Abstract
The expression of CD10 has long been used to define human lymphoid commitment. We report a unique lymphoid-primed population in human bone marrow that was generated from hematopoietic stem cells (HSCs) before the onset of CD10 expression and B cell commitment. This subset was identified by high expression of the homing molecule L-selectin (CD62L). CD10−CD62Lhi progenitors possessed full lymphoid and monocytic potential, but lacked erythroid potential. Gene expression profiling placed the CD10−CD62Lhi population at an intermediate stage of differentiation between HSCs and lineage-negative (Lin−) CD34+CD10+ progenitors. L-selectin was expressed on immature thymocytes and its ligands were expressed at the cortico-medullary junction, suggesting a possible role in thymic homing. These studies identify the earliest stage of lymphoid priming in human bone marrow.
Although much is known about the identity of progenitor stages in murine lymphopoiesis, considerably less is understood about the critical stages of lymphoid commitment of human hematopoietic cells. Early models developed from murine studies assumed strictly dichotomous pathways of lineage commitment1. These concepts have evolved more recently into models of gradual loss of lineage potential that can occur via multiple alternative pathways, although the physiological relevance of lineage potential revealed in certain in vitro assays continues to be debated2–5. A stage in which murine bone marrow (BM) progenitors are “lymphoid primed” prior to complete loss of myeloid potential has been defined based on expression of the FLT3 cell surface receptor, and termed the Lymphoid-primed Multipotent Progenitor (LMPP)2.
Critical species-specific differences create challenges when translating knowledge of cellular hierarchies derived from murine studies to the specifics of human hematopoiesis6. In addition, the source and stage in ontogeny of human hematopoiesis can influence the functional capacity, surface immunophenotype and transcriptional profiles of the cells under study6–8. Most studies of the earliest progenitor stages in human hematopoiesis have used neonatal umbilical cord blood as the source of hematopoietic cells. However, to understand how lymphopoiesis is regulated during steady-state adult hematopoiesis it is necessary to directly study hematopoietic stem cells and progenitors from postnatal human BM8,9.
The stepwise process of lymphoid differentiation from multipotent hematopoietic stem cells (HSCs) in human BM has been assumed to begin with the expression of the cell surface antigen CD10 (aka CALLA, MME) on CD34+ cells10. However, while CD34+lin−CD10+ cells can give rise to cells of all lymphoid lineages, subsequent work has shown that CD10 expression on progenitors is associated with a strong bias toward B cell potential and minimal T and natural killer (NK) cell potential11,12. CD34+lin−CD10+ cells that lack expression of CD24 are precursors of the CD34+lin− CD10+CD24+ population, but nonetheless show molecular evidence of B cell commitment with expression of PAX5, EBF1 and VPREB12. Therefore, to understand the progenitor hierarchy of human lymphoid commitment, we sought to identify a stage of lymphoid priming that precedes B lymphoid commitment, either prior to or independent of CD10 expression.
L-selectin (CD62L) is expressed on lymphocytes and mediates homing to peripheral lymphoid organs13. Recent studies have reported that up-regulation of CD62L expression on c-Kit+lin−Sca1+ murine BM cells correlates with loss of erythroid and megakaryocyte potential and efficient thymic engraftment14–16. In this study we have identified a CD34+lin−CD10− progenitor subpopulation in human BM that expressed high amounts of L-selectin and was devoid of clonogenic myeloid or erythroid potential. In stromal cultures these cells were able to generate B, NK and T cells as well as monocytic and dendritic cells, similar to the previously described LMPPs in murine BM2. CD34+lin−CD10−CD62Lhi (“CD10−CD62Lhi”) cells rapidly engrafted immune-deficient mice, producing B and myeloid cells. Despite evidence of lymphoid skewing, comprehensive molecular analysis revealed that CD10−CD62Lhi cells not only lacked B cell specific transcripts, but also had not initiated DNA recombination based on absent RAG1, RAG2 and minimal DNTT expression. Genome-wide expression and functional analysis placed the CD10−CD62Lhi progenitor population as a developmental intermediate between the multi-potent CD34+lin−CD38− population and the CD34+lin−CD10+ lymphoid progenitor.
We also find that primitive lymphoid-restricted CD34+CD1a− progenitors in human thymus expressed CD62L, and that the vasculature at the cortico-medullary junction of human thymus expressed ligands for CD62L, suggesting the possibility that L-selectin may play a role human thymic homing. We propose that the CD10−CD62Lhi progenitor in BM represents the earliest stage at which adult human progenitors become lymphoid-primed. The identification of this progenitor population will facilitate a more complete understanding of the regulation of lymphoid commitment from HSCs during normal and aberrant human hematopoiesis.
RESULTS
CD7 expression does not define lymphoid commitment
In view of previous studies by our group and others linking CD7 expression to early stages of lymphoid commitment in umbilical cord blood17–20, we first investigated if expression of CD7 was sufficient to identify human lymphoid commitment in bone marrow, independent of CD10 expression. Examination of lineage-depleted cells revealed that the CD34+lin−CD38−CD7+ population previously identified in umbilical cord blood17 was not detectable in human BM (Supplementary Fig. 1a). However, as previously noted7, low expression of CD7 was detected on a small (2.8 ± 0.6%, n = 5) population of CD34+lin−CD38+ human BM cells, most of which did not co-express CD10 (Fig. 1a). Clonogenic assays demonstrated that CD7 expression alone was insufficient to define lymphoid restriction within the CD34+lin−CD10− population of BM; non-lymphoid clonogenic cells, particularly erythroid progenitors, were readily detectable in the CD34+lin−CD10−CD7+ population by Colony Forming Unit-Cell (CFU-C) assay (Fig. 1b). Consistent with previous studies in BM and umbilical cord blood7,10–12,21, CD34+lin−CD10+ progenitors were devoid of clonogenic myeloid and erythroid progenitors (Fig. 1b).
Figure 1. Identification of BM progenitors that lack myeloid and erythroid clonogenic potential.
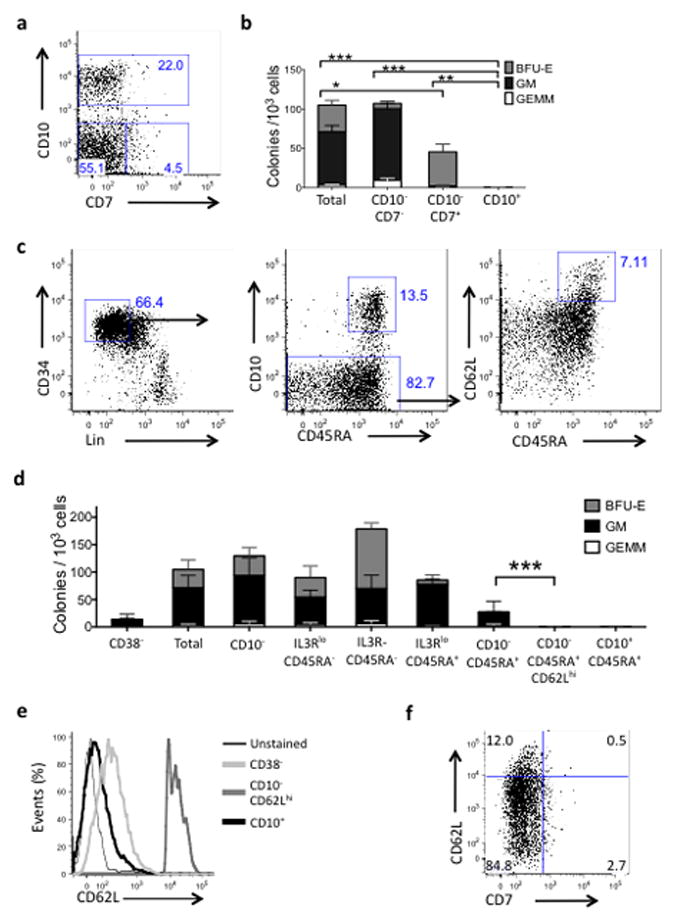
(a) CD7 and CD10 expression on CD34+lin−Bone Marrow (BM) cells (representative of 7 independent experiments). Error bars represent SEM. * = p<0.050, ** = p<0.010, *** = p<0.001 (b) Myeloid and erythroid clonogenic output in methylcellulose assay of the CD34+lin− subsets shown. (Total n=4, CD10−CD7− and CD10−CD7+ n=2, CD10+ n=5) (c) Flow cytometry isolation strategy of CD34+lin−CD45RA+10+ (“CD10+”) and CD34+lin−CD45RA+CD10−CD62Lhi (“CD10−CD62Lhi ”). Over 30 independent BMs examined (see also Fig S3). (d) Myeloid and erythroid clonogenic capacity of each subset shown. All populations shown, including “total” are CD34+lin−. (p<0.001 comparing CD10−CD45RA+ and CD10−CD45RA+CD62Lhi; frequency was also significantly decreased in CD10− CD62Lhi and CD10+ relative to all other populations shown, see Table S1a, b). IL3Rlo CD45RA−(“CMP”), IL3R−CD45RA−(“MEP”), and IL3Rlo CD45RA+ (“GMP”). (e) CD62L expression on CD34+lin−populations shown, representative of over 20 independent BMs. (f) Flow cytometry of gated CD34+lin−CD10− cells showing lack of CD7 expression on CD62Lhi cells, representative of 5 independent BM. [For all phenotypes, lin− is defined as negative for CD3, CD14, CD15 (aka FUT4), CD19, CD56 (aka NCAM1), and CD235a (aka GYPA)].
L-selectinhi progenitors do not possess CFU potential
CD45RA has previously been shown to be expressed on various lymphoid progenitors10,17–19 and granulocyte-macrophage progenitors (GMPs)22. Analysis of the CD34+lin−CD10− subpopulation demonstrated the presence of both CD45RA− and CD45RA+ fractions; in contrast all CD34+lin−CD10+ cells expressed CD45RA (Fig. 1c). Erythroid potential was depleted, but clonogenic myeloid progenitors (CFU-GM) were still readily detectable, in the CD34+lin−CD10−CD45RA+ population (Fig. 1d). As expected, erythroid potential was high in CFU-C from megakarocytic-erythroid progenitors (MEPs) and common myeloid progenitors (CMPs) (Fig. 1d), neither of which express CD45RA.
Further refinement of the CD10−CD45RA+ population was necessary to identify those cells that lacked clonogenic myeloid potential. L-selectin (CD62L) is a cell surface receptor that mediates lymphocyte homing to peripheral nodes13 and which is expressed on certain murine BM progenitors that lack erythroid or megakaryocytic potential14. Analysis of the CD34+lin−CD10− CD45RA+population demonstrated that although most cells dimly expressed CD62L, a distinct subpopulation (9 ± 1.5%, n = 14) of CD34+lin−CD10−CD45RA+ cells in normal human BM highly expressed CD62L (Fig. 1c). Functional screening of CD34+lin− fractions in CFU-C assay demonstrated that only the CD34+lin−CD10−CD45RA+CD62Lhi (“CD10−CD62Lhi ”) population and the CD34+lin−CD10+(“CD10+”) population were devoid of clonogenic myelo-erythroid potential (Fig. 1d, Supplementary Table 1). Of note, the CD34+lin−CD10−CD45RA+ population that expressed intermediate amounts of CD62L contained low but detectable CFU-C potential, suggesting that progressive loss of multi-potency correlates with increasing CD62L expression (population B, Supplementary Fig. 1b,c).
CD10+ cells expressed low or undetectable amounts of CD62L, and the CD34+lin−CD38− population, which is highly enriched for HSC and multipotent progenitor cells (MPPs), showed intermediate expression of CD62L (Fig. 1e, Supplementary Fig. 1b). Notably, CD10− CD62Lhi cells did not express CD7 (Fig. 1f). Thus, the progenitor subset with highest CD62L expression expressed neither CD10 nor CD7, markers previously relied upon for the isolation of human lymphoid progenitors. Analysis of BM from 20 different individuals from infancy to adulthood consistently showed the presence of CD10− CD62Lhi cells (Supplementary Fig. 2a–c).
Lymphoid and monocyte potential of CD10−CD62Lhi cells
Culture in lymphoid conditions demonstrated that the CD10− CD62Lhi population robustly generated both B and NK cells (Fig. 2a). Consistent with previous studies, CD10+ cells (all of which were CD19− through lineage depletion) generated mostly B cells with relatively weak NK potential11. Cell output under B-NK lymphoid conditions tended to be higher in cultures initiated with CD10− CD62Lhi than with CD10+ cells (Fig. 2b). Following in vitro culture under T cell conditions, CD10−CD62Lhi cells generated cells with the immunophenotype typical of thymocytes (expressing CD1A, CD7, CD4, CD8, CD3, TCRαβ)23 (Fig. 2c,d and Supplementary Fig. 3), and expressing the T cell associated genes TCF7, GATA3, DNTT and RAG1 (Supplementary Fig. 3) as well as CD56+ NK cells (some of which co-expressed CD8). Cell output was significantly higher in T cell cultures initiated with CD10−CD62Lhi than with CD10+ cells (P = 0.038) (Fig. 2e).
Figure 2. Lympho-myeloid potential of bone marrow progenitors.
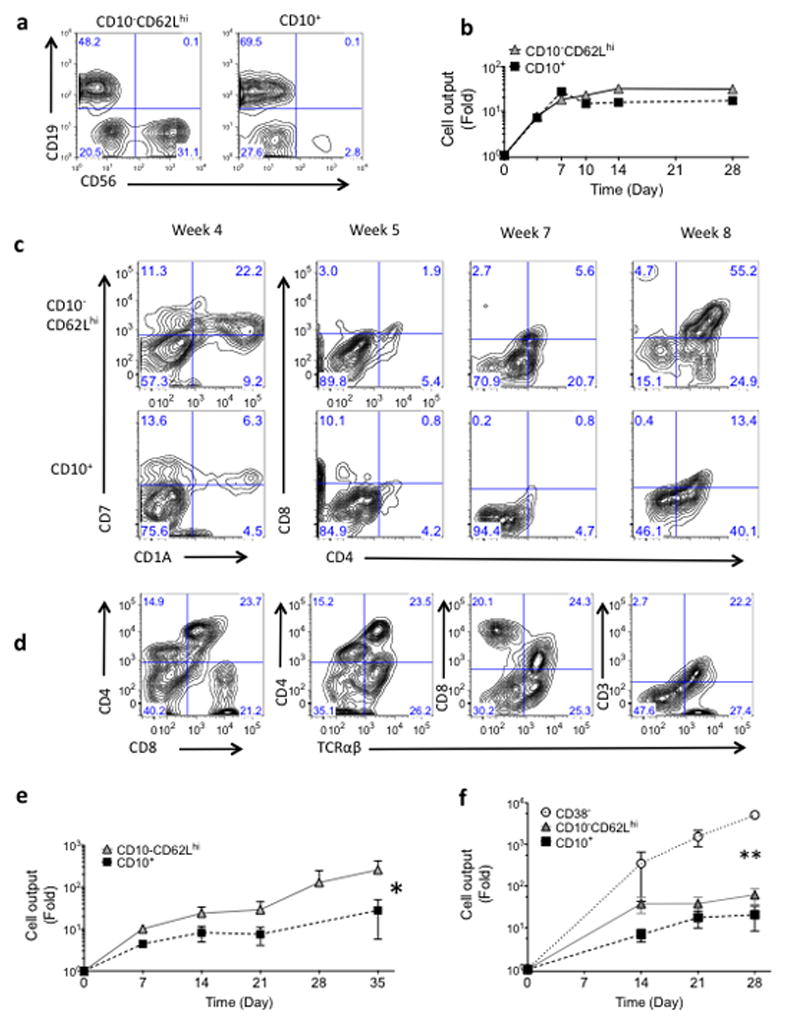
Analysis of B-NK cultures (a,b) and T cell cultures (c,d,e) initiated with CD10−CD62Lhi or CD10+ cells. (a) Flow Cytometry analysis of 4 week B-NK lymphoid cultures (OP9 with SCF, FLT-3, TPO); CD19 (B lymphoid) CD56 (NK cells); (one representative of 10 independent experiments). (b) Cell output (fold increase from Day 0) of CD34+lin− populations in B-NK lymphoid conditions. (c,d) Flow Cytometry analysis of T Lymphoid cultures (OP9-DL1 stroma with SCF, FLT-3, and IL7.) (e) CD10− CD62Lhi cells generate significantly more cells in T cell conditions than do CD10+ cells. Shown is fold increase from Day 0 of total cells generated from bulk cultures of CD34+lin−populations shown (* p <0.038, n=6 experiments). (f) Cell growth (fold increase from Day 0) of BM CD34+lin− populations in myelo-erythroid co-culture (OP9 stroma with IL-3, TPO, SCF, EPO, and FLT-3). HSC (CD34+lin−CD38− cells) generate significantly more cells in myelo-erythroid conditions than either CD10−CD62Lhi or CD10+ cells (** p <0.0001 for CD38− vs either CD10− CD62Lhi or CD10+. p = 0.49 for CD10− CD62Lhi vs CD10+, n=3 independent experiments).
Although clonogenic myeloid cells were not detected in CFU-C assay, both the CD10+ and CD10− CD62Lhi subsets were able to generate relatively low numbers of myeloid cells when cultured on stromal layers; however cell output from both progenitor types was significantly reduced relative to HSC-MPPs (P < 0.0001) (Fig. 2f). The majority of the non-lymphoid cells generated in stromal co-culture from the CD10+ and CD10−CD62Lhi populations were CD14+CD33+ monocyte-macrophages or CD209+CD1a+ dendritic cells (Supplementary Fig. 4); CD66b+ granulocytes were uncommon. Erythroid differentiation was rarely seen from CD10+ or CD10−CD62Lhi cultures but was robust in cultures from CD38− HSC-MPPs.
Cloning efficiency of CD10−CD62Lhi cells in lymphoid cultures initiated with single cells (~11%) and by limiting dilution analysis (1 in 5.3 for B-NK and 1 in 5.6 for T cell cultures) (Fig. 3a,b) was similar to that of CD10+ cells (~12% from single cells). However lineage analysis of clones demonstrated that the CD10−CD62Lhi population contained bi-potent B-NK progenitors whereas the CD10+ population contained predominantly unipotent B cell progenitors (Fig. 3c). Myeloid cells were detected in 86% of clones that could be assigned lineages in B-NK conditions (Fig. 3d) and 97% of all clones assayed from T cell cultures (Fig. 3e).
Figure 3. Lineage potential of CD10− CD62Lhi cells by in vitro clonal analyses and in vivo transplantation studies.
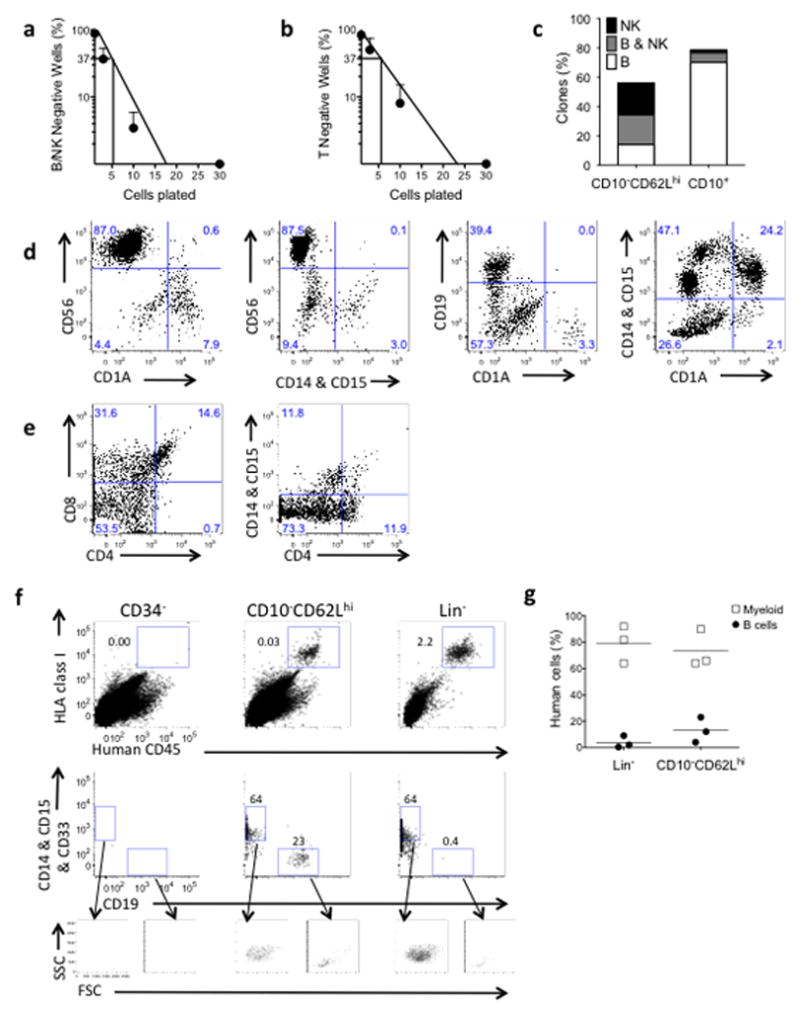
(a,b) Limiting dilution analysis of CD10− CD62Lhi cells grown in either : (a) B-NK conditions (cloning efficiency 1 in 5.3, 95% confidence interval 1 in 4.4–6.4, n=3 experiments), or (b) T cell conditions (cloning efficiency 1 in 5.6, 95% confidence interval 1 in 4.6–6.9, n=3 experiments). (c) Lineage analysis of clones from single CD10− CD62Lhi or CD10+ cells in B-NK lymphoid co-culture. Shown is percentage of wells with clonal growth containing B cells, NK cells or both. (d) FACS analysis of clones generated in B-NK conditions from 1–3 CD10− CD62Lhi cells showing NK (CD56+), myeloid (CD14 & CD15) and dendritic (CD1a) potential from one clone (first two panels); B (CD19+) and dendritic (CD1a+) (third panel); co-expression of myeloid and dendritic markers from single cell clone (panel far right). (e) FACS analysis of a single clone generated in T conditions showing T (CD4+CD8+) and myeloid (CD4dimCD14+&CD15+) potential. (f) FACS analysis of NSG mouse BM analyzed 2wk post transplant with 30,000 CD34+lin−CD10− CD62Lhi cells (center) or 150,000 CD34+lin− cells (right). Negative control mouse (left) received 100,000 irradiated CD34− carrier cells only. Human engraftment shown top row as hCD45+HLA-Class 1+ cells, middle row shows B (CD19+) cells and myeloid (CD14, CD15, & CD33+) cells from gated human cells, bottom row shows backgating of B and Myeloid cells shown in each panel above. (g) Compiled data from transplant experiments (n=3 mice each group).
Consistent with the in vitro assays of lineage potential, intratibial transplantation of CD10−CD62Lhi progenitors into immune-deficient (NOD–SCID–IL-2Rγ−/−) mice produced rapid marrow engraftment of both myeloid and B lymphoid cells (Fig. 3f,g and Supplementary Fig. 5). T lymphoid differentiation from non-self renewing progenitors would not be expected in this xenogeneic adult mouse model.
In summary, functional assays showed that the CD10−CD62Lhi population possessed full T, B and NK lymphoid potential, was less skewed toward the B lineage than the CD10+ population, and had greater T potential than CD10+ population. Although depleted of clonogenic myelo-erythroid potential, some myeloid (mostly monocyte-macrophage and dendritic cell) differentiation could be induced in stromal co-cultures from the CD10−CD62Lhi population, and in short term engraftment assays. However, myeloid potential was markedly decreased relative to that of HSC-MPPs and erythroid potential was absent.
Differentiation stages of HSCs and lymphoid progenitors
In view of the lineage potential shown in the functional studies, we next explored the relative stages of differentiation of the CD10−CD62Lhi and CD10+ populations when compared to the most primitive CD34+lin−CD38− (“CD38-”) HSC-MPP population. Expression of the differentiation marker CD38 rose progressively from the CD34+CD38− to the CD10−CD62Lhi population and was maximal in the CD10+ population (n = 14) (Fig. 4a). Expression of the stem cell-associated receptors KIT, FLT3, ITGA6 (aka CD49f) and PROM1 (aka CD133) was similar in CD38− and CD10−CD62Lhi populations but down-regulated in CD10+ cells; THY1 (aka CD90) was most highly expressed on CD38− cells. HLA-DR was up-regulated in both CD10−CD62Lhi and CD10+ progenitors (Fig. 4a). After one week in lymphoid cultures, CD10+ cells differentiated and lost CD34 expression faster than CD10−CD62Lhi cells (Fig. 4b). In addition, CD10−CD62Lhi cells were able to generate CD34+CD10+ cells in vitro, suggesting that CD10−CD62Lhi cells are precursors to the CD10+ population (Fig. 4b).
Figure 4. CD10−CD62Lhi cells represent an intermediate stage of differentiation between HSC and CD10+ progenitors.
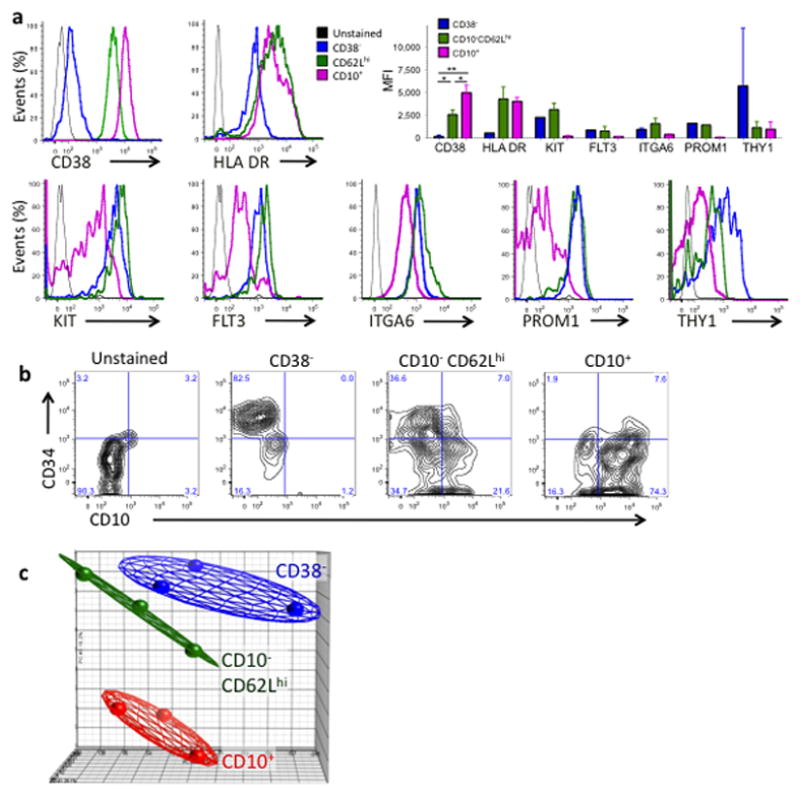
(a) Flow Cytometry showing co-expression of key cell surface markers on three CD34+lin−populations. Bar graph shows Mean Florescent Intensity (MFI) summarized from 2 or 3 independent samples for each marker, n=14 for CD38. (* = p<0.010, ** = p<0.001) (b) Flow Cytometry analysis of one week B-NK lymphoid cultures initiated with CD38−, CD10−CD62Lhi or CD10+ cells. (c) Unsupervised whole genome Principal Component Analysis of three independent BM samples.
Principal components analysis performed on global gene expression data from microarrays on three different BM samples also placed the CD10−CD62Lhi progenitors in an intermediate position between the CD38− HSC-MPPs and the CD10+ progenitors (Fig. 4c). Gene expression of CD10−CD62Lhi progenitors clustered hierarchically with CD38− HSC-MPPs rather than with CD10+ progenitors (Supplemental Fig. 6a). In pairwise comparison to HSC-MPPs, similar numbers of genes were up-regulated in CD10−CD62Lhi and CD10+ populations; approximately half of these up-regulated genes were common to both progenitor types (Supplemental Fig. 6b). More than twice as many genes were down-regulated in the CD10+ population than were down-regulated in the CD10−CD62Lhi population, and most downregulated genes in CD10−CD62Lhi cells were also downregulated in CD10+ cells (Supplemental Fig. 6b). Thus differentiation of HSC-MPPs involves many shared molecular pathways but additional transcriptional modulation appears to occur after the CD10−CD62Lhi stage during the generation of CD10+ cells.
Downregulation of HSC-associated genes in CD10−CD62Lhi cells
Analysis of expression patterns of genes known to regulate critical hematopoietic stages of differentiation was then performed by microarray and quantitative PCR (qPCR) to dissect the molecular relationships amongst CD38−, CD10−CD62Lhi, and CD10+ populations. All genes included in the heatmaps were at least 2-fold differentially expressed (P <0.05), and belonged to one of 6 different expression patterns (clusters 1–6) (Fig. 5a,b). Known HSC-related transcription factors (TAL1, GATA2, and PRDM16) were significantly down-regulated in both the CD10−CD62Lhi and CD10+ cells relative to the CD38− population (cluster 1, Fig. 5a). HOXB genes were also downregulated during the transition of the CD38− HSC-MPP to the CD10−CD62Lhi LMPP stage with no significant further change at the CD10+ stage (cluster 1). In contrast, expression of HOXA genes decreased later in differentiation at the CD10+ progenitor stage (cluster 2 and 3). Reciprocal patterns of expression were seen for members of the polycomb repressive complexes PRC1 (PCGF2, PHC2, and SCML4) (cluster 1) and PRC2 (SUZ12, EZH2 and EED) (cluster 5)24. These analyses reveal a highly coordinated program of transcriptional regulation as HSC lose multipotency, become lymphoid-primed and then commit to B lymphopoiesis.
Figure 5. CD10−CD62Lhi cells represent a distinct progenitor population with a unique expression profile that combines hematopoietic stem cell and early lymphoid genes.
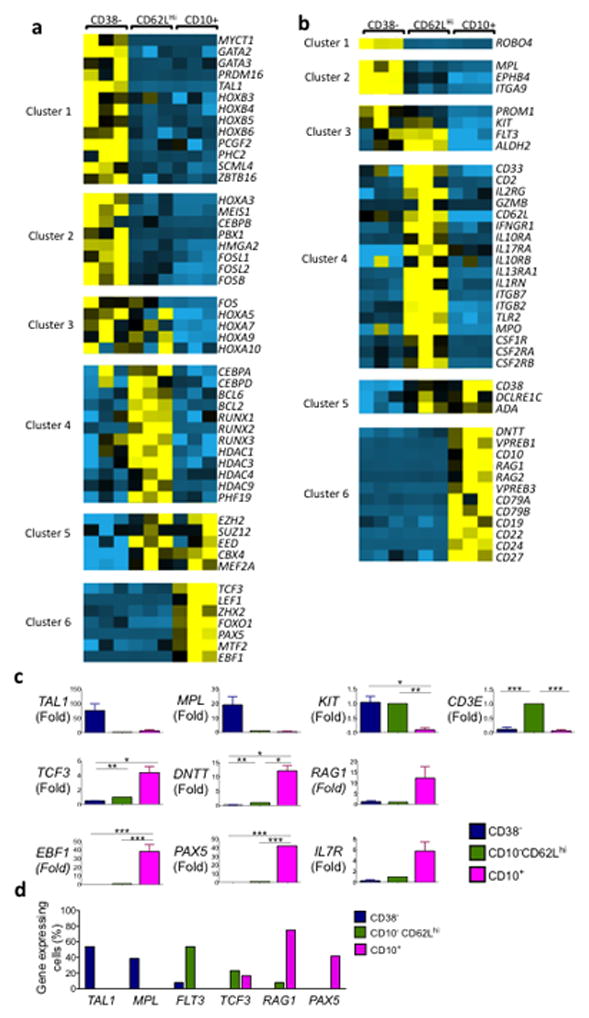
(a) Gene expression of transcription factors or (b) cytoplasmic and cell surface molecules. Genes included in cluster designations of heatmap were all more than 2 fold differentially regulated in pair-wise comparisons (p <0.05), and based on statistical analysis (not heatmap appearance) defined as Cluster 1: upregulated only in CD38−, and other 2 populations equivalent i.e. CD38− > (CD10−CD62Lhi = CD10+); Cluster 2: CD38− >CD62Lhi >CD10+; Cluster 3: (CD38− = CD10−CD62Lhi) >CD10+; Cluster 4: CD10−CD62Lhi > (CD38− =CD10+); Cluster 5: (CD10−CD62Lhi = CD10+) >CD38−; Cluster 6: CD10+ > (CD10−CD62Lhi = CD38−). (c) qPCR for selected genes, each normalized to CD10−CD62Lhi (n= 3 biological replicates, * = p≤0.050, ** = p<0.010, *** = p<0.001) mean ± SEM (d) qPCR assay of expression of selected genes in single cells using Fluidigm Biomark 48.48 analyzer (bars represent percentage of single cells tested expressing gene transcript, n=13 cells analyzed per each gene).
Lymphoid differentiation stages of CD10-CD62Lhi and CD10+ cells
Analyses of genes up-regulated only in the CD10− CD62Lhi population (Cluster 4) revealed a profile consistent with the dual lymphoid and monocyte potential of this population. Specifically, early T and NK lineage-associated genes (CD2 and CD3E)14,25–27 and lymphoid cytokine receptors (IL2RG, IL10RA, IL10RB, IL17RA, IFNGR1) were up-regulated, as were myeloid associated genes (MPO, CSF1R, and CSF2R) (Fig 5b, c). Consistent with its cell surface expression, FLT3 was expressed in both HSC-MPP and CD10− CD62Lhi cells but not CD10+ cells (cluster 3, Fig 5b).
Consistent with the B cell skewed differentiation potential of the CD10+ population, genes known to be expressed specifically during B cell commitment (EBF1, PAX5, IL-7R, CD79A, CD79B, VPREB1, VPREB3, CD19, CD22, CD24, CD27) were highly expressed in CD10+ cells (Cluster 6, Fig. 5a–c). Notably, none of these B cell specific genes were expressed in either the CD34+CD38− or CD10− CD62Lhi cells.
A detailed analysis by qPCR showed that although expression of genes essential for lymphoid commitment was highest in CD10+ cells, up-regulation of certain early lymphoid genes began at the CD10− CD62Lhi stage. TCF3 (aka E2A) expression was 2.1-fold increased during the transition from CD38− to CD10− CD62Lhi (P = 0.003) and 4.4-fold increased in the transition from CD10− CD62Lhi to CD10+ (P = 0.058) cells (Fig. 5c). Similarly DNTT (aka TDT) was 8.0-fold increased in CD10−CD62Lhi (P = 0.002) and 12.0-fold further increased in CD10+ cells (P = 0.027) (Fig. 5c). In contrast, RAG1 expression was limited to CD10+ cells, demonstrating that the mechanisms of DNA rearrangement for T cell receptor and immunoglobulin are not fully initiated in the CD10− CD62Lhi population (Fig. 5c).
To investigate further the degree of heterogeneity within the three populations, the expression of key genes was assayed in single cells (Fig. 5d). These analyses showed that the HSC genes TAL1 (aka SCF) and MPL were expressed exclusively in CD38− cells, and RAG1 and PAX5 expression was limited to CD10+ cells. TCF3 was detected at a similar frequency in CD10− CD62Lhi and CD10+ cells. Detectable FLT3 expression in single cells was limited almost exclusively to the CD10− CD62Lhi population (Fig. 5d). Thus the CD38− HSC-MPP, CD10− CD62Lhi and CD10+ populations have distinct molecular profiles, consistent with their functional readout in vitro. Whereas the CD10+ population is committed to B lymphopoiesis, the CD10− CD62Lhi population contains cells with evidence of early lymphoid priming but no expression of B lineage commitment genes (Supplemental Fig. 7).
L-selectin and ligand expression in human thymus
Co-expression of receptor-ligand pairs, previously reported in murine studies as important in thymic homing and settling, were analyzed by flow cytometry and gene expression. The chemokine receptor CXCR4 was expressed at similar abundance in CD10− CD62Lhi and CD10+ populations (Fig. 6a). However PSGL-1, the ligand for P-selectin, and CD44, were both expressed in CD10− CD62Lhi cells at higher amounts than in CD10+ cells (Fig. 6a). In addition, the gene encoding the chemokine receptor CCR7, which is expressed on murine early thymic progenitors and mediates migration of early thymocytes28–30, was significantly up-regulated in the CD10− CD62Lhi population relative to both CD10+ cells (P< 0.001) and CD38− population (P < 0.001) (Fig. 6b). No consistent differences between the populations were seen in expression of the chemokine CCR9 (data not shown).
Figure 6. High CD62L expression and recruitment to human thymus.
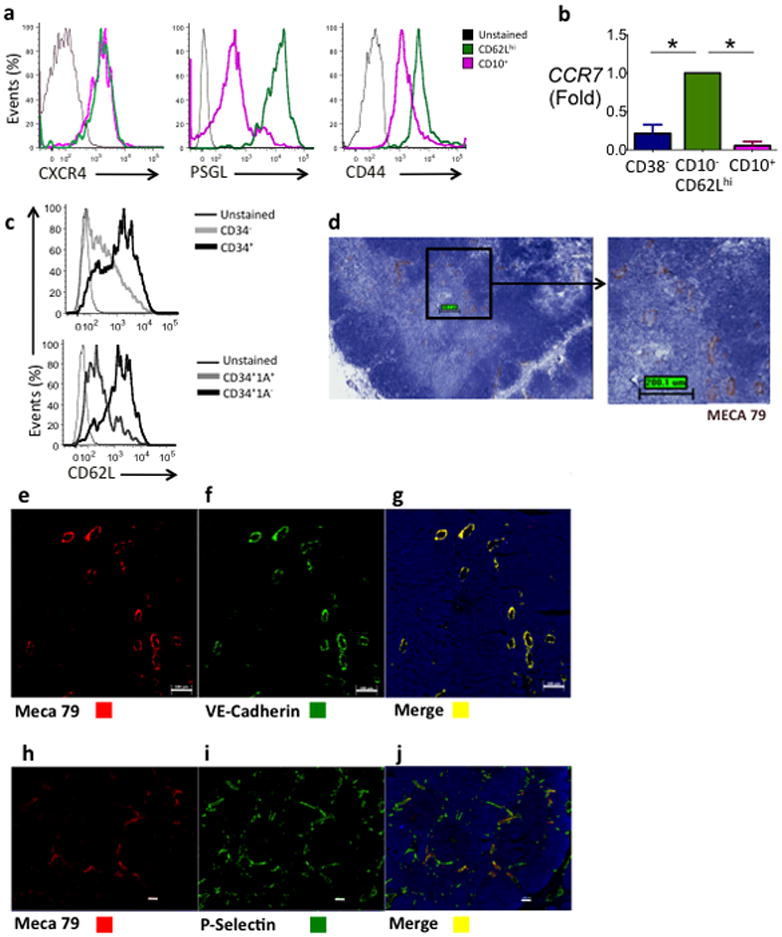
(a) Relative expression of homing molecules on CD10−CD62Lhi cells (green) and CD10+ cells (red) by FACS (gated as CD34+lin−). Unstained (black) (b) RNA expression of CCR7 in CD38−, CD10−CD62Lhi and CD10+ cells by qPCR (n=3 biological replicates, * = p≤0.050, ** = p<0.010, *** p<0.001) mean ± SEM. (c) CD62L expression in human CD34+ and CD34− thymocytes (upper) and CD34+CD1A+ and CD34+ CD1A− thymocytes (lower) by flow cytometry. (d) Chromagen immunohistochemistry showing MECA-79 staining at the cortico-medullary junction. (e, f, g) Fluorescence immunohistochemistry of same region as in (d) showing (e) MECA79 co- staining with (f) VE-Cadherin+ blood vessels. (h, i, j) Fluorescence immunohistochemistry showing (h) MECA79 co-staining in a subset of (i) P-Selectin blood vessels at the cortico-medullary junction.
The expression of CD62L in progenitor populations from human thymus was next examined. CD62L expression was higher in CD34+ thymic progenitors than the more mature CD34− thymocytes (which represent >95% of all thymocytes) (Fig. 6c). Upon further dissection of the CD34+ thymocyte population, the majority of CD62L expressing cells were within the CD34+CD1a− subset rather than the more mature CD34+CD1a+ subset (Fig. 6c). MECA79 detects a carbohydrate epitope that is found on a family of CD62L ligands known as peripheral node addressins (PNAds)31. MECA79 staining was detected in the thymic vasculature specifically in a subset of P-selectin+ endothelial cells at the cortico-medullary junction, the site of entry into the thymus of marrow-derived precursors (Fig. 6d–j), suggesting a possible role for L-selectin in homing to human thymus.
DISCUSSION
The studies presented here demonstrate for the first time that “lymphoid priming” in human BM begins prior to the onset of CD10 expression, in a subset of CD34+ progenitors that highly express the homing molecule L-selectin. Several pieces of evidence argue strongly that the CD10− CD62Lhi population is a precursor of the more B cell-restricted, CD10+ stage of lymphopoiesis. First, it is widely assumed that all human B cell differentiation passes through a CD10+ progenitor stage, and cultures initiated with CD10− CD62Lhi cells were able to generate CD10+ progenitors prior to differentiating into CD19+ B cells. In addition, although the CD10− CD62Lhi population contained greater NK potential, the number of B cells generated in culture was at least equivalent to those from CD10+ cultures. Patterns of gene and cell surface antigen expression were also consistent with the model that positions the CD10− CD62Lhi population prior to CD10 expression.
A recent study described a CD10+ subset within the CD34+CD38−/lo population with lymphoid, monocytic and dendritic cell, but no erythroid potential21. However, this CD10+ “multi-lymphoid progenitor” (MLP) also expressed the B cell specific gene PAX5. It should be noted that the MLP isolation strategy included cells with intermediate expression of CD38, similar to those expressed in the CD10− CD62Lhi population, and higher than in the most primitive HSC fraction. We propose that lymphoid priming begins with upregulation of CD38 (relative to HSCs), and B cell commitment is initiated with the onset of CD10 and further upregulation of CD38 expression.
The vast majority of human hematopoietic studies have used umbilical cord blood (UCB), largely because this source of human cells is more readily accessible than BM. BM progenitors have substantially lower proliferative output than their immunophenotypic homologs in UCB7,17,32–34, or than HSC from either source35, making in vivo assessment of rare, non-self renewing BM progenitor populations difficult and sometimes unfeasible. However, UCB does not represent steady-state postnatal hematopoiesis and substantial differences in immunophenotype and function are known to exist between progenitors from UCB and BM6. Interestingly, we note that functional and molecular profiles in CD10− CD62Lhi BM progenitors (which do not express CD7) are similar to CD34+CD38− CD7+ UCB progenitors 20. We also note that the CD10− CD62Lhi immunophenotype described here is less reliable for the identification of a pure lymphoid-primed population in UCB than in BM; a clear CD62Lhi population is difficult to detect in UCB, and CD34+lin− CD10− CD45RA+CD62L+ cells in UCB contain small but readily detectable numbers of CFU (Q-L.H. & G.M.C., unpublished data). The differences in lineage potentials of cells with similar immunophenotypes in UCB and BM, as well as the intrinsic functional differences that would be expected between cells that are detected transiently in the postnatal circulation and those that are generated throughout life in the BM microenvironment, highlight the critical need for studies that focus on human BM.
A large amount of elegant data has been generated in murine studies to argue both for and against the classical concept that the lymphoid and the myelo-erythroid pathways emerge separately from a multipotent progenitor stage1–4,36. The “lymphoid-primed” LMPP in murine BM retain full lymphoid and some myeloid potential but have lost erythro-megakaryocytic potential, whereas common lymphoid progenitors (CLPs) represent a more mature, lymphoid-restricted progenitor population. FLT3 cell surface expression has been used to isolate LMPPs from a subpopulation of cKit+Lin−Sca1+ cells in murine BM2 and IL-7Rα is used to define murine CLPs within the cKit−Lin−Sca1lo population1. Based on our functional and molecular data, the CD10−CD62Lhi human BM progenitor appears most similar to the murine LMPPs, and the CD10+ progenitor is more analogous to the murine CLPs. However, despite FLT3 upregulation at the transcriptional level, we and others21 have not found the cell surface expression of FLT3 to be useful as a marker to discriminate between human HSCs and LMPPs. Interestingly, recent studies have reported that up-regulation of CD62L expression in cKit+Lin−Sca1+ murine BM cells correlates with high expression of FLT3 and loss of erythroid and megakaryocyte potential, thus suggesting that CD62L expression might be used as an alternative marker to discriminate between murine multipotent progenitors and LMPPs14.
The myeloid potential of the CD10−CD62Lhi population consisted mostly of monocyte-macrophage and dendritic cells. The absence of clonogenic myeloid-erythroid potential in CFU assays suggests strongly that the CD10−CD62Lhi population does not represent a precursor to the major myelo-erythroid pathways that are initiated by CMPs and GMPs. Rather we favor the concept that the CD10−CD62Lhi cells are “lymphoid-primed” progenitors that precede CD10 expression and which are able to generate limited numbers of monocyte-macrophage and dendritic cells. This type of residual myeloid and dendritic potential has been reported by several groups using even more lymphoid-committed progenitors10, 17,21. An earlier paper noted that murine IL-7Rα+ CLPs, despite their complete lack of either CFU activity or in vivo myeloid potential, could generate myeloid cells in stromal co-cultures, suggesting that myeloid differentiation may be an alternative pathway revealed in certain in vitro conditions5. Nonetheless, it is clear that the capacity for myeloid differentiation in vitro progressively wanes as lymphoid commitment proceeds and that this residual, mostly monocytic, potential is retained after erythroid potential is lost.
We note both differences and similarities in gene expression between the CD10−CD62Lhi cells and the previously described murine LMPPs2,36. In both murine LMPPs and human CD10−CD62Lhi cells, genes encoding the transcription factor TAL-1 and the cytokine receptor MPL are significantly down-regulated relative to HSCs, while KIT expression is retained36,37. E2A expression, which is essential for the development of murine LMPPs38, is also up-regulated during generation of the CD10−CD62Lhi population from HSC-MPPs, but B cell-specific genes such as EBF1 and PAX5 are not. In contrast, the molecular machinery required for DNA recombination appears to be highly expressed in murine LMPPs36, but in our human studies, RAG1 and RAG2 were expressed at the CD10+ stage and DNTT expression in the CD10+ cells was significantly higher than in CD10−CD62Lhi cells.
The identification of a lymphoid primed precursor to the previously described CD10+ “CLP”, begs the question of whether the CD10− CD62Lhi cells are recruited to the thymus to initiate T cell differentiation. Controversy regarding the identity of precursors that seed the murine thymus has continued for over a decade, and it seems likely that more than one type of BM progenitor may be able to initiate thymopoiesis. Experimental restrictions make it impossible to definitively prove the identity of the BM precursors that normally seed the human thymus. The CD10+CD24− population in BM is likely to represent a lymphoid progenitor that seeds the human thymus, based on finding a similar immunophenotypic subset within human thymocytes12. Thymocyte data presented here provides evidence that the CD10−CD62Lhi cells may be an additional or alternative thymic precursor population. It should be noted that, although CD62L expression was highest on CD34+CD1a− progenitors, CD10−CD62Lhi BM cells are clearly not precursors of the most primitive (CD7−) subset of CD34+CD1a− thymocytes. CD34+CD1a−CD7− thymocytes have high myeloid and erythroid potential in clonogenic assays39 and do not express CD62L. It is not clear at this stage whether CD62L becomes up-regulated as CD34+CD1a−CD7− MPPs differentiate into CD34+CD1a−CD7+ thymocytes, or that CD7 is rapidly up-regulated when CD7−CD62Lhi LMPPs engage with the thymic microenvironment. PSGL-1–P-selectin interactions are critical mediators of homing to the murine thymus40. As PSGL-1 was abundantly expressed on both HSC-MPP and CD10−CD62Lhi BM cells, it is possible that homing to human thymus involves the same mechanism. However, the high expression of L-selectin in the primitive CD34+CD1a− thymocyte population and the endothelial expression of L-selectin ligands in the human thymus, specifically in the cortico-medullary region, raises the intriguing possibility that in addition to lymphocyte homing to peripheral lymphoid organs, L-selectin may have a role in progenitor homing to human thymus. We have noted that CD62L is expressed in a subset of CD34+lin−CD10−cells (but not CD34+CD10+ cells) in mobilized peripheral blood (data not shown), but the physiological relevance and lineage potential of this mobilized population is as yet unclear. Of note, although L-selectin interactions are not described in murine thymus homing, CD62L expression has been used to identify a population of murine BM progenitors that efficiently and rapidly reconstitutes the murine thymus upon transplantation15,16, and an identical immunophenotypic population of cKit+Lin−Sca-1+CD62L+Rag1-deficient progenitors can be detected in the murine thymus16.
The reliance on CD10 expression as a marker of lymphoid commitment in previous studies of hematopoietic progenitors in human BM has until now meant that states of differentiation could only be compared between multipotent progenitors and B committed progenitors. The identification of a progenitor in human BM primed for full lymphoid differentiation, and prior to B cell commitment, now allows us to dissect the molecular regulation of the first stages of lymphoid commitment in human hematopoiesis and to understand how these processes are affected during aberrant hematopoiesis in disease states.
METHODS
Bone marrow cell isolation
Normal human BM and thymic cells were obtained from healthy donors via the UCLA Pathology Tissue Core, Cincinnati Children’s Hospital, or ALLCELLS according to guidelines approved by UCLA Institutional Review Board. CD34+ cells were enriched using the magnetic activated cell sorting (MACS) system (Miltenyi Biotec).
CD34+ enriched cells were incubated with combinations of the following anti-human–specific monoclonal antibodies CD34-APC-Cy7 (581) (Biolegend), or CD45RA-PE-Cy5 (HI100), CD38-APC (HIT2), CD10-PE-Cy7 (HI10a), CD62L-PE (DREG-56), CD7-PE & CD7-Pe-Cy5 (M-T701), and FITC-labeled lineage depletion antibodies: CD3 (SK7), CD14 (M2E2), CD19 (4G7), CD56 (MY31), and CD235a (GA-R2)(Becton Dickinson [BD]). 4’,6-diamidino-2-phenylindole (DAPI) was added as a viability dye. A no-antibody control defined negative gates. Additional analyses used: CD127-Alexa 647 (HIL-7R-M21), CD117-APC (YB5.B8), CD184-APC (aka CXCR4) (12G5), PSGL1-APC (aka CD162, or SELPLG) (KPL-1), FLT3-PE (aka CD135) (4G8), CD44-APC (G44-26), CD62L-APC (DREG-56), CD90-PECy5 (5E10), HLA-DR-PE (L234) (BD). Cells were isolated on a FACSAria (355, 405, 488, 561 and 633 nm lasers) (BD Immunocytometry Systems).
B-NK Lymphoid Cultures
Flow cytometry isolated cells were plated in bulk on OP9 stroma in 48-well plates, or as single cells or limiting dilution on OP9 or MS5 stroma in 96-well plates using the Automated Cell Deposition Unit (ACDU). Cells were cultured in lymphoid medium [RPMI 1640 (Irvine Scientific) with 5% FCS (Biowhittaker), 50 μM 2-mercaptoethanol (Sigma), penicillin-streptomycin, L-glutamine (Gemini Bio Products)] with IL-7 (5 ng/mL), FLT-3Ligand (FL) (5 ng/mL) and Thrombopoietin (TPO)(5 ng/mL), (+/− IL-3 (5ng/mL) for first 3–5 days of culture) (R&D Systems). Clones were recorded as positive if greater than 100 cells. Cloning efficiency from single cells was defined as (# positive wells/total wells) × 100. (see Supplementary Table 3 for limiting dilution plating information)
T Lymphoid Cultures
Cells were plated in bulk on 6-well or 96-well plates, or as single cells or in limiting dilution (via ACDU) on established OP9-DL1 stroma in lymphoid medium with IL-7 (5 ng/mL), FL (5 ng/mL) and Stem Cell Factor (SCF) (1 ng/mL) (R&D)41.
Myelo-Erythroid Cultures and CFUs
Populations were plated on OP9 stroma in DMEM with 10% FBS, with IL-3 (5 ng/mL), FL (5 ng/mL), SCF (5 ng/mL), TPO (50 ng/mL), and Erythropoietin (4 U/mL) (R&D). CFU assays were performed as described39.
Lineage-specific analysis
Flow cytometry of cultured cells and cells harvested from transplanted mice was performed on a Fortessa or LSRII (BD) after staining with human specific monoclonal antibodies: CD45 (HI30) (pan-human hematopoietic); HLA-A, B, C (G46-2.6)(pan-human); CD19 (4G7 and SJ25C1) (B lymphoid); CD56 (MY31)(NK cells); CD209 (DCN46)(dendritic); CD1A (HI149), CD3 (SK7), CD4 (RPA-T4), CD7(M-T701), CD8 (RPA-T8), TCR α/β (WT31)(T-lymphoid); CD235a (GA-R2) (erythroid), CD14 (M5E2), CD11B (ICRF44)(monocytic), CD14(M5E2), CD15 (W6D3), CD33 (WM53) (myeloid), CD66B (G10F5) (granulocytic) (all antibodies from BD). Data was analyzed using FlowJo software. T cell differentiation was assessed by RT-PCR of human CD45+ & mouse CD29− cells isolated at 4–5 weeks from T lymphoid cultures.
In vivo studies
Adult Nonobese diabetic/severe combined immunodeficiency Interleukin 2 receptor gamma chain knock out (NSG) mice (Jackson Laboratories, Bar Harbor, ME), were used for in vivo experiments according to protocols approved by the Institutional Animal Care and Use Committee of University of California Los Angeles. Adult NSG mice were irradiated (375 cGy) prior to intra-tibial injection of 3 × 104 CD10−CD62Lhi cells (n = 3) or 2–15 × 104 CD34+lin− BM cells (n = 3), each with 1 × 105 “carrier” cells [irradiated (3,000 cGy) CD34− UCB cells], and euthanized 2 weeks later for flow cytometry analysis. Total human engraftment was defined as cells positive for HLA-A, B, C and humanCD45. Negative control mice received only irradiated carrier cells.
Quantitative PCR analysis
After FACSAria isolation, RNA was extracted with the Qiagen RNAEsay Microkit (Qiagen) and reverse-transcribed using Omniscript RT, OLIGO DT, and RNAguard (Pharmacia Biotech). ABI Viia7 was used for real-time PCR with Taqman Mastermix and TaqMan probe based gene expression analysis assays. (List of probes, Supplementary Table 2) (Applied Biosystems). Reactions were done in technical and biological triplicates. Nine candidate reference genes were analyzed with geNormplus software to determine optimal reference genes.42 Using the ΔΔCt method, qPCR expression was normalized to the geometric mean of reference targets ACTB and B2M.
Single cell qPCR was performed on the Fluidigm Biomark 48.48 gene expression chip with Taqman probes, and analyzed with Fluidigm’s Real-Time Software v3.0.2. β2microglobulin was used as a positive control for presence of cDNA.
Microarray Analysis
RNA from BM from 3 different individuals was extracted using Microkit (Qiagenand hybridized onto Affymetrix U133 Plus 2.0 Array (Affymetrix).
Robust Multichip Average (RMA)43 method was used to obtain normalized expression levels from the three populations. MAS5 algorithm44 was used to make Present, Marginal, or Absent calls for all replicates.
Replicate arrays from the three populations were hierarchically clustered using Spearman rank correlation (distance metric) and average linkage (agglomeration) method. Only probe sets called as "Present" by MAS5 method in all replicates in any of the three populations (24067 probes) were used for hierarchical clustering.
The number of differentially expressed genes in Venn diagrams was calculated using the R/Bioconductor45 package Limma46 at P-value < 0.01 and fold change threshold +/−2. For genes with multiple probe sets, the probe set with the lowest P -value was chosen. Probe sets not mapped to a gene with official symbol were excluded.
Genes were considered for inclusion to the heatmap only if they were differentially expressed at a fold change of +/− 2 and significant at a P -value <0.05 when compared with the other population of cells in at least one condition. Gene Set Enrichment Analysis was performed as described47.
For drawings, Cluster 3.0 (clustering)48 and Java TreeView (dendrograms, heatmaps)49 software was used.
Immunohistochemistry
Human thymi were frozen at −80°C and OCT embedded (Tissue-Tek) and 5 μm sections were stained with hematoxylin and eosin. For immunohistochemistry (IHC), sections were fixed in 10% Neutral Buffered Formalin, then incubated with primary antibody PNad (MECA79/sc-19602, 1:83, Santa Cruz Biotechnology, Inc.) and/or VE-cadherin (BV6, 1:83, Chemicon International), followed by incubation with secondary antibody anti-rat and/or anti-mouse peroxidase antibody (Vector). For fluorescence IHC, TSA Alexa 594 and/or TSA Alexa 488 was applied (Molecular Probes). For chromagen staining, DAB was applied, followed by hematoxylin (Jackson Immunoresearch). Sections were viewed with Axioimager with Apotome Imagining System (10x); images were captured with Axiocam MRm (florescence) or HRc (chromagen) (Zeiss).
Statistical analysis
Prism version 5 (GraphPad Software Inc) was used for statistical analysis. Two-way ANOVA compared growth potential. Total CFU output of populations, MFI and q-PCR were analyzed for mean, SEM calculation, and one-way ANOVA with a Tukey post-test. Limiting Dilution analysis used ELDA software http://bioinf.wehi.edu.au/software/elda/50.
Supplementary Material
Supplementary Figure 1. Relationship of CD7, CD38, CD10 and CD62L expression on CD34+lin− cells and CFU potential of progenitor subsets.
Representative experiment of (a) CD38 and CD7 expression on CD34+lin- cells from BM (n=6), (b) CD62L and CD10 expression on CD34+lin− BM cells (% of each subset within CD34+lin- population is shown) (n=30). (c) Table of CFUs for populations gated as in (a) (n=2–9). CFUs are given in # of colonies/103 BM cells plated.
Supplementary Figure 2. CD10−CD62Lhi profiles amongst different bone marrow donors
(a) Gating strategy for isolation of CD10−CD62Lhi, CD10+ and CD38− cell populations (b) Flow cytometric analysis of CD62L vs CD45RA of 20 independent human BMs. Cells were previously gated as DAPI− CD34+ Lin− CD10− as shown in (a). (c) Relationship of frequency of CD10−CD62Lhi cells within the CD34+lin−CD45RA+ population to age. 17 BM donors in which age was known are shown (1 year to adult; n = 7 adults > 25y). Note: at 1 year of age, CD34+lin− cells are predominantly CD10+.
Supplementary Figure 3. T cell cultures generated from CD10−CD62Lhi cells.
Analysis of T cell cultures initiated with CD10−CD62Lhi cells (a,b) Unstained Flow Cytometry analysis of T Lymphoid cultures in companion to (a) Figure 2c (Week 4 CD7-FITC, CD1A-PE, Weeks 5, 7, 8 CD8-PE, CD4-FITC) (n=6) or (b) Figure 2d (TCRαβ-FITC, CD4-APC-Cy7, CD8-PE-Cy7, CD3-APC) (n=2). (c) Semi-quantitative RT-PCR analyses of T cell associated genes from total cells harvested at 4–6 weeks from 2 independent experiments. 1kb ladder shown at left of each transcript.
Supplementary Figure 4. The CD10-CD62Lhi population has monocytic potential in myeloid stromal co-cultures
(a) Flow Cytometry analysis of 2 week myelo-erythroid co-cultures (OP9 with SCF, FLT-3, TPO, IL3 and EPO) initiated with CD38-, CD10-CD62Lhi or CD10+ cells. Far left panels show unstained controls. Monocytes/macrophages (CD14) and dendritic cells (CD209) were detected after initial gating on CD45+ cells (not shown). FSC and SSC gating (top row) are shown to analyze small (middle row) and large (bottom row) cells separately. Glycophorin and CD66b expression were rarely detected. Data representative of 3 independent experiments. (b) Co-expression of myeloid (CD14, CD15 and CD33) and dendritic (CD1A+) markers in myelo-erythroid cultures initiated with CD10-CD62Lhi cells. Data representative of 3 independent experiments.
Supplementary Figure 5. Additional in vivo reconstitution data.
Flow cytometry analysis of NSG mice transplanted with human BM populations and analyzed 2wk post transplant. (a-d) Data from 5 additional animals not included in Fig 3f. Mouse #4 received 100,000 irradiated CD34− carrier cells only (negative control), Mice #5 and #6 received 30,000 CD34+lin−CD10− CD62Lhi cells, Mouse #7 received 150,000 CD34+lin− cells, and Mouse #8 received 30,000 CD34+lin− cells. (a) Unstained controls for PE (hCD45) and PE-Cy7 (HLA-class 1), (b, c) Fluorescence minus one (FMO) controls from composite of cells from animals #7 and #8, (b) is gated from FSC vs SSC as shown in (a), (c) is gated on CD45+HLA+ cells as shown in (d). (d) Human engraftment shown top row as hCD45+ & HLA-class 1+ cells, bottom row shows B (CD19+) cells and myeloid (CD14, CD15, & CD33+) cells from gated human cells.
Supplementary Figure 6. Global gene expression analysis places CD10−CD62Lhi population in intermediate position between CD38−and CD10+,with similar profile to umbilical cord blood CD34+CD38−CD7+ lymphoid progenitors
Microarray datasets from three independent BM samples. (a) Hierarchical clustering of global gene expression using Spearman rank correlation and average linkage method. (b) Venn Diagram showing pair-wise comparisons of global gene expression. Shown are number of genes that are > 2 fold differentially up-regulated or down-regulated relative to CD38−, p<0.01. (c) Gene Set Enrichment Analysis (GSEA) comparing gene expression between BM CD10−CD62Lhi and BM CD38− and published CD34+CD38−CD7+ UCB20. Overall the set of genes down-regulated in BM CD10−CD62Lhi (LMPP) compared to BM CD38− (HSC-MPP) was similar to the set of genes down-regulated in UCB CD34+CD38−CD7+ (MLP) compared to UCB CD34+CD38−CD7− (HSC-MPP) from Hoebeke et al. (FDR q-value 1.6180314E-4, Normalized Enrichment Score −2.398794).
Supplementary Figure 7. Stages of lymphoid priming and B cell commitment in human bone marrow.
(a, b) Changes in relative expression of key cytokine receptors in BM progenitor populations listed above, (a) protein expression by flow cytometry and (b) gene expression by array and/or qPCR. Note, although FLT3 transcripts are increased in the CD10−CD62Lhi LMPP population, expression by flow cytometry is similar in HSC-MPP and LMPP. (c) Proposed model of progenitor relationships during early stages of lymphoid commitment in human bone marrow. CD34+lin− CD10+ cells are predominantly B cell progenitors (BCP) whereas CD10-CD62Lhi cells readily generate all three lymphoid lineages; dashed lines represent less prominent lineage differentiation pathways.
Supplementary Table 1. Colony Forming Unit assay raw data
(a) Raw data for Colony Forming Unit Assays. All data is from subsets shown of human BM CD34+ Lin- cells. Each experiment represents duplicate plates from one flow cytometry sorted BM. (b) P-values of unpaired t-tests comparing CFU output from CD10− CD62Lhi to each population shown (N=number of times that cell population was tested using a different donor, all in duplicate).
Supplementary Table 2. List of qPCR probes
List of ABI Taqman Probes and amplicon size used for qPCR analysis.
Supplementary Table 3. Set up for limiting dilution experiments
Number of wells seeded at each cell number for limiting dilution analysis on (a) MS5 stroma (n=3 independent samples) (see Fig 3a) or (b) OP9DL1 stroma (n=3 independent samples) (see Fig 3b)
Acknowledgments
We would like to thank J. Scholes and F. Codrea from the Broad Stem Cell Research Center FACS Core; X. Li, Director of JCCC Gene Expression Shared Resource/Pathology Clinical Microarray Core Laboratories, UCLA and S. Dandekar at the UCLA Genseq Core. Thanks to D. Kohn, G. Dravid, S. Sandoval and M. Corselli for helpful advice on the manuscript.
This work was supported by grants from the NIH (P01 HL073104 and RO1 HL077912), and California Institute of Regenerative Medicine (RC1-00108 and RM1-01717) (GMC), and California Institute for Regenerative Medicine New Faculty Award (RN1-00557-1) (HKAM).
Footnotes
Accessions
The microarray data were deposited in NCBI’s Gene Expression Omnibus database (Accession# GSE35685) and are accessible through the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ztenfowesgyagjk&acc=GSE35685
Author contributions
LAK designed, performed and analyzed experiments and wrote the paper, Q-LH designed, performed and analyzed experiments, RS performed bioinformatics on microarray data, SG and YZ assisted in experiments, CP performed experiments, HKAM supervised bioinformatics analysis, GMC designed and analyzed experiments and wrote the paper.
The authors have no competing financial interests to declare.
References
- 1.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–310. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–2624. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payne KJ, Crooks GM. Immune-cell lineage commitment: translation from mice to humans. Immunity. 2007;26:674–677. doi: 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Rossi MI, et al. B lymphopoiesis is active throughout human life, but there are developmental age-related changes. Blood. 2003;101:576–584. doi: 10.1182/blood-2002-03-0896. [DOI] [PubMed] [Google Scholar]
- 8.Mold JE, et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010;330:1695–1699. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leavy O. Haematopoiesis: Baby tolerance. Nat Rev Immunol. 2011;11:78. doi: 10.1038/nri2923. [DOI] [PubMed] [Google Scholar]
- 10.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 11.Ichii M, et al. The density of CD10 corresponds to commitment and progression in the human B lymphoid lineage. PLoS One. 2010;5:e12954. doi: 10.1371/journal.pone.0012954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Six EM, et al. A human postnatal lymphoid progenitor capable of circulating and seeding the thymus. J Exp Med. 2007;204:3085–3093. doi: 10.1084/jem.20071003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 14.Cho S, Spangrude GJ. Enrichment of functionally distinct mouse hematopoietic progenitor cell populations using CD62L. J Immunol. 2011;187:5203–5210. doi: 10.4049/jimmunol.1102119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry SS, et al. L-selectin defines a bone marrow analog to the thymic early T-lineage progenitor. Blood. 2004;103:2990–2996. doi: 10.1182/blood-2003-09-3030. [DOI] [PubMed] [Google Scholar]
- 16.Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao QL, et al. Identification of a novel, human multilymphoid progenitor in cord blood. Blood. 2001;97:3683–3690. doi: 10.1182/blood.v97.12.3683. [DOI] [PubMed] [Google Scholar]
- 18.Storms RW, Goodell MA, Fisher A, Mulligan RC, Smith C. Hoechst dye efflux reveals a novel CD7(+)CD34(−) lymphoid progenitor in human umbilical cord blood. Blood. 2000;96:2125–2133. [PubMed] [Google Scholar]
- 19.Haddad R, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104:3918–3926. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- 20.Hoebeke I, et al. T-, B- and NK-lymphoid, but not myeloid cells arise from human CD34(+)CD38(−)CD7(+) common lymphoid progenitors expressing lymphoid-specific genes. Leukemia. 2007;21:311–319. doi: 10.1038/sj.leu.2404488. [DOI] [PubMed] [Google Scholar]
- 21.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 22.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Motte-Mohs RN, Herer E, Zuniga-Pflucker JC. Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–1439. doi: 10.1182/blood-2004-04-1293. [DOI] [PubMed] [Google Scholar]
- 24.Majewski IJ, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116:731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- 25.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 26.Biassoni R, Ferrini S, Prigione I, Moretta A, Long EO. CD3-negative lymphokine-activated cytotoxic cells express the CD3 epsilon gene. J Immunol. 1988;140:1685–1689. [PubMed] [Google Scholar]
- 27.Lanier LL, Chang C, Spits H, Phillips JH. Expression of cytoplasmic CD3 epsilon proteins in activated human adult natural killer (NK) cells and CD3 gamma, delta, epsilon complexes in fetal NK cells. Implications for the relationship of NK and T lymphocytes. J Immunol. 1992;149:1876–1880. [PubMed] [Google Scholar]
- 28.Misslitz A, et al. Thymic T cell development and progenitor localization depend on CCR7. J Exp Med. 2004;200:481–491. doi: 10.1084/jem.20040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Forster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 30.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streeter PR, Rouse BT, Butcher EC. Immunohistologic and functional characterization of a vascular addressin involved in lymphocyte homing into peripheral lymph nodes. J Cell Biol. 1988;107:1853–1862. doi: 10.1083/jcb.107.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arakawa-Hoyt J, et al. The number and generative capacity of human B lymphocyte progenitors, measured in vitro and in vivo, is higher in umbilical cord blood than in adult or pediatric bone marrow. Bone Marrow Transplant. 1999;24:1167–1176. doi: 10.1038/sj.bmt.1702048. [DOI] [PubMed] [Google Scholar]
- 33.Kim DK, et al. Comparison of hematopoietic activities of human bone marrow and umbilical cord blood CD34 positive and negative cells. Stem Cells. 1999;17:286–294. doi: 10.1002/stem.170286. [DOI] [PubMed] [Google Scholar]
- 34.De Smedt M, et al. T-lymphoid differentiation potential measured in vitro is higher in CD34+CD38−/lo hematopoietic stem cells from umbilical cord blood than from bone marrow and is an intrinsic property of the cells. Haematologica. 2011;96:646–654. doi: 10.3324/haematol.2010.036343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci U S A. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Luc S, et al. Down-regulation of Mpl marks the transition to lymphoid-primed multipotent progenitors with gradual loss of granulocyte-monocyte potential. Blood. 2008;111:3424–3434. doi: 10.1182/blood-2007-08-108324. [DOI] [PubMed] [Google Scholar]
- 38.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao QL, et al. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7- lympho-myeloid thymic progenitors. Blood. 2008;111:1318–1326. doi: 10.1182/blood-2007-08-106294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi FM, et al. Recruitment of adult thymic progenitors is regulated by P-selectin and its ligand PSGL-1. Nat Immunol. 2005;6:626–634. doi: 10.1038/ni1203. [DOI] [PubMed] [Google Scholar]
- 41.Holmes R, Zuniga-Pflucker JC. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb Protoc. 2009:pdb prot5156. doi: 10.1101/pdb.prot5156. [DOI] [PubMed] [Google Scholar]
- 42.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Liu WM, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 45.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth GK. Bioinformatics and Computational Biology Solutions using R and Bioconductor, R. Springer; New York: 2005. [Google Scholar]
- 47.Subramanian ATP, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43) doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 49.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 50.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Relationship of CD7, CD38, CD10 and CD62L expression on CD34+lin− cells and CFU potential of progenitor subsets.
Representative experiment of (a) CD38 and CD7 expression on CD34+lin- cells from BM (n=6), (b) CD62L and CD10 expression on CD34+lin− BM cells (% of each subset within CD34+lin- population is shown) (n=30). (c) Table of CFUs for populations gated as in (a) (n=2–9). CFUs are given in # of colonies/103 BM cells plated.
Supplementary Figure 2. CD10−CD62Lhi profiles amongst different bone marrow donors
(a) Gating strategy for isolation of CD10−CD62Lhi, CD10+ and CD38− cell populations (b) Flow cytometric analysis of CD62L vs CD45RA of 20 independent human BMs. Cells were previously gated as DAPI− CD34+ Lin− CD10− as shown in (a). (c) Relationship of frequency of CD10−CD62Lhi cells within the CD34+lin−CD45RA+ population to age. 17 BM donors in which age was known are shown (1 year to adult; n = 7 adults > 25y). Note: at 1 year of age, CD34+lin− cells are predominantly CD10+.
Supplementary Figure 3. T cell cultures generated from CD10−CD62Lhi cells.
Analysis of T cell cultures initiated with CD10−CD62Lhi cells (a,b) Unstained Flow Cytometry analysis of T Lymphoid cultures in companion to (a) Figure 2c (Week 4 CD7-FITC, CD1A-PE, Weeks 5, 7, 8 CD8-PE, CD4-FITC) (n=6) or (b) Figure 2d (TCRαβ-FITC, CD4-APC-Cy7, CD8-PE-Cy7, CD3-APC) (n=2). (c) Semi-quantitative RT-PCR analyses of T cell associated genes from total cells harvested at 4–6 weeks from 2 independent experiments. 1kb ladder shown at left of each transcript.
Supplementary Figure 4. The CD10-CD62Lhi population has monocytic potential in myeloid stromal co-cultures
(a) Flow Cytometry analysis of 2 week myelo-erythroid co-cultures (OP9 with SCF, FLT-3, TPO, IL3 and EPO) initiated with CD38-, CD10-CD62Lhi or CD10+ cells. Far left panels show unstained controls. Monocytes/macrophages (CD14) and dendritic cells (CD209) were detected after initial gating on CD45+ cells (not shown). FSC and SSC gating (top row) are shown to analyze small (middle row) and large (bottom row) cells separately. Glycophorin and CD66b expression were rarely detected. Data representative of 3 independent experiments. (b) Co-expression of myeloid (CD14, CD15 and CD33) and dendritic (CD1A+) markers in myelo-erythroid cultures initiated with CD10-CD62Lhi cells. Data representative of 3 independent experiments.
Supplementary Figure 5. Additional in vivo reconstitution data.
Flow cytometry analysis of NSG mice transplanted with human BM populations and analyzed 2wk post transplant. (a-d) Data from 5 additional animals not included in Fig 3f. Mouse #4 received 100,000 irradiated CD34− carrier cells only (negative control), Mice #5 and #6 received 30,000 CD34+lin−CD10− CD62Lhi cells, Mouse #7 received 150,000 CD34+lin− cells, and Mouse #8 received 30,000 CD34+lin− cells. (a) Unstained controls for PE (hCD45) and PE-Cy7 (HLA-class 1), (b, c) Fluorescence minus one (FMO) controls from composite of cells from animals #7 and #8, (b) is gated from FSC vs SSC as shown in (a), (c) is gated on CD45+HLA+ cells as shown in (d). (d) Human engraftment shown top row as hCD45+ & HLA-class 1+ cells, bottom row shows B (CD19+) cells and myeloid (CD14, CD15, & CD33+) cells from gated human cells.
Supplementary Figure 6. Global gene expression analysis places CD10−CD62Lhi population in intermediate position between CD38−and CD10+,with similar profile to umbilical cord blood CD34+CD38−CD7+ lymphoid progenitors
Microarray datasets from three independent BM samples. (a) Hierarchical clustering of global gene expression using Spearman rank correlation and average linkage method. (b) Venn Diagram showing pair-wise comparisons of global gene expression. Shown are number of genes that are > 2 fold differentially up-regulated or down-regulated relative to CD38−, p<0.01. (c) Gene Set Enrichment Analysis (GSEA) comparing gene expression between BM CD10−CD62Lhi and BM CD38− and published CD34+CD38−CD7+ UCB20. Overall the set of genes down-regulated in BM CD10−CD62Lhi (LMPP) compared to BM CD38− (HSC-MPP) was similar to the set of genes down-regulated in UCB CD34+CD38−CD7+ (MLP) compared to UCB CD34+CD38−CD7− (HSC-MPP) from Hoebeke et al. (FDR q-value 1.6180314E-4, Normalized Enrichment Score −2.398794).
Supplementary Figure 7. Stages of lymphoid priming and B cell commitment in human bone marrow.
(a, b) Changes in relative expression of key cytokine receptors in BM progenitor populations listed above, (a) protein expression by flow cytometry and (b) gene expression by array and/or qPCR. Note, although FLT3 transcripts are increased in the CD10−CD62Lhi LMPP population, expression by flow cytometry is similar in HSC-MPP and LMPP. (c) Proposed model of progenitor relationships during early stages of lymphoid commitment in human bone marrow. CD34+lin− CD10+ cells are predominantly B cell progenitors (BCP) whereas CD10-CD62Lhi cells readily generate all three lymphoid lineages; dashed lines represent less prominent lineage differentiation pathways.
Supplementary Table 1. Colony Forming Unit assay raw data
(a) Raw data for Colony Forming Unit Assays. All data is from subsets shown of human BM CD34+ Lin- cells. Each experiment represents duplicate plates from one flow cytometry sorted BM. (b) P-values of unpaired t-tests comparing CFU output from CD10− CD62Lhi to each population shown (N=number of times that cell population was tested using a different donor, all in duplicate).
Supplementary Table 2. List of qPCR probes
List of ABI Taqman Probes and amplicon size used for qPCR analysis.
Supplementary Table 3. Set up for limiting dilution experiments
Number of wells seeded at each cell number for limiting dilution analysis on (a) MS5 stroma (n=3 independent samples) (see Fig 3a) or (b) OP9DL1 stroma (n=3 independent samples) (see Fig 3b)


