1). These findings indicate that UNG reacts directly with this site with no requirement for rate-limiting local or global conformational rearrangements of the NCP (see molecular docking studies below). In contrast, buried site U−32N1 and exposed site U+44N1 both showed multi-exponential single turnover excision rates that were independent of UNG concentration in the range 100 to 400 nM (Fig. 5c,d,e,f). Both sites displayed similar rapid phases (10–20 s−1) and a slower phase (0.1 – 0.2 s−1), but site U+44N1 also showed a very slow third phase (0.003 s−1) necessitating the use of a log time scale to display the data (Fig. 5c, 5d)(Table 1). The similar rates for the first two phases with U−32N1 and U+44N1 suggest that similar conformational processes may be rate-limiting for reaction of both of these sites even though they are distant from each other in the linear DNA sequence NCP. A common process is a reasonable proposal because these sites are located on adjacent tiers of the DNA chain as it wraps around the histones. The absence of an UNG concentration dependence for excision of U−32N1 and U+44N1 in the range 100 to 400 nM UNG (Fig. 5e,f), indicates that rate-limiting conformational changes in the NCP (kconf) are required to place these sites in a transient reactive state. Accordingly, UNG must react with these transient states more rapidly than they revert back to an unreactive conformation (i.e k−conf < k´sto[UNG]). A similar trapping mechanism by UNG is plausible in the human nucleus because UNG is an abundant enzyme (~200,000 copies/cell)2.
Mechanism of Site Exposure
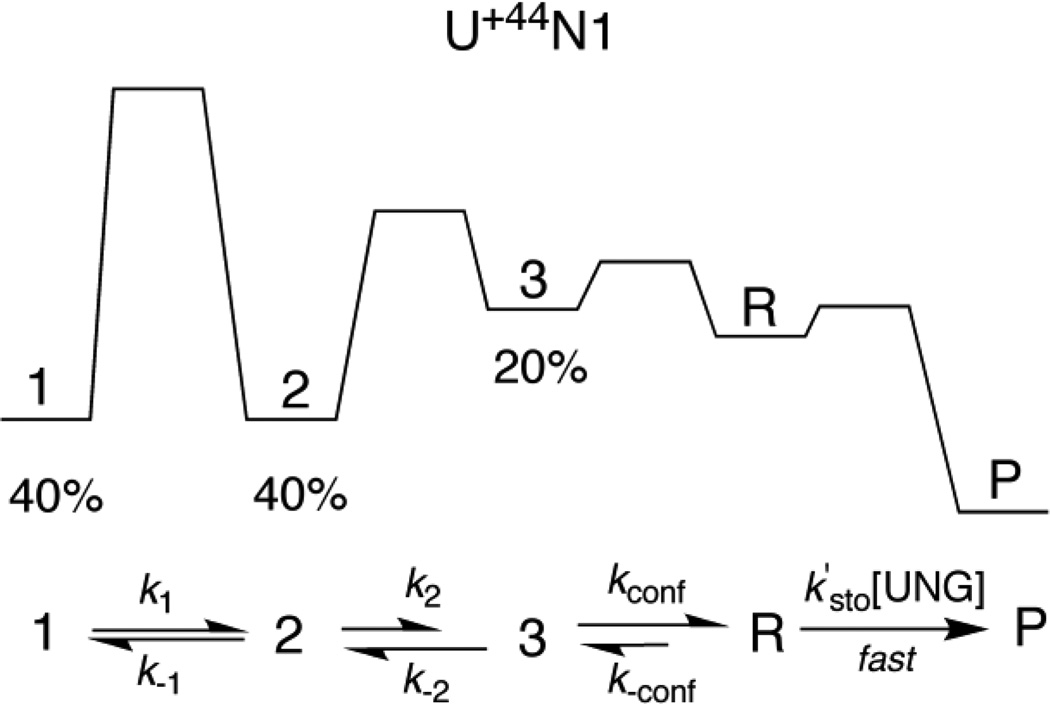
At least two related models can be envisioned to explain the multi-exponential kinetic reactivity of sites U−32N1 and U+44N1. First, it is formally possible that these sites exist in multiple stable ground-state conformations, giving rise to different activation barriers for achieving a single UNG reactive state. In this case, the fractional amplitudes of the individual kinetic phases would represent the populations of the ground state conformations at the initiation of the reaction. For this simple model to hold, each ground state conformation should not interconvert during the time frame of the UNG reaction. However, if interconversion of the ground state conformations does occur (a more plausible scenario), then excision would proceed via a pathway that preferentially funnels through the conformation with the lowest activation barrier for transition to the UNG reactive state (Fig. 6). Despite rapid interconversion, when the reverse rates for the conformational changes are slow compared to the forward steps, this mechanism would also show fractional amplitudes corresponding to different ground state conformations (see legend to Fig. 6). Although more complicated models are possible, a general mechanism involving a rate-limiting conformational change followed by rapid reaction of UNG is required to satisfactorily account for the excision of uracil from sites U−32N1 and U+44N1.
Figure 6.
Kinetic mechanism for excision of uracil from site U+44N1. The single-turnover kinetic measurements for excision of uracil from this site show three kinetic transients (Figure 5d), suggesting that three distinct conformational states of the NCP exist with relative populations of 40% (state 1), 40% (state 2) and 20% (state 3) as depicted in the qualitative free energy reaction coordinate diagram. The rates for conversion of these individual states to the reactive conformation (R) that is competent for uracil excision will depend on the forward rate constants k1, k2 and kconf, respectively. In this minimal mechanism, all excision events occur by funneling through the conformation that has the lowest barrier for conversion to the reactive R conformation. The individual rate constants in Table 1 will approximate k1, k2 and kconf under the conditions where (i) k1 << k2 and kconf, and (ii) k2 and kconf exceed the corresponding reverse rate constants k−1 and k−2. The single turnover kinetic data are independent of UNG concentration requiring that k´sto[UNG] (for [UNG] > 100 nM) is faster than these NCP conformational changes.
Do the observed rates for uracil excision from sites U-32N1 and U+44N1 match the conformational transitions previously observed for nucleosomal DNA? Previous studies by the Widom group using ensemble-based stopped-flow fluorescence measurements and FRET-fluorescence autocorrelation spectroscopy have led to a model for site exposure that involves successive unwrapping of the DNA duplex from the histone core9, 41, 42. Since the histones tend to contact the DNA about once every turn of the helix, each 10 bp of unwrapping from the DNA entry/exit sites leads to an increased activation barrier largely due to the increasingly unfavorable thermodynamics of breaking multiple histone-DNA interactions. Thus, duplex site exposure rates were around 4 s−1 for sites one or two turns from the entry/exit sites, and many orders of magnitude lower for sites three and four turns from the entry/exit sites (~0.02 and 0.002 s−1, respectively)42, 43. In contrast, thermodynamically favorable rewrapping rates were rapid and only moderately dependent on position from the entry/exit sites. For instance, the wrapping rate for a site at the dyad axis was 1.4 s−1, as compared to 21 s−1 for a site one turn from the entry/exit sites43. These kinetic values for full duplex unwrapping show little if any agreement with the single-turnover kinetic constants for sites U+22N1, U−32N1 and U+44N1 (Table 1). For site U+22N1 (~ five turns from the nearest edge of the nucleosome), the reaction rate is six-orders of magnitude greater than the duplex unwrapping rate, and for sites U−32N1 and U+44N1, located about three to four turns from the nearest DNA ends, the rates are ~ 100 to 10,000 times greater than the expected unwrapping rates. Only the slowest phase with site U+44N1 is in the range expected for duplex unwrapping. These differences are most likely attributed to the DNA binding mode of UNG that does not require full duplex unwrapping. Instead, the reaction of this enzyme is likely to occur directly with some uracil sites (middle pathway, Fig. 1) or through local DNA breathing events that are distinct from complete duplex unwrapping (upper pathway, Fig. 1). These conclusions are supported by the molecular docking studies described below.
Molecular Docking of UNG to Uracil Sites U+22N1, U−32N1 and U+44N1 in the NCP
The above single-turnover kinetic data suggested that UNG could directly dock to uracil site U+22N1, but that conformational transitions were required for sites U−32N1 and U+44N1. We tested these kinetic predictions using a molecular docking approach21. To obtain good restraints for the initial docking model, we employed the crystal coordinates of the complex of hUNG with undamaged duplex DNA (2OXM)31. This is a reasonable model for these purposes because it reflects a very early step along the reaction coordinate where the enzyme has formed nascent interactions with a U:A base pair, but has not yet severely bent the DNA duplex as observed in later stage Michaelis complexes31, 44. Using this model, attempts to dock UNG to sites U−32N1 and U+44N1 were unsuccessful due to severe steric clashes with the histone core (site U−32N1), or with histone N-terminal tail side chains that were present in the DNA minor groove (site U+44N1, see above)(Supplemental Figs. S8 and S9). These docking attempts clearly indicated the need for conformational transitions in order for UNG to react with these sites as required from the single-turnover kinetic measurements above. Although the detailed aspects of these transitions cannot be discerned from these studies, they are likely to involve transient release of the histone tail, and/or breaking of local ionic interactions releasing the duplex partially or completely from the histone surface. An additional implication from these findings is that histone tail modifications could have a large effect on reactivity of certain sites (i.e. site U+44N1).
In contrast with the unreactive sites, it was straightforward to dock UNG on site U+22N1 using the same 2OXM model (Figure 7a, b). This docking procedure was restrained using several well-studied interactions of UNG with the DNA phosphate backbone (Ser247, Ser270 and Ser 273)32, 45, and the requirement for positioning of the wedge residue Leu272 in the minor groove24, 43. Using these restraints, converged models were readily obtained with good docking scores. The model provides structural information that helps explain the high reactivity of this site. First, the highly perturbed base pair parameters of twist, roll, buckle, tip and incline (Fig. S5), likely promote breathing of the base pair and generate motions that are on the trajectory for base extrusion from the duplex (Fig. 7b). Second, the wide minor groove could facilitate entry of the enzyme loop that contains the wedge residue Leu272. The unhindered reactivity of this site indicates how the DNA binding mode of UNG, which emphasizes a limited number of strand interactions immediately adjacent to the damage site, is ideally suited for recognition in diverse contexts of duplex DNA, single-stranded DNA and nucleosomes.
Conclusions
These measurements extend our current understanding of damaged base recognition in the context of nucleosomes. Although previous work has established the importance of rotational orientation of damaged sites on the nucleosome in their reaction with enzymes such as UNG3, 5, 6, the current findings provide additional structural and dynamic insights into reactivity of several sites embedded in NCPs. Our findings establish the importance of local site dynamics, rather than slow global duplex unwrapping, in the recognition of buried or sterically occluded uracil bases, and define the time scale of dynamic events at several sites. The data also suggest that the kinetics of damage recognition will be affected by the modification of histone tails (acetylation, methylation), and that the epigenetic status of chromatin may influence DNA base excision repair. Finally, structural models now demonstrate how direct recognition of a damaged base in the context of an NCP can rapidly occur in the absence of any conformational rearrangement of the histones or DNA.
Supplementary Material
Abbreviations
- UNG
uracil DNA glycosylase
- NCP
nucleosome core particles
Footnotes
This work was supported by NIH grants GM056834 (J.T.S), GM078221 (J.J.G). and GM084192 (G.D.B.)
Supporting Information Available. Nine supporting figures further describing the kinetic and structural analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. J. Biol. Chem. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker JB, Stivers JT. Dynamics of uracil and 5-fluorouracil in DNA. Biochemistry. 2011;50:612–617. doi: 10.1021/bi101536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 11.Polach KJ, W J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 12.Anderson JD, Thastrom A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol. Cell. Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 14.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 16.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 17.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol. Cell. Biol. 2011;31:4746–4759. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 19.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with Image J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 20.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. Benchmarking and analysis of protein docking performance in rosetta v3.2. PLoS One. 2011;6:e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhury S, Lyskov S, Gray JJ. PyRosetta: A script-based interface for implementing molecular modeling algorithms using rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 25.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Bishop EP, Rohs R, Parker SC, West SM, Liu P, Mann RS, Honig B, Tullius TD. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem. Biol. 2011;6:1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez AG, Anderson JN. Nucleosome positioning determinants. J. Mol. Biol. 2007;371:649–668. doi: 10.1016/j.jmb.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 29.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: Protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 31.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang YL, Stivers JT. Mutational analysis of the base-flipping mechanism of uracil DNA glycosylase. Biochemistry. 2002;41:11236–11247. doi: 10.1021/bi026226r. [DOI] [PubMed] [Google Scholar]
- 33.Jiang YL, Kwon K, Stivers JT. Turning on uracil-DNA glycosylase using a pyrene nucleotide switch. J. Biol. Chem. 2001;276:42347–42354. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang YL, Stivers JT, Song F. Base-flipping mutations of uracil DNA glycosylase: Substrate rescue using a pyrene nucleotide wedge. Biochemistry. 2002;41:11248–11254. doi: 10.1021/bi026227j. [DOI] [PubMed] [Google Scholar]
- 35.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davey GE, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosomefacilitatesalkylation by an intercalatingantitumouragent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 38.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-state analysis of a single catalytic turnover by escherichia coli uracil-DNA glycosylase reveals a "pinch-pull-push" mechanism. J. Biol. Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 39.Parker JB, Stivers JT. Uracil DNA glycosylase: Revisiting substrate-assisted catalysis by DNA phosphate anions. Biochemistry. 2008;47:8614–8622. doi: 10.1021/bi800854g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grogan BC, Parker JB, Guminski AF, Stivers JT. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tims HS, Widom J. Stopped-flow fluorescence resonance energy transfer for analysis of nucleosome dynamics. Methods. 2007;41:296–303. doi: 10.1016/j.ymeth.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirier MG, Oh E, Tims HS, Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase-DNA substrate and product structures: Conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner RM, Jiang YL, Gordley RG, Jagadeesh GJ, Ladner JE, Xiao G, Tordova M, Gilliland GL, Stivers JT. Stressing-out DNA? the contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase. Biochemistry. 2000;39:12585–12594. doi: 10.1021/bi001532v. [DOI] [PubMed] [Google Scholar]
- 46.Varrazzo D, Bernini A, Spiga O, Ciutti A, Chiellini S, Venditti V, Bracci L, Niccolai N. Three-dimensional computation of atom depth in complex molecular structures. Bioinformatics. 2005;21:2856–2860. doi: 10.1093/bioinformatics/bti444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.