Abstract
The transcription factor Foxp3 is indispensible for the differentiation and function of regulatory T cells (Treg cells). To gain insights into the molecular mechanisms of Foxp-mediated gene expression we purified Foxp3 complexes and explored their composition. Biochemical and mass-spectrometric analyses revealed that Foxp3 forms multi-protein complexes of 400–800 kDa or larger and identified 361 associated proteins, ~30% of which are transcription-related. Foxp3 directly regulated expression of a large proportion of the genes encoding its co-factors. Reciprocally, some transcription factor partners of Foxp3 facilitated its expression. Functional analysis of Foxp3 cooperation with one such partner, GATA-3, provided further evidence for a network of transcriptional regulation afforded by Foxp3 and its associates to control distinct aspects of Treg cell biology.
The X-chromosome encoded forkhead domain containing transcription factor Foxp3 is a lineage-specifying factor responsible for the differentiation and function of regulatory T cells (Treg cells). This subtype of CD4+ T cells is indispensible for control of autoimmunity and excessive inflammation caused by the immune response to pathogens and commensal microorganisms1, 2. Mutations in the human FOXP3 gene are associated with fatal early onset autoimmune syndrome IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked). Likewise, in mice loss of Foxp3 function is associated with an early onset widespread autoimmunity3–5. Furthermore, continued expression of Foxp3 in mature Treg cells is essential to maintain the gene expression program enabling suppressive function of Treg cells6.
Despite its central role in Treg biology, the molecular basis of Foxp3 function has been poorly understood. Genome-wide analyses of Foxp3 target genes using a chromatin immunoprecipitation (ChIP)-on-chip approach, a combination of ChIP with a genome-wide DNA array, coupled to the analyses of differential gene expression in Treg cells expressing functional Foxp3GFP reporter allele and Treg “wannabe” cells, which express Foxp3 reporter null allele (Foxp3gfpko) and resemble Treg precursor cells, revealed that Foxp3 induced both activation and repression of its target genes7–10. However, examination of the identified functional Foxp3 binding sites failed to reveal an obvious sequence motif, likely a result of a relatively low affinity of the Foxp3 forkhead domain for DNA. Yet, multiple mutations in the DNA binding forkhead domain, including those leading to its loss, were identified in IPEX patients11–13. These results suggested that capacity of Foxp3 to bind DNA is critical for its functionality and that Foxp3-DNA interactions are likely assisted by Foxp3 co-factors and by multimerization. Indeed, Foxp3 polypeptides homodimerize and likely form higher-order complexes14,15; growing numbers of sequence-specific transcription factors interacting with Foxp3 have been identified and some have been implicated in the Treg cell specific gene expression program. Among these factors, Foxp1 forms heterodimers with Foxp3 through interactions with its leucine dimerization domain15, 16. Besides Foxp1, recent studies suggested that Foxp3 functionality is dependent on its cooperation with the Nuclear Factor of Activated T cells (NFAT), Ikaros family member Eos, and Runx1-Cbfβ complex17–19. In addition to sequence-specific transcription factors, histone acetyl transferases (HATs) and histone deacetylases (HDACs) have also been implicated in Foxp3 mediated gene expression. In particular, the Tat-interacting protein 60 kDa (TIP60) and the type II HDACs (HDAC7 and HDAC9) have been suggested to bind the N-terminal region of Foxp3 and contribute to Foxp3-mediated repression, presumably because acetylation by TIP60 is prerequisite for Foxp3 function14,20. Although these data imply that multiple Foxp3 partners play an important role in Foxp3 function, protein composition of Foxp3 transcriptional complexes and relationships between Foxp3 and its partners remained largely unknown as each of the previous studies focused on a single or at best very few partners at a time.
In the current study we employed unbiased proteomic approach to comprehensively analyze composition of Foxp3 complexes. Our mass-spectrometric analyses identified 361 partners of Foxp3. Unexpectedly, a high proportion of the genes that encode Foxp3 partners, which include transcription factor GATA-3, served as direct targets of Foxp3. Functional analyses of GATA-3 cooperation with Foxp3 suggested that reciprocal close-circuit networks regulate expression of Foxp3 itself and its interacting partners and the downstream Foxp3-dependent transcriptional program.
Results
Biotinylation based isolation of Foxp3 protein complexes
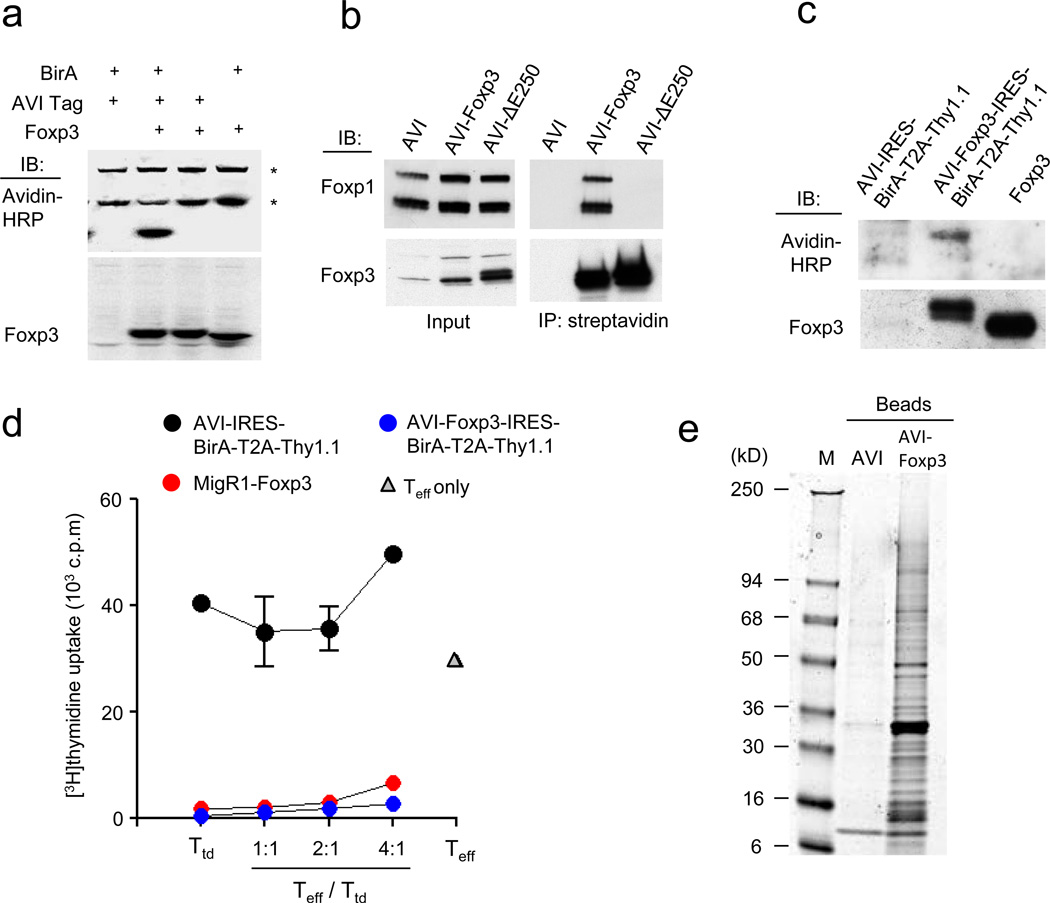
To identify Foxp3 interacting partners using a proteomic approach, we generated a T cell line expressing a biotin-tagged Foxp3 protein to allow for its efficient purification. Previously characterized T cell hybridoma TCli (ref. 21) was retrovirally transduced with a construct encoding a peptide containing BirA ligase biotinylation site fused to the N-terminus of the Foxp3 protein (AVI-Foxp3) and the prokaryotic biotin ligase BirA. The AVI peptide biotinylated at a lysine residue by the BirA enzyme allows for efficient purification of a tagged protein and its associated factors by affinity chromatography using streptavidin conjugated magnetic beads22. As expected, biotinylated Foxp3 protein was detected only in TCli cells harboring both AVI-Foxp3 and the BirA biotin ligase (Fig. 1a). Importantly, Foxp3 expression in TCli cells was comparable to that of endogenous Foxp3 protein in primary Treg cells (Supplementary Fig. 1). Efficient pull-down of the protein complexes containing biotinylated Foxp3 protein using streptavidin-conjugated magnetic beads was confirmed by immunoblotting for Foxp3 and its previously identified partner Foxp1. In agreement with previous reports, Foxp3-Foxp1 interactions were completely abolished in a TCli cell line harboring a biotinylated form of the ΔE250 mutant of Foxp3 (ref. 16) suggesting that at least this well-documented interaction was unperturbed and retained its specificity after biotinylation of Foxp3 (Fig. 1b). To confirm that expression of the biotinylated AVI-Foxp3 protein is sufficient to confer suppressor function, we generated a tri-cistronic retroviral construct encoding AVI-Foxp3 fusion protein along with an internal ribosomal entry site (IRES) preceding BirA coding sequence that was followed by “self-cleaving” 2A peptide from picornavirus and Thy1.1 as a reporter23. The latter was expressed on the surface of transduced cells after cleavage of T2A peptide upon translation of a single transcript. A similar construct containing the AVI tag alone was used as a control. Co-expression of AVI-Foxp3 and BirA from a single retroviral vector in primary CD25−Foxp3−CD4+T cells (Tconv cells) resulted in efficient biotinylation of the AVI-Foxp3 protein (Fig. 1c). Transduced T cells expressing biotinylated AVI-Foxp3 protein and Thy1.1 reporter were purified by flow cytometry and their capacity to curb proliferation of conventional T (Tconv) cells was assessed using a standard in vitro suppression assay. Tconv cells transduced with retroviruses expressing AVI-Foxp3-IRES-BirA-T2A-Thy1.1 or wild-type Foxp3-IRES-GFP (MigR1-Foxp3) used as a positive control exhibited comparable suppressive capacity (Fig. 1d). In contrast, the negative control vector (AVI-IRES-BirA-T2A-Thy1.1) failed to impart suppressive properties. Thus, these data indicate that biotinylated AVI-Foxp3 and wild-type Foxp3 protein were similarly functional.
Figure 1.
Strategy for purification of Foxp3-associated proteins. (a) Immunoblot analysis of biotinylated AVI-Foxp3 in nuclear lysates prepared from TCli cells expressing AVI-Foxp3 and BirA. AVI-tag and BirA expressing cells were used as a control. * indicates endogenously biotinylated nuclear proteins. Data are representative of three independent experiments. (b) Immunoprecipitation of AVI-Foxp3 or AVI-ΔE250 Foxp3 mutant protein from nuclear lysates of TCli cells using streptavidin-conjugated magnetic beads. The presence of Foxp3 and its known partner Foxp1 was determined by immunoblot analysis. Data are representative of two independent experiments. (c) Immunoblot analysis of nuclear extracts prepared from activated CD4+CD25− T cells that were transduced with the indicated retroviral vectors. Data are representative of two independent experiments. (d) In vitro suppressor activity of the transduced T cells (Ttd) described in (c). Transduced T cells were co-cultured with CD4+Foxp3− (GFP−) responder T cells (Teff) from Foxp3GFP mice at indicated ratios for 72 h in the presence of anti-CD3 and irradiated (2000 rads) T cell-depleted splenocytes. The data are shown as mean [3H]-thymidine incorporation in triplicate cultures and are representative of two independent experiments. (e) Fractionation of affinity purified Foxp3 protein complexes in SDS PAGE followed by Coomassie staining. Data are representative of three independent experiments.
Proteomic analysis of Foxp3 protein complexes
To identify Foxp3 binding partners we isolated Foxp3-containing protein complexes using streptavidin magnetic bead chromatography from nuclear lysates of BirA expressing TCli cells harboring AVI-Foxp3 (TCli-AVI-Foxp3) and analyzed purified protein complexes by SDS-PAGE. In contrast to negative control protein preparation from TCli-AVI cell lysates, biotinylated Foxp3 was bound to a large number of proteins of different molecular weights (Fig. 1e). Individual bands were excised and subjected to in-gel trypsin digestion and sequencing by micro-liquid chromatography-mass spectrometry/mass spectrometry (µLC-MS/MS). The Foxp3 partner list was compiled from 361 protein hits repeatedly identified in at least three of four independent experiments using TCli-AVI-Foxp3 cells and lacking or present at relatively low abundance in two parallel experiments using negative control TCli-AVI cells (Supplementary Table 1). The mass-spectrometry data were validated by co-immunoprecipitation (co-IP) of a select panel of the identified Foxp3 interacting proteins, mainly nuclear factors related to transcriptional regulation, and immunoblot analysis. Among confirmed Foxp3-interacting partners we found several proteins including Runx1, YY1, Ikzf1 and Smarca5 that were also detected by mass spectrometry in negative control samples, albeit with a lower yield, presumably due to high sensitivity of the technique. For all proteins tested association with Foxp3 was observed both in the presence and absence of DNase suggesting that the interactions were preserved in the absence of DNA (Supplementary Fig. 2).
To test whether protein partners of biotinylated Foxp3 in TCli-AVI-Foxp3 cells bind endogenous Foxp3 protein in primary Treg cells, we prepared nuclear lysates of 100 × 106 CD4+CD25+ Treg cells isolated from C57BL/6 mice using magnetic bead sorting. Foxp3 complexes were isolated from nuclear lysates using affinity-purified rabbit Foxp3 antibody conjugated to tosyl-activated magnetic beads and fractionated by SDS-PAGE. In agreement with comparable ability of endogenous Foxp3 protein and biotinylated AVI-Foxp3 to confer suppressor function, mass-spectrometric analysis of affinity purified Foxp3 complexes revealed a substantial number of Foxp3-binding proteins identified in TCli-AVI-Foxp3 cells (Supplementary Table 1). As a negative control we performed mass-spectrometric analysis of nuclear proteins from Foxp3-deficient CD4+ T cells binding non-specifically to anti-Foxp3 beads. The interactions between Foxp3 and a large panel of its protein partners in primary Treg cells were also confirmed by co-immunoprecipitation and immunoblot analysis (Supplementary Fig. 2). Incomplete overlap between the two data sets was most likely due to smaller amounts of endogenous Foxp3 complexes and higher non-specific background in antibody vs. streptavidin-biotin-based affinity purification procedures.
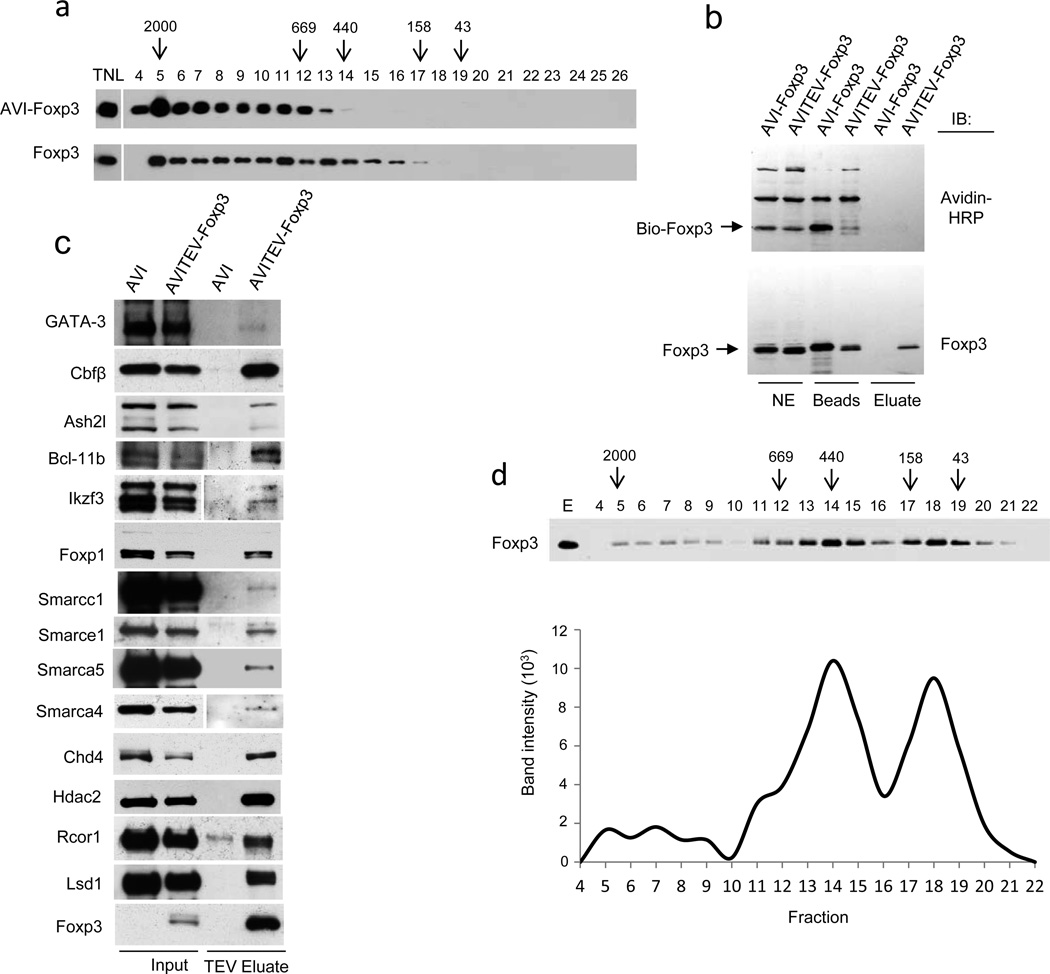
Mass-spectrometric, co-IP and immunoblot analyses raised the possibility that Foxp3 and its multiple protein partners formed very large protein complexes. Indeed, chromatographic analyses of nuclear lysates of TCli-AVI-Foxp3 cells using fractionation on a Superose 6 FPLC gel-filtration column revealed that the bulk of Foxp3 protein was present in large 400–2000 kDa complexes (Fig. 2a, top). DNase and RNase treatment of the nuclear lysate did not alter the fractionation pattern (data not shown). Furthermore, analyses of nuclear lysates prepared from ex vivo purified primary Treg cells demonstrated a similar separation pattern of endogenous Foxp3 complexes suggesting that their large size was not a result of an artifact unique to Foxp3 transduced transformed cell line (Fig. 2b, bottom).
Figure 2.
Foxp3 forms large protein complexes with its partners. (a) Total nuclear lysates (TNL) prepared from TCli-AVI-Foxp3 cells (top) and magnetic bead purified Treg cells (bottom) were subjected to fractionation on a Superose 6 FPLC column and distribution of Foxp3 complexes in the resulting fractions was assessed by western blot analysis after ethanol precipitation. Fraction numbers and molecular weights of complexes (in kD) are indicated. Data are representative of two independent experiments. (b) Immunoblot analysis of biotinylated AVITEV-Foxp3 protein released from streptavidin-conjugated magnetic beads upon cleavage with TEV protease. AVI-Foxp3 protein lacking a TEV cleavage site was used as a control. Foxp3 proteins were visualized using anti-Foxp3. Data are representative of at least three independent experiments. (c) Immunoblot analysis of the TEV eluted Foxp3 complexes to confirm the presence of Foxp3 co-factors identified by mass-spectrometric analyses. Data are representative of two to three independent experiments. (d) Fractionation of TEV-cleaved Foxp3 complexes in a Superose 6 FPLC column. The intensities of the bands in different fractions were determined by the “Image J” software and shown in the lower panel. Data are representative of three independent experiments.
To directly test the size of purified Foxp3 associated protein complexes; we introduced a cleavage site for the Tobacco Etch Virus (TEV) protease between Foxp3 and the AVI tag peptide and co-expressed the AVI-TEV-Foxp3 protein and BirA biotin ligase in TCli cells. Treatment with the TEV protease of biotinylated AVI-TEV-Foxp3 protein bound to streptavidin beads efficiently released intact Foxp3 protein complexes from the beads under non-denaturing conditions (Fig. 2b). Mass-spectrometric and immunoblot analysis of the TEV eluted Foxp3 complexes revealed composition similar to that described above for biotinylated AVI-Foxp3 complexes eluted from streptavidin beads upon boiling in 1% SDS containing buffer (Fig. 2c; data not shown). To assess the apparent size of TEV eluted Foxp3 complexes we subjected TEV eluted Foxp3 complexes on a Superose 6 gel-filtration column. In addition to 40–200 kDa fractions likely containing monomeric and multimeric Foxp3 and intermediate 400–800 kDa fractions, immunoblot analysis revealed a subset of Foxp3 complexes in very heavy fractions with an apparent molecular mass of 1,500–2,000 kDa (Fig. 2d). The difference in migration pattern of purified Foxp3 complexes after TEV elution from streptavidin beads (Fig. 2d) to that observed upon the fractionation of nuclear lysates (Fig. 2a) is likely due to the loss of higher-order complexes during the purification and elution procedure. Thus, our biochemical analysis indicates that Foxp3 forms very large protein complexes with numerous protein partners.
Functional annotation of Foxp3-associated proteins
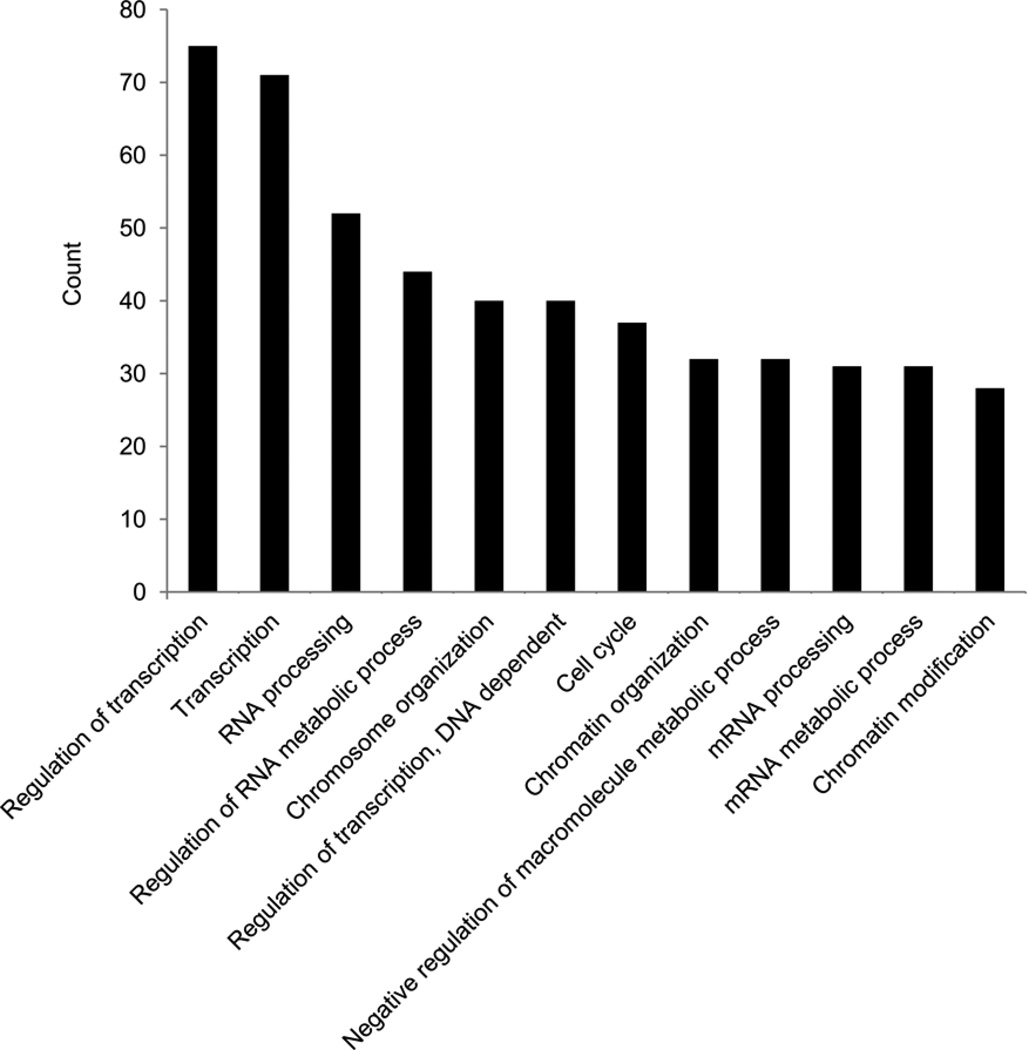
To account for compound functionality and interconnectivity of the vast set of Foxp3 interacting proteins, it was functionally annotated using DAVID 6.7 software package (http://david.abcc.ncifcrf.gov/home.jsp). In addition to expected enrichment for the gene ontology (GO) terms, categories or biological process such as DNA binding, transcription regulator activity, chromatin binding, regulation of transcription, chromosome organization and chromatin modification, we found statistically significant enrichment in RNA binding, processing, splicing and metabolism categories suggesting a yet unexplored RNA-associated role of Foxp3 (Fig. 3; Supplementary Table 2a,b). Indeed, 23% of identified Foxp3 protein partners are implicated in RNA binding and regulation and their function in Foxp3 complexes remain to be explored.
Figure 3.
Functional annotation of Foxp3 associated proteins. (a) Analysis of GO term enrichment of the “biological process” category of Foxp3-associated proteins. Top 12 GO terms ranked according to the number of counts are plotted; full list of GO terms is shown in Supplementary Table 2b.
For further in-depth analyses we focused on a subset of transcription-related Foxp3 associated proteins, comprised of 94 proteins (27%), which were manually assigned to several known protein complexes or functional categories (Table 1). The observed association of Foxp3 with a number of its transcription-related partners, including a component of SWI/SNF complex Baf57, remained when co-immunoprecipitation experiments were performed in the presence of both DNase and RNase (Supplementary Fig. 3). Of note, the SWI/SNF complex binds snRNP proteins and mRNA splicing complex and associated RNA24,25. Therefore, while a role for DNA and RNA templates in the formation of Foxp3 complexes in vivo remains likely, these results suggest that after complex formation the depletion of nucleic acid templates leaves Foxp3 protein complexes intact (Supplementary Fig. 3). Analysis of domain composition of the transcription-related partners revealed an enrichment of C2H2 and PHD-type zinc-finger domains, winged helix DNA binding, DEAD-like and ATP-binding helicase domains, SNF2-related, chromo-, bromo- and SANT domains, all of which are implicated in influencing gene expression either by heterodimerization with transcription factors, ATP-dependent chromatin remodeling or binding to modified chromatin (Supplementary Table 2c). By employing STRING 9.0 (http://string-db.org/), a modular interactome building tool, we constructed a map of the Foxp3 interactome based on previously characterized interactions among the identified Foxp3 partners. Interaction modules of previously known nuclear protein complexes affecting transcription including NURD, MLL, SWI/SNF, ISWI, NCoR and PolII transcription complex were readily identifiable within the Foxp3 protein complex network (Supplementary Fig. 4).
Table 1.
Foxp3-associated factors implicated in transcription regulation and their assignment to known protein complexes and functional categories.
| Complex/ Functional classification |
IP id |
Gene name | Average percentage coverage |
Average number of unique peptides |
Description |
|---|---|---|---|---|---|
| Bait | IPI00115123 | Foxp3* | 33.175 | 15.5 | forkhead box P3 |
| Transcription factors | |||||
| IPI00121608 | Bcl11b* | 13.225 | 6 | B-cell leukemia/lymphoma 11B | |
| IPI00169477 | Bclaf1* | 8.35 | 3.75 | BCL2-associated transcription factor 1 | |
| IPI00229487 | Cbfb* | 18.425 | 3 | Cbf beta-2 | |
| IPI00120304 | Elf1* | 8.625 | 3.25 | spectrin beta 2 | |
| IPI00323239 | Foxp1* | 14.8 | 6.25 | forkhead box P1 | |
| IPI00120267 | Foxp4* | 20.525 | 8.5 | forkhead box P4 | |
| IPI00130283 | Ikzf1* | 7.675 | 2.25 | IKAROS family zinc finger 1 | |
| IPI00117144 | Ikzf3* | 7.3 | 2 | IKAROS family zinc finger 3 | |
| IPI00131214 | Runx1* | 18.325 | 6.75 | runt related transcription factor 1 | |
| IPI00311892 | Yy1* | 11.775 | 3 | YY1 transcription factor | |
| IPI00314507 | Zfp326* | 4.35 | 1 | zinc finger protein 326 | |
| IPI00222330 | Zfp771* | 4.025 | 1 | zinc finger protein 771 | |
| IPI00123565 | Aatf | 10.7 | 3.75 | apoptosis antagonizing transcription factor | |
| IPI00122594 | Ahctf1 | 6.35 | 3.25 | AT hook containing transcription factor 1 | |
| IPI00120684 | Bax | 12.375 | 2.25 | NK-3 transcription factor, locus 1 (Drosophila) | |
| IPI00123159 | Cdca7l | 5.425 | 2.25 | cell division cycle associated 7 like | |
| IPI00132439 | Cebpz | 10 | 9.25 | DNA-damage inducible transcript 3 | |
| IPI00169667 | Cnot3 | 6.175 | 3.25 | CCR4-NOT transcription complex, subunit 3 | |
| IPI00117727 | Dpf2 | 11.325 | 3 | D4, zinc and double PHD fingers family 2 | |
| IPI00131410 | Elf4 | 2.9 | 1 | E74-like factor 4 (ets domain transcription factor) | |
| IPI00322297 | Etv3 | 7.95 | 1.5 | ets variant gene 3 | |
| IPI00135883 | Gata3 | 9.9 | 3.5 | GATA binding protein 3 | |
| IPI00310962 | Max | 9.525 | 1 | Max protein | |
| IPI00378026 | Nacc1 | 4.15 | 1.75 | nucleus accumbens associated 1, BEN and BTB (POZ) domain containing | |
| IPI00229456 | Nfatc2 | 8.5 | 4 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | |
| IPI00400329 | Nkrf | 2.625 | 1.25 | NF-kappaB repressing factor | |
| IPI00125443 | Nr3c1 | 6.875 | 3.75 | nuclear receptor subfamily 3, group C, member 1 | |
| IPI00125656 | Patz1 | 4.7 | 2 | POZ (BTB) and AT hook containing zinc finger 1 | |
| IPI00377843 | Sp110 | 14.45 | 1.75 | peripheral myelin protein 22 | |
| IPI00469258 | Sp2 | 4 | 1.25 | zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 | |
| IPI00341944 | Ssbp3 | 6.725 | 1.75 | single-stranded DNA binding protein 3 | |
| IPI00227814 | Stat3 | 9.25 | 5.25 | signal transducer and activator of transcription 3 | |
| IPI00406306 | Trp53 | 6.35 | 1.5 | transformation related protein 53 | |
| IPI00309068 | Zbtb33 | 11.475 | 5 | zinc finger and BTB domain containing 33 | |
| IPI00123531 | Zfp148 | 10.45 | 5.5 | zinc finger protein 148 | |
| IPI00127286 | Zfp346 | 4.15 | 0.75 | zinc finger protein 346 | |
| IPI00222382 | Zfp512 | 6.45 | 2.25 | zinc finger protein 512 | |
| MLL complex | |||||
| IPI00420997 | Mllt10 | 6.9 | 5 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | |
| IPI00131513 | Ash2l | 6.875 | 2.5 | ash2 (absent, small, or homeotic)-like (Drosophila) | |
| IPI00117168 | Dpy30* | 22.25 | 2 | dpy-30 homolog (C. elegans) | |
| IPI00226384 | Rbbp5 | 6.125 | 2 | retinoblastoma binding protein 5 | |
| IPI00407590 | Chd8 | 5.025 | 6.75 | chromodomain helicase DNA binding protein 8 | |
| IPI00462502 | Las1l | 14.9 | 3.75 | LAS1-like (S. cerevisiae) | |
| IPI00109326 | Senp3* | 7 | 2.25 | SUMO/sentrin specific peptidase 3 | |
| NURD-LSD1 complex | |||||
| IPI00396802 | Chd4 | 13.825 | 20.25 | chromodomain helicase DNA binding protein 4 | |
| IPI00229784 | Gatad2a* | 16.1 | 6.5 | GATA zinc finger domain containing 2A | |
| IPI00453837 | Kdm1a* | 13.2 | 7 | lysine (K)-specific demethylase 1A | |
| IPI00265217 | Rcor1 | 8.075 | 2.25 | REST corepressor 1 | |
| IPI00114232 | Hdac1 | 15.95 | 5 | histone deacetylase 1 | |
| IPI00137668 | Hdac2 | 16.125 | 4.75 | histone deacetylase 2 | |
| SWI/SNF complex | |||||
| IPI00127131 | Arid1a | 3.375 | 3.75 | AT rich interactive domain 1A (SWI-like) | |
| IPI00381019 | Smarcc2 | 2.65 | 2.25 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | |
| IPI00119892 | Smarce1 | 17.15 | 4.75 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | |
| IPI00125662 | Smarcc1 | 4.95 | 3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | |
| IPI00460668 | Smarca4 | 9.675 | 10 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | |
| IPI00133099 | Brd7* | 9 | 3.5 | bromodomain containing 7 | |
| ACF-ISWI chromatin remodelling complex | |||||
| IPI00396739 | Smarca5 | 20.825 | 23.25 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | |
| IPI00229432 | Bptf | 2.9 | 4.5 | bromodomain PHD finger transcription factor | |
| IPI00461396 | Baz1a* | 10.925 | 15.5 | bromodomain adjacent to zinc finger domain 1A | |
| NCoR complex | |||||
| IPI00308283 | Tbl1xr1* | 10.5 | 4 | transducin (beta)-like 1X-linked receptor 1 | |
| IPI00135456 | Hdac3 | 7.05 | 1.25 | histone deacetylase 3 | |
| Polycomb repressive complexes | |||||
| IPI00116041 | Cbx4 | 6.375 | 2.5 | chromobox homolog 4 (Drosophila Pc class) | |
| IPI00133917 | Phc2 | 2.95 | 0.75 | polyhomeotic-like 2 (Drosophila) | |
| IPI00396676 | Suz12 | 5.25 | 2 | suppressor of zeste 12 homolog (Drosophila) | |
| NSD family histone methyl transferases | |||||
| IPI00107975 | Whsc1* | 8.375 | 6.5 | Wolf-Hirschhorn syndrome candidate 1 (human) | |
| IPI00353681 | Whsc1l1* | 8.725 | 3.25 | Wolf-Hirschhorn syndrome candidate 1-like 1 (human) | |
| Components of HAT and HDAC complexes | |||||
| IPI00110256 | Msl1 | 5.225 | 1.5 | male-specific lethal 1 homolog (Drosophila) | |
| IPI00320317 | Ccdc101 | 10.1 | 2 | coiled-coil domain containing 101 | |
| IPI00115831 | Sap18 | 23.7 | 2.75 | Sin3-associated polypeptide 18 | |
| Co-activator or co-repressor activity | |||||
| IPI00108427 | Lims1* | 2.7 | 0.75 | LIM and senescent cell antigen-like domains 1 | |
| IPI00317722 | Dmap1 | 9.775 | 3 | DNA methyltransferase 1-associated protein 1 | |
| IPI00130670 | Strap | 13.6 | 3.5 | serine/threonine kinase receptor associated protein | |
| Histones | |||||
| IPI00404590 | H1f0 | 6.725 | 1.25 | H1 histone family, member 0 | |
| IPI00230264 | H2afx | 28.175 | 3.5 | H2A histone family, member X | |
| Transcription machinery | |||||
| IPI00330385 | Taf1 | 6.55 | 4 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor | |
| IPI00313515 | Pold1* | 1.725 | 1.5 | polymerase (DNA directed), delta 1, catalytic subunit | |
| IPI00136207 | Polr2a* | 5.525 | 5.5 | polymerase (RNA) II (DNA directed) polypeptide A | |
| IPI00153874 | Gtf2b* | 17.475 | 4.5 | general transcription factor IIB | |
| IPI00277858 | Gtf3c5* | 6.025 | 2.5 | general transcription factor IIIC, polypeptide 5 | |
| IPI00125670 | Med15* | 2.5 | 1.5 | mediator complex subunit 15 | |
| IPI00124520 | Rpa1 | 7.5 | 3.75 | polymerase (RNA) I polypeptide A | |
| IPI00113070 | Ercc3 | 6.8 | 3.75 | excision repair cross-complementing rodent repair deficiency, complementation group 3 | |
| IPI00132417 | Gtf2e1 | 6.7 | 2 | general transcription factor II E, polypeptide 1 (alpha subunit) | |
| IPI00403414 | Gtf3c1 | 4.85 | 6.25 | general transcription factor III C 1 | |
| IPI00308807 | Gtf3c2 | 6.95 | 3 | general transcription factor IIIC, polypeptide 2, beta | |
| IPI00224399 | Med19 | 16.825 | 2 | mediator of RNA polymerase II transcription, subunit 19 homolog (yeast) | |
| Miscellaneous | |||||
| IPI00380790 | Sirt7 | 15.475 | 4 | sirtuin 7 (silent mating type information regulation 2, homolog 7 (S. cerevisiae) | |
| IPI00466859 | Chd1l | 8.475 | 6.25 | chromodomain helicase DNA binding protein 1-like | |
| IPI00130218 | Kif11 | 12.3 | 9.5 | kinesin family member 11 | |
| IPI00471431 | Wac | 8.9 | 2.75 | WW domain containing adaptor with coiled-coil | |
| IPI00153212 | Ccdc137 | 11.375 | 2.25 | coiled-coil domain containing 137 | |
| IPI00125382 | Noc2l* | 3.725 | 2.5 | nucleolar complex associated 2 homolog (S. cerevisiae) | |
| IPI00227152 | Tdrd3 | 11.75 | 4.75 | tudor domain containing 3 | |
| IPI00229571 | Sltm | 4.675 | 3 | SAFB-like, transcription modulator | |
“*” indicates proteins that were also identified as partners of Foxp3 in ex vivo isolated Treg cells. A complete list of all Foxp3-associated proteins is provided in Supplementary Table 1.
Foxp3 partners as transcriptional targets of Foxp3
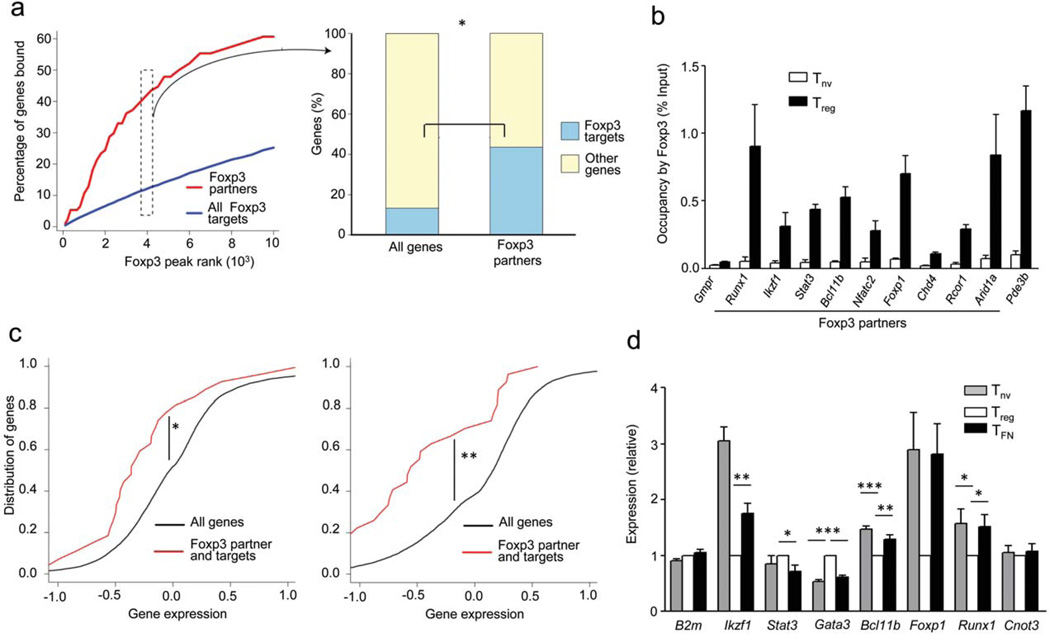
Although the majority of Foxp3-associated transcription-related nuclear factors are expressed in all T cell types, it was possible that Foxp3 modulates expression of its own partners as a means to specifically tune their functionality in Treg cells. Indeed, genome-wide analysis of Foxp3 target genes in primary Treg cells using ChIP sequencing (ChIP-Seq) revealed that regulatory regions of ~ 50% genes encoding transcription-related Foxp3 co-factors were bound by Foxp3 (ref. 7, 8 and 49) (Fig. 4a,b, Supplementary Table 3). Comparison of expression of this gene subset in naïve T cells (Tnv), CD4+ T cells expressing a functional Foxp3GFP reporter allele (Treg) and expressing a non-functional Foxp3GFPKO reporter allele (TFN) revealed statistically significant Foxp3-dependent shift in expression of genes encoding Foxp3 partners (Fig. 4c,d). Interestingly, a shift in the cumulative gene expression curve towards the left (red line vs. black line) indicates that although some Foxp3 target genes are up-regulated in a Foxp3-dependent manner, the majority is down-regulated. This observation suggests a possibility that Foxp3 fine-tunes the expression of many associated transcriptional regulators for functional benefits of Treg cells. Remarkably, several Foxp3 interacting partners including Runx, NFAT, and GATA-3, whose gene expression was affected by Foxp3, mirror its regulatory role by reciprocally contributing to regulation of the Foxp3 gene26–33. These results imply a close-circuit connectivity of reciprocal regulation of expression and cooperation between Foxp3 and several sequence-specific transcription factors which serve as its principal partners (Supplementary Fig.5).
Figure 4.
A large proportion of its associated factors are also transcriptional targets of Foxp3. (a) Analysis of genome-wide Foxp3 ChIP-seq data demonstrates that genes encoding Foxp3–transcription-related interaction partners are enriched as targets of Foxp3. The left panel shows percentage of genes bound by Foxp3 (Y-axis) sorted by Foxp3 peak read count in a 200 bp window (X-axis). The right panel shows the statistical enrichment in the top 4000 Foxp3 peaks, which correspond to 2705 out of 21289 refseq annotated genes and 41 out of 94 Foxp3 transcription-related cofactors. *P < 10−8 (Fisher’s exact test). (b) ChIP-qPCR analysis to demonstrate the occupancy of Foxp3 on the regulatory regions of genes encoding several of its interacting partners. Pde3b and Gmpr serve as positive and negative controls of Foxp3 occupied genes, respectively. Data represent two independent experiments. (c) Cumulative distribution analysis of the difference in expression of genes encoding Foxp3-associated factors between indicated cell types. *P < 0.0015 and **P < 0.00002 (One-tailed Kolmogorov-Smirnov test). (d) Real-time PCR analysis of mRNA encoded by genes of selected Foxp3 partners from indicated cell types isolated from heterozygous Foxp3GFPKO/WT female mice. B2m is an unrelated negative control gene and Cnot3 is a partner, but its encoding gene is not a significant target of Foxp3 according to ChIP-seq analysis. Data represents six to nine replicates from two to three independent experiments. *P < 0.05, **P < 0.005 and ***P < 0.0001 (Student’s t-test).
Foxp3-GATA-3 regulatory module in Treg cells
Recent studies suggested a prominent role for GATA-3 in Treg cell function and homeostasis29,30. Our mass-spectrometric analysis identified GATA-3 as a prominent Foxp3-associated factor (Fig.2c, Supplementary Fig. 3; Table 1). Thus, as a “case study” of a regulon formed by Foxp3 and one of its co-factors in Treg cells we investigated mechanistic aspects of cooperation between GATA-3 and Foxp3. To aid the analysis of genes co-regulated by Foxp3 and GATA-3 we leveraged the datasets from a recent genome-wide analysis of GATA-3 binding genes in various CD4+ T cell types, including Treg cells34.
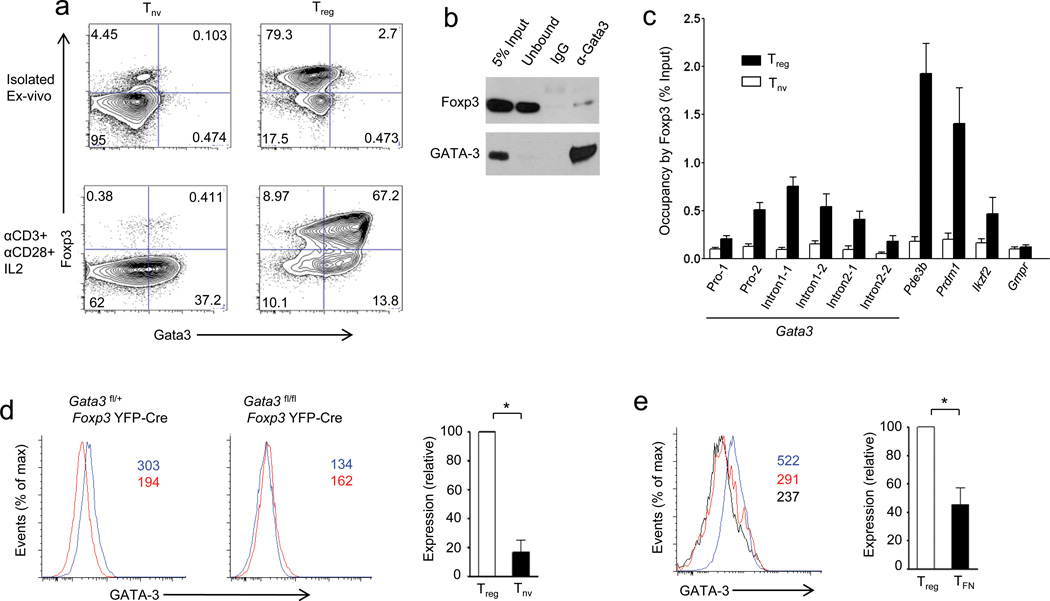
First, we revisited the expression pattern of GATA-3 in activated and resting Treg cells. In agreement with other reports, only a small proportion of ex vivo isolated Treg cells expressed GATA-3 under steady-state conditions. However, upon in vitro TCR stimulation in the presence of interleukin 2 (IL-2), a large proportion of Treg cells up-regulated GATA-3 (Fig. 5a). To determine if Foxp3 interacts with GATA-3 in primary Treg cells, we performed co-immunoprecipitation experiments from nuclear lysates prepared from Treg cells that had been activated for 24 h in vitro in the presence of IL-2 and observed efficient interaction between Foxp3 and GATA-3 under these conditions (Fig. 5b).
Figure 5.
Foxp3-Gata3 gene regulatory module in Treg cells. (a) Purified CD4+CD25+ (Treg) and CD4+CD25− (Tnv) T cells were stimulated by plate-bound CD3 and CD28 antibodies for 24 h in the presence of IL-2 followed by flow cytometric analysis of intracellular Foxp3 and GATA-3 expression. (b) Co-immunoprecipitation of GATA-3 and Foxp3 from nuclear extracts of activated Treg cells (described in (a)) followed by immunoblot analysis of the indicated proteins. A representative of two independent experiments is shown. (IgG: immunoprecipitation with IgG, α-GATA-3: immunoprecipitation with anti-GATA-3). (c) Immunoprecipitation of Foxp3-bound chromatin isolated from purified CD4+CD25+ Treg cells and control CD4+CD25− T cells using rabbit anti-Foxp3. Foxp3 occupancy of the Gata3 locus was determined by qPCR. Foxp3 binding to Pde3b, Prdm1 and Helios served as positive and to Gmpr as negative controls, respectively. A representative of two independent experiments is shown. (d) GATA-3 expression in CD4+Foxp3+Treg cells (blue line) and CD4+Foxp3− naïve T cells (red line) in mice harboring GATA-3 -sufficient (left) and -deficient (right) Treg cells. (e) GATA-3 expression in Treg (blue line) and TFN (red line) cells from heterozygous female Foxp3GFPKO/WT mice compared to Treg cells lacking GATA-3 (black line). Absolute mean fluorescent intensity (MFI) values are indicated in red and blue in the histogram plots or relative expression (MFI of GATA-3 relative to GATA-3-deficient Treg cells) is shown in the bar graphs. Data are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Next, we sought to show using ChIP combined with quantitative PCR (ChIP-qPCR) that Foxp3 and GATA-3 occupy regulatory sequences within Gata3 and Foxp3 genes as a potential means to augment their expression. We found that Foxp3 binds to the promoter and intronic regions of the Gata3 gene, which is expressed at a higher level in Treg cells than in naïve T cells or Foxp3-deficient T cells expressing Foxp3GFPKO (TFN) cells (Fig. 4d, 5c–e). Conversely, in agreement with recent reports we found GATA-3 binds the conserved non-coding sequence 2 (CNS2) of the Foxp3 gene; Treg cell-specific ablation of Gata3 resulted in reduced Foxp3 expression (Supplementary Fig. 6a,b)29, 30. Thus, Foxp3 and GATA-3 not only interact with each other in Treg cells, but also reciprocally increase the expression of each other at least in part through direct binding to the corresponding genetic loci. It must be noted that Foxp3 CNS2 deletion does not result in a decrease in Foxp3 abundance in Treg cells, suggesting that GATA-3 binding elsewhere either within or outside the Foxp3 locus contributes to regulation of its expression35.
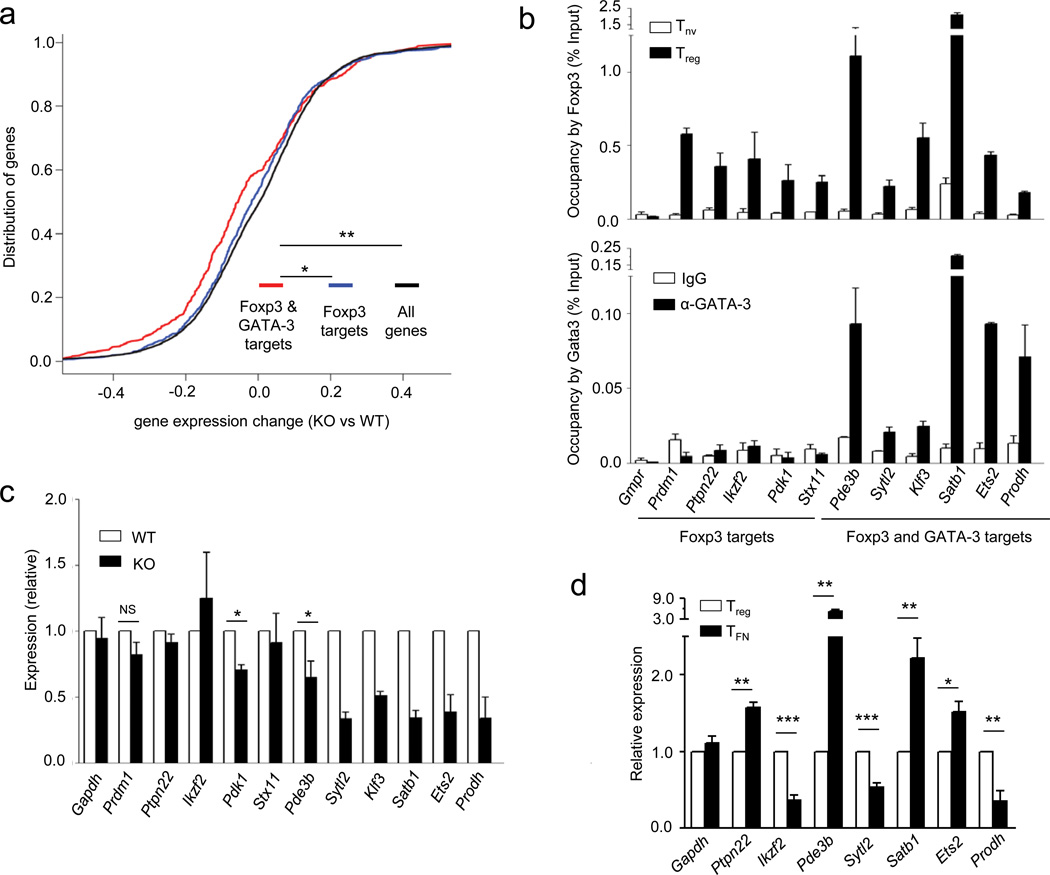
To identify additional genes bound and potentially regulated by Foxp3 and GATA-3 we cross-referenced Treg cell GATA-3 and Foxp3 ChIP-Seq datasets (Refs. 7, 8 and 49) and observed that GATA-3 co-occupies regulatory sites in a sizable group of Foxp3 target genes (Supplementary Table 4). Thus, a subset of Foxp3-occupied genes in Treg cells are also bound by GATA-3, suggesting a role for the Foxp3-GATA-3 complex in modulating their expression and Treg cell function. To test this idea we defined a GATA-3-dependent transcriptome in Treg cells by performing Affymetrix 430A 2.0 gene array of GATA-3-deficient and -sufficient Treg cells sorted from young Gata3fl/flFoxp3YFP-Cre or littermate control Gata3fl/wtFoxp3YFP-Cre mice, respectively. Indeed, expression of a subset of genes, which are bound by both Foxp3 and GATA-3, was significantly altered in the absence of GATA-3 suggesting a role for GATA-3 partnership with Foxp3 in regulation of a subsection of the Treg cell transcriptome (Fig. 6a). ChIP-qPCR and cDNA real-time experiments on a select set of candidate genes further confirmed these results by demonstrating specific changes in expression of genes that are occupied by both Foxp3 and GATA-3 (Fig. 6b–d).
Figure 6.
A subset of genes regulated by Foxp3 and Gata3 in Treg cells. (a) Cumulative distribution analysis of the differential gene expression in Gata3-sufficient and -deficient Treg cells for Foxp3- and Gata3-bound and Foxp3 only bound genes. *P < 0.00026 and **P < 5.4×106 (One-tailed Kolmogorov-Smirnov test). (b) ChIP-qPCR analysis demonstrating overlapping occupancy of Foxp3 and Gata3 on some of the target genes identified by genome wide ChIP-sequencing. Data represents two to three independent experiments. (c) Real-time PCR analysis of mRNA encoded by representative genes co-occupied by Foxp3 or Foxp3 and Gata3 in Treg cells purified by flow cytometry from Gata3fl/+ Foxp3YFP-Cre (WT) or Gata3fl/flFoxp3YFP-Cre (KO) mice. Relative expression is calculated by dividing the Hprt normalized expression values for each mRNA in KO over those in WT Treg cells. Data are shown as averages of three independent experiments. *P < 0.05 (Student’s t-test). (d) Real-time PCR analysis of mRNA encoded by representative genes from indicated cells sorted from heterozygous female Foxp3GFPKO/WT mice. Data represents six to nine replicates from two to three independent experiments.*P < 0.05, **P < 0.005 and ***P < 0.005 (Student’s t-test).
Next, we explored the functional significance of GATA-3 and Foxp3 co-regulation in Treg cells in unmanipulated Gata3fl/flFoxp3YFP-Cre mice. Although healthy at young age, Gata3fl/flFoxp3YFP-Cre mice developed intestinal pathology and dermatitis after 6 months (Supplementary Fig. 7a–c) accompanied by a marked Foxp3−CD4+T cell activation and increase in IL-4-, IL-5-, IL-13-producing and GATA-3+ TH2 cells whereas proportion of interferon-γ (IFN-γ)-producing T-bet+ cells was reduced (Supplementary Fig. 7d–f). Furthermore, in agreement to recent reports29, 30, we observed significant increase in IL-17 production primarily in the Foxp3+ Treg cells in Gata3fl/flFoxp3YFP-Cre mice (Supplementary Fig. 7g). The numbers of Treg cells in Gata3fl/flFoxp3YFP-Cre mice was increased compared to littermate controls, suggesting that the observed phenotype was not due to reduced Treg cell numbers (Supplementary Fig. 7h). Since the TH2 cytokine production was most significant in the mesenteric lymph nodes of the Gata3fl/flFoxp3YFP-Cre mice, we analyzed the large intestine-associated lamina propria lymphocytes (LI-LPL) where we observed a more pronounced selective increase in the magnitude of IL-4, IL-5 and IL-13 production by T effector cells (Supplementary Fig. 8a,b). Thus, in addition to reciprocal regulation of expression, Foxp3 and GATA-3 form a complex in activated Treg cells and lack of GATA-3 restricts the ability of Treg cells to control TH2 responses and restrict Treg cell-specific IL-17 production in unmanipulated mice.
Discussion
Lineage specification factors play a pivotal role in cellular differentiation by modulating expression of a core set of genes whose expression patterns define functional and phenotypic properties of a given cell type. Although the transcriptional programs guided by many lineage-specification factors and mechanisms of their expression have been extensively studied, comprehensive analysis of the complexes they form and relationships between their partners has been lacking. Foxp3 represents a rare example of a lineage specification factor with a specialized role in supporting differentiation and function of a single cell type, regulatory T cells.
Our biochemical and mass-spectrometric studies showed that Foxp3 forms unexpectedly large transcriptional complexes comprised of several hundred partners. In addition to a large number of newly identified partners, several Foxp3 interacting proteins (e.g. Runx, NFAT, SWI/SNF components, and Foxp1) previously identified by other groups, largely through the use of transfection experiments, have been confirmed by our analysis although we failed to observe interactions with others (e.g. TIP60, HDAC7, HDAC9, Eos, Irf4 and Hif1α)14, 15, 17–19, 36, 37, 38. Although two recent studies suggested that N-terminal fusion of GFP with Foxp3 protein modulates of Foxp3 protein complex composition in Foxp3GFP reporter mice, it seems unlikely that the aforementioned results are due to the use of biotin tagged Foxp3 protein as we have compared AVI-Foxp3 and endogenous Foxp3 complexes39, 40. Thus, the lack of a signal in our mass-spectrometry experiments from some factors expressed in Treg cells or associated with Foxp3 in an inducible manner is more likely due to dynamically modulated composition of Foxp3 complexes in response to various environmental cues (e.g. induction of GATA-3 and Foxp3 and their association with Foxp3 in response to TCR stimulation or induction of Hif1a in response to hypoxia). The largest group of Foxp3 partners consisted of proteins that have been implicated in regulation of transcription including a large number of sequence specific transcription factors including NFATc2, Runx1, Bcl11b, Foxp1, Foxp4, GATA-3, STAT3, Ikaros (Ikzf1), Aiolos (Ikzf3), Ets, and Cnot3. The latter observation is consistent with our recent finding that the majority of Foxp3 binding sites within the genome lack an identifiable forkhead-binding motif in Treg cells and suggests that to a large degree Foxp3 co-factors facilitate Foxp3 binding to a given site either through direct recruitment of Foxp3-containing complexes or through facilitating interactions with Foxp3-bound sites containing forkhead motif via loop formation (Refs.7, 8 and 49).
Importantly, examination of Foxp3 ChIP-Seq datasets and Foxp3-dependent changes in gene expression revealed that approximately 50% of Foxp3 partners within the “transcriptional control” category serve as direct targets of Foxp3 (Ref. 49). A high degree of enrichment in Foxp3 target genes among Foxp3 partners in comparison to the rest of the genome highlights Foxp3 mediated transcriptional control over a tightly regulated protein network it forms with its partners. Analysis of gene expression in Treg cells in knock-in mice expressing a Foxp3GFP reporter allele and CD4+T cells expressing Foxp3GFPKO reporter null allele or naive CD4+ T cells revealed that while many of Foxp3-bound factors, like GATA-3 and STAT3, are up-regulated in a Foxp3-dependent manner, some are down-regulated (e.g., Foxp1, Runx1, Bcl11b)9.These observations raise possibility that Foxp3-mediated up- or down-regulation of its binding partners allows for tuning their relative amounts associated with Foxp3 and, thereby, gene expression in Treg cells.
Another feature of a highly integrated regulatory network formed by Foxp3 and its partners was that some of the Foxp3-bound transcription factors are not only controlled by Foxp3, but also regulate Foxp3 gene expression by binding to its promoter and intronic enhancers in both thymus and extrathymically generated Treg cells. Indeed, targeted ablation of Runx1 or its essential co-factor Cbfβ in a Treg- or T cell-specific fashion results in a decreased expression of Foxp3 mRNA and protein on a per cell basis26, 27, 41. Furthermore, Runx1 and Foxp3 cooperatively facilitate stability of Foxp3 lineage upon Runx-dependent recruitment of Foxp3 to CNS2, an intronic Foxp3 enhancer with a non-redundant role in the heritable maintenance of Foxp3 gene expression in the progeny of dividing Treg cells35. NFAT1, an important partner of Foxp3, whose mouse homologue Nfatc2 was identified in our study, partakes in a multi-protein complex that forms an enhanceosome facilitating TGFβ-dependent Foxp3 induction28. Likewise, the Ets transcription factor family, two members of which were identified in our study (Elf1, Elf4), facilitates Foxp3 expression in thymic Treg cells32. Another identified partner STAT3 interacts with Foxp3 in an activation-dependent manner and contributes to control of pathogenic TH17 mediated inflammation; on the other hand, STAT3 limits Foxp3 induction in peripheral CD4+ T cells, by restricting access of Smad3 to Foxp3 enhanceosome 33, 42. Finally Bcl11b and GATA-3, two prominent partners of Foxp3, promote Foxp3 gene expression by direct binding to its regulatory elements29–31.
The large number of Foxp3 partners identified by mass-spectrometry and confirmed by co-immunoprecipitation and the broad distribution of Foxp3 during fractionation in a gel-filtration column suggested that Foxp3 complexes are heterogeneous. This notion is consistent with the fact that Foxp3 interactome encompasses proteins involved in both repression and activation of gene expression including the aforementioned NuRD, Lsd1 containing CoREST and N-CoR repressor complexes, Polycomb complex and its associated factor YY1, histone deacetylases HDAC1 and HDAC2, as well as nucleosome remodelers SWI/SNF, ISWI and MLL complexes. In agreement with the presence of Foxp3-associated key component Suz12 of the Polycomb complex, which mediates tri-methylation of K27 in histone H3, we previously reported enrichment in H3K27me3 at Foxp3 binding sites within the Foxp3-repressed genes8.
In addition to heterogeneity based on recruitment of chromatin modifiers with opposing functions, some of the sequence-specific transcription factors, such as STAT3 and GATA-3, partner with Foxp3 in an activation-dependent manner. Our previous studies showed that STAT3 as well as STAT1 and STAT4 are recruited into Foxp3 complexes upon activation in a particular cytokine environment leading to their phosphorylation and nuclear translocation of these transcription factors (ref. 42; AC and AR unpublished). Likewise, we found that GATA-3 was upregulated and formed complexes with Foxp3 upon TCR stimulation of Treg cells. These data suggest that some of the Foxp3 partners are activated in distinct inflammatory and tissue environments and their recruitment into large Foxp3 containing transcriptional complexes allows for integration of environmental cues and adaptive changes in Treg cell homeostasis, homing and functional capabilities.
It must be noted that the sheer size of Foxp3 complexes containing multiple interacting proteins and their likely heterogeneity complicate mechanistic causal studies of a role of individual components of these complexes using a site-directed mutagenesis approach as mutations in putative interaction sites between Foxp3 and its given partner might result in a loss or gain of other interacting proteins. Future studies involving biochemical isolation of individual Foxp3 complexes, their in vitro reconstruction and solution of their structures will inform mutagenesis efforts for selective elimination of Foxp3 partners under control of mass-spectrometric analysis. Nevertheless, our comparative analysis of binding sites of Foxp3 and one of its co-factors GATA-3 combined with the analysis of changes in gene-expression and function associated with the selective loss of GATA-3 in Treg cells revealed a subset of genes regulated by GATA-3-containing Foxp3 complexes. A relatively modest change in the cumulative expression of these genes upon deletion of Gata3 was likely due to its expression in a minor subset of cells within the total peripheral Treg population analyzed in these experiments. Interestingly, our real-time PCR analyses of Foxp3-deficient TFN cells and GATA-3-deficient Treg cells revealed that while for some genes (e.g., Sytl2 and Prodh) Foxp3 and GATA-3 act in a cooperative fashion, for others (e.g., Pde3b, Satb1 and Ets2) the effect appears to be antagonistic. It seems likely that in such cases Foxp3 counteracts the effect of GATA-3, which otherwise acts as a transcriptional activator for these genes. These observation was reminiscent of recent study where partnership of glucocorticoid receptor with STAT3 led to either up- or down-regulation of cooperatively controlled genes43. The observed increase in IL-4 and IL-5 producing GATA-3+ Foxp3− effector T cells and unprovoked autoimmunity associated with Gata3 ablation in Treg cells are consistent with the notion of “symmetric” requirements for some transcription factors (like T-bet, IRF4, and STAT3) involved in differentiation of effector T cells and their suppression by Treg cells 30, 37, 42, 44. In addition, increased production of IL-17 in GATA-3-deficient Treg cell is in agreement with recent findings suggesting a role for GATA-3 in suppressing RORγt-mediated IL-17 production in Treg cells29. Furthermore, impaired homeostasis of GATA-3-deficient Treg cells was observed in peripheral tissues during inflammatory response possibly due to a requirement for GATA-3 for maintaining high Foxp3 expression in rapidly dividing Treg cells30. The discrepancy between the observed phenotype and a much more severe pathology recently reported in mice harboring GATA-3-deficient Treg cells was likely due to a difference in expression of a knock-in Foxp3Cre allele and a Foxp3 BAC transgene-driven Cre recombinase employed in our and the other study29, respectively.
In conclusion, our biochemical and mass-spectrometric analyses revealed that Foxp3 forms transcriptional complexes of 400–800 kDa and larger and identified 361 associated proteins, many of which are involved in the regulation of transcription. Notably, Foxp3 binds and directly regulates expression of a large proportion of the genes that serve as its co-factors. Reciprocally, some of the sequence-specific transcription factors which serve as Foxp3 partners facilitate Foxp3 expression. Several distinguishing characteristics of Foxp3 complexes, specifically, association with NuRD repressor complexes and SWI/SNF nucleosome remodeling complexes, large number of protein partners, and most notably pronounced regulation of expression of its components, were reminiscent of features recently reported for Oct4, a principal transcription factor defining stem cell identity45. This notable similarity suggests that the observed principal features of Foxp3 transcriptional complexes are likely common to cell lineage specification transcription factors operating during early and late cellular differentiation and defining a particular cell fate. It is likely that multiple activating and inhibitory complexes with constitutive and inducible membership formed by a single lineage specification factor like Foxp3 enable multi-faceted organization of a specialized functional genome and confer a wide range of functions associated with a given state of cellular differentiation.
Methods
Experimental animals
Foxp3YFP-Cre mice have been described elsewhere47. Gata3fl/fl mice were generated and kindly provided by I-C. Ho (Harvard University 48. Mice were housed and bred under specific pathogen–free conditions in accordance with guidelines of the Institutional Animal Care Committee of Memorial Sloan-Kettering Cancer Center.
Generation of retroviral constructs and their expression
MigR1-AVI-Foxp3 and MigR1-AVITEV-Foxp3 constructs were generated by PCR amplification of the Foxp3 cDNA with forward primers “Primer1” or “Primer2”, respectively, and reverse primer “Primer3”. The PCR products were subcloned within BglII and EcoRI sites of the MigR1 vector. For constructing MigR1-AVI vector, the MigR1-AVI-Foxp3 construct was digested with the Hpa1 restriction enzyme, whose recognition site was introduced into “Primer1” and “Primer3”, and re-ligated. The ΔE250 mutant was generated using Stratagene Quickchange kit, “Primer4” and “Primer5” as mutagenesis primers, and MigR1-Foxp3 as a template. MigR1-AVI-Foxp3-IRES-BirA-T2A-Thy1.1 construct was generated by first performing a two part ligation of the PCR products of primers “Primer 6”, “Primer 7” (amplifying 5′-NcoI-BirA-BamHI-3′) and “Primer 8”, “Primer 9” (amplifying 5′-BamHI-T2A-Thy1.1-SalI-3′ from T2A-Thy1.1 containing vector) and sub-cloning the resulting fragment into NcoI and SalI digested MigRI (that releases GFP-coding sequence from the parent vector). AVI-Foxp3 was subsequently cloned into this construct as described above. Primer sequences are listed in Supplementary Table 5.
For retroviral transduction, CD25−CD4+ T cells were isolated from C57BL/6 mice by magnetic bead sorting (Invitrogen) and stimulated in 24-well plates (2 × 106 cells per well) pre-coated with 1 µg/ml of CD3 and CD28 antibodies in the presence of 50 IU/ml of recombinant IL-2 in DMEM supplemented with 10% (vol/vol) FCS, 200 mM L-glutamine, 1 mM sodium pyruvate, 50 µM 2-mercaptoethanol and antibiotics. T cell cultures were 'spin-infected' twice over a 48-h period with viral supernatants collected from the Phoenix packaging cell line transfected with retroviral constructs as described46. After infection, cells were expanded for additional three days and Thy1.1 and GFP expressing cells were sorted by flow cytometry for immunoblotting and in vitro suppression assays. For transduction of TCli cell line, 0.3 × 106 cells were transfected with retroviral constructs (MigR1-AVI-Foxp3, MigR1-AVI-ΔE250, MigR1-AVITEV-Foxp3 and MigR2-BirA) as described above.
Nuclear extract preparation, Foxp3 complex purification and fractionation
TCli cells (2 × 109) were resuspended in 15 ml cytoplasmic lysis buffer (10 mM HEPES, pH 7.9, 1.5mM MgCl2, 10 mM KCl, 0.1 mM DTT, plue Proteinase inhibitors) and incubated on ice for 30 min. Cells lysed in Down’s homogenizer and nuclei were pelleted by centrifugation at 550g for 30 min. The nuclear pellet was resuspended in 10 mM HEPES, pH 7.9, 100mM KCl, 3mM MgCl2, 0.1mM DTT, 20% glycerol in the presence of protease inhibitor cocktail (nuclear resuspension buffer), and nuclear extracts were prepared by drop wise addition of high salt containing extraction buffer 10 mM HEPES, pH 7.9, 2.2M KCl, 1.5mM MgCl2, 0.25mM EDTA, 20% glycerol until the final KCl concentration reached 300 mM. The nuclear lysates were treated with DNase (Invitrogen #18047–019; 20 Kunitz units/ml), and in some experiments with both DNase and RNase (Promega A7973; 10 µg/ml). The nuclear lysates were spun for 30 min at 15000g for 30 min and protein concentration was quantified. For immunoprecipitation, nuclear lysate was diluted in nuclear resuspension buffer without KCl to 150 mM KCl final concentration, supplemented with 0.1% NP40 and incubated with M-280 streptavidin magnetic beads (Invitrogen) pre-blocked with 200 µg/ ml chicken egg albumin for 4–5 h. The beads were washed six times with 10 mM HEPES pH 7.9, 250mM KCl, 1.5mM MgCl2, 0.25mM EDTA, 0.1% NP40 and resuspended in 2x Laemmli buffer. For TEV cleavage, beads were resuspended in 10 mM Tris-HCl, 150 mM NaCl, pH 7.5 buffer containing 0.1% NP40, 0.5 mM EDTA, and 1 mM DTT and cleaved with TEV enzyme upon overnight incubation at 4 °C.
Fractionation of TEV cleaved Foxp3 complexes
TEV eluted Foxp3 complexes were loaded onto a Superose 6 [GE Healthcare) equilibrated in 200 mM KCl, 20 mM HEPES pH 7.9. 1.0 mM MgCl2 0.5mM EGTA and 10% Glycerol and fractionated at a flow rate of 400 µl/min. Collected fractions were immediately precipitated with TCA and the pellets were washed 2X with ice-cold acetone, dried and resuspended in 2x Laemmli buffer.
Mass-spectrometric analysis
Proteins released from the beads were separated by 4–20% gradient SDS polyacrylamide gel electrophoresis, stained with colloidal Coomassie (Pierce) and washed with HPLC-grade water prior to cutting bands from the gel lanes and digesting with sequencing grade modified trypsin (Promega). Peptide digests were analyzed by electrospray ionization in the positive ion mode on a hybrid linear ion trap-Fourier transform-ion cyclotron resonance mass spectrometer (LTQ-FT; Thermo Electron Corp.). A Michrom Bioresources, Paradigm MS4B MDLC HPLC was used to separate peptides on a home-made 75 micron analytical fused silica capillary column packed with ~ll cm of 5 micron C18 beads (C18AQ; Michrom Bioresources) with gravity-pulled tapered tip run at a flowrate of 200 nl/min. Peptides were eluted by acetonitrile gradient consisting of three mobile phases: A, H20; B, CH3CN; and C, 1% (v/v) formic acid. The gradient program was: 0–5 min, A (85%), B (5%), C (10%); 60 min, A (55%), B (35%), C (10%); 65–74 min, A (10%), B (80%); C (10%), 75min, A (85%), B (5%), C (10%). For MS, ICR resolution was set to 100,000 (M/Z 400) and ICR ion populations were held at 1e6 through the use of automatic gain control. For MS/MS in the linear ion trap the ion population was set to 1e4, the precursor isolation width was set to 2Da and the collision energy was set to 35%. Data was acquired using an MS “survey” scan in the ICR followed by MS/MS data-dependent selection of the 5 most abundant precursors from the survey scan in the linear ion trap. Data was acquired using Xcalibur, Version 1.4 (Thermo) and analyzed using SEQUEST data analysis program.
Co-immunoprecipitation and immunoblot analysis
For co-immunoprecipitation studies demonstrating interaction between Foxp3 and candidate proteins in primary Treg cells, nuclear complex co-IP kit (Active Motif) was used according to the manufacturer’s instructions. Briefly, cells were washed with PBS supplemented with phosphatase inhibitors, lysed in hypotonic buffer and nuclei were separated from cytoplasm by a short spin and nuclear extract was prepared by re-suspending the nuclear pellet in digestion buffer and incubated in 4 °C with enzymatic cocktail for 90 min. Immune complexes were captured by protein A-conjugated magnetic beads, washed thoroughly, and resuspended in Laemmli buffer for SDS-PAGE fractionation and immunoblot analysis. The following antibodies were used for immunoblotting: anti-HDAC1 (clone 2E10, Millipore #05-614), anti-HDAC2 (clone 3F3, Millipore #05-814), anti-Bcl11b (clone 25B6, Abcam #18465), anti-YY1 (clone 13G10, Cell Signaling #2185), anti-Kdm1a (clone C69G12, Cell Signaling #2184S), anti-Foxp1 (Cell Signaling #2005S), anti-Ash2l (clone D93F6, Cell Signaling 5019S), anti-H3 (Cell signaling #9715S), anti-RCOR1 (Lifespan Biosciences #LS-C14592), anti-SMARCC1 (Bethyl Laboratories #A301-019A), anti-Snf2h (Bethyl laboratories A301-017A), anti-SMARCA4 (Lifespan Biosciences #LS-C89131), anti-CHD4 (Lifespan Biosciences #LS-C79110), anti-GATA-3 (clone L50-823, BD Pharmingen #558686); Cbfβ antibody was provided by Ichiro Taniuchi, RUNX1 antibody was provided by Dan Littman, Ikzf1 (clone 4E9) and Ikzf3 (clone 9010) antibodies were provided by Katia Georgopoulos.
cDNA real-time PCR (qPCR) and ChIP-qPCR analyses
Total RNA was isolated and prepared from sorted populations of cells using Trizol reagent and cDNA was synthesized using oligo(dT) primers and a SuperScript III First-Strand Synthesis system (Invitrogen).
Foxp3 and GATA-3 ChIP were performed using purified rabbit anti-Foxp3 and mouse anti-GATA-3 (BD Pharmingen) as described before26. The relative abundance of regions of interest in precipitated DNA was measured by qPCR using Power SYBR Green PCR master mix (Applied Biosystems). Real-time PCR primer sequences are listed in Supplementary Table 5.
Processing of ChIP-seq data
ChIP of Foxp3 and GATA-3 were obtained from ref. 49 and ref. 32, respectively. Peaks were called by the R SPP package and ranked by number of reads aligned after strand-specific shift of 75 nt. For analyses that called for a fixed number of Foxp3 peaks, the top 4000 peaks were used. For analyses that called for a fixed number of GATA-3 peaks, the overlapping top ranked peaks in both nTreg (n = 5283) and iTreg (n = 2495) ChIP-seq experiments resulting in 1299 peaks were used. In all analyses, binding sites were linked to most proximal gene.
Statistical analysis
Significance of Foxp3 binding of Foxp3 partners was established by comparing to all genes bound (21289 Refseq annotated transcripts) using a two-tailed Fisher’s exact test (Fig. 4a). Significance of expression shift in Foxp3 partners and targets was determined by one-tailed Kolmogorov-Smirnov test relative to expression change of all expressed genes.
Supplementary Material
Acknowledgments
We would like to thank S. Lee, A. Bravo, J. Herlihy and J. Gerard for help with the mouse colony management and technical assistance; I-Cheng Ho for Gata3f/f mice, Dan Littman for Runx1 antibody, Ichiro Taniuchi for Cbfβ andtibody and K. Georgopoulos for Ikzf1 and Ikzf3 antibodies. This work was supported by NIAID R37 AI034206 grant (A.Y.R.). D.R was supported by Arthritis Foundation postdoctoral fellowship. A.C. is supported by the Irvington Institute Fellowship Program of the Cancer Research Institute. R.E.N is supported by NIH MSTP grant GM07739 and NINDS grant 1F31NS073203-01. R.M.S. is supported by NIH DK091968 and MSTP grant GM07739. A.Y.R. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Accession codes. GEO: gene expression data, # pending.
Author contributions
D. R. designed and performed the majority of the experiments, analyzed the data and wrote the manuscript, experiments; P. D. performed chromatography and protein purifications experiments, and was involved in mass-spectrometric analysis; A. C. was involved in co-immunoprecipitation studies; R. N. was involved in functional analysis of Gata3flFoxp3YFP-Cre mice; R. M. S. performed Foxp3 ChIP-Seq and gene expression analyses in Treg and TFN cells; A. A. and C. L. performed computational and statistical analysis of ChIP-Seq and gene expression datasets; S. A. S. and D. R. G. assisted with mass-spectrometric analyses; A. Y. R. directed the project and was involved in design of the experiments, data analysis and interpretation, and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T Cells: Mechanisms of Differentiation and Function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol. 1991;138:1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 5.Wildin RS, Freitas A. IPEX and FOXP3: clinical and research perspectives. J Autoimmun. 2005;25(Suppl):56–62. doi: 10.1016/j.jaut.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 7.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 9.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 11.van der Vliet HJ, Nieuwenhuis EE. IPEX as a result of mutations in FOXP3. Clinical & developmental immunology. 2007;2007:89017. doi: 10.1155/2007/89017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Bras S, Geha RS. IPEX and the role of Foxp3 in the development and function of human Tregs. J Clin Invest. 2006;116:1473–1475. doi: 10.1172/JCI28880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 14.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 16.Chae WJ, Henegariu O, Lee SK, Bothwell AL. The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc Natl Acad Sci USA. 2006;103:9631–9636. doi: 10.1073/pnas.0600225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 20.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 21.Morkowski S, et al. T cell recognition of major histocompatibility complex class II complexes with invariant chain processing intermediates. J Exp Med. 1995;182:1403–1413. doi: 10.1084/jem.182.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Boer E, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotech. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 24.Chen YI, et al. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucl Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000470. e1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitoh A, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Ruan Q, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wohlfert EA, et al. GATA3 controls Foxp3 regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanvalkenburgh J, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208:2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouly E, et al. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, et al. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33:313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Song X, Li B, Greene MI. FOXP3 and its partners: structural and biochemical insights into the regulation of FOXP3 activity. Immunol Res. 2008;42:19–28. doi: 10.1007/s12026-008-8029-x. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darce J, et al. An N-terminal mutation of the foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bettini ML, et al. Loss of epigenetic modification driven by the foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klunker S, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhry A, et al. CD4+ Regulatory T Cells Control TH17 Responses in a Stat3-Dependent Manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR Interaction Code: Predictive Value of Direct/Indirect DNA Recruitment for Transcription Outcome. Molecular Cell. 2012;47:38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 44.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardo M, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell stem cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubtsov YP, et al. IL-10 produced by regulatory T cells contributes to their suppressor function by limiting inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 48.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 49.Samstein RM, et al. Foxp3 exploits a preexistent enhancer landscape for regulatory T cell lineage specification. Cell. doi: 10.1016/j.cell.2012.06.053. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.