Abstract
As a light-driven water-plastoquinone oxidoreductase, Photosystem II produces molecular oxygen as an enzymatic product. Additionally, under a variety of stress conditions, reactive oxygen species are produced at or near the active site for oxygen evolution. In this study, Fourier-transform ion cyclotron resonance mass spectrometry was used to identify oxidized amino acid residues located in several core Photosystem II proteins (D1, D2, CP43 and CP47) isolated from spinach Photosystem II membranes. While the majority of these oxidized residues (81%) are located on the oxygenated solvent-exposed surface of the complex, several residues on the CP43 protein (354E, 355T, 356M and 357R) which are in close proximity (<15 Å) to the Mn4CaO5 active site are also modified. These residues appear to be associated with putative oxygen/reactive oxygen species exit channel(s) in the photosystem. These results are discussed within the context of a number of computational studies which have identified putative oxygen channels within the photosystem.
Photosystem II (PS II) functions as a light-driven, water-plastoquinone oxidoreductase. In higher plants and cyanobacteria at least six intrinsic proteins appear to be required for O2 evolution. These are CP47, CP43, D1, D2, and the α and β subunits of cytochrome b559. Deletion of these subunits uniformly results in the loss of PS II function and assembly (1, 2). Additionally, in higher plants, three extrinsic proteins, PsbO, PsbP and PsbQ are also required for maximal rates of O2 evolution under physiological inorganic cofactor concentrations (3). Of these three proteins, the PsbO protein appears to play a central role in the stabilization of the manganese cluster, is essential for efficient and stable O2 evolution and is required, along with PsbP, for photoautotrophic growth and PS II assembly in higher plants propagated under normal growth conditions (4–6). Under low light growth conditions the PsbQ component is also required for photoautotrophy (7). Over the past eleven years, moderate resolution crystal structures of cyanobacterial PS II have significantly enhanced our understanding of the molecular organization of the constituent polypeptides of the photosystem and the active site for oxygen evolution, the Mn4O5Ca cluster (8–12). Recently, a high resolution 1.9 Å crystal structure of cyanobacterial PS II has been presented (13). It should be noted that no crystal structures for higher plant PS II are available. While there are differences between the higher plant and the cyanobacterial photosystems, particularly with respect to the identity and organization of the extrinsic proteins (3), the amino acid sequences of the intrinsic components (D1, D2, CP43 and CP47) are nearly identical (>85% similarity, (14)). Consequently, one would expect that the structural organization of these components within PS II would be highly homologous between higher plants and cyanobacteria.
The production of molecular oxygen by PS II is accompanied by the unavoidable possibility of oxidative modification of amino acid residues within the PS II complex. While one would predict that amino acid residues located at the surface of the complex which are in contact with the bulk, oxygenated solvent would be susceptible to such modification, buried amino acid residues in the vicinity of the Mn4CaO5 cluster, the active site for water oxidation, might also be particularly susceptible to oxidative modification. This is exacerbated by the production of reactive oxygen species (ROS), such as H2O2, O2•− and OH• at, or in the vicinity of, the Mn4CaO5 cluster under a variety of stress conditions (15). Indeed, earlier studies have identified the double oxidation of the CP43 residue 365W to form an N-formylkynurenine derivative (16, 17). This residue is located 17 Å from Mn2 of the Mn4CaO5 cluster.
Given the possibility that oxidative modification of PS II could lead to damage of the photosystem, several groups have hypothesized that channels present in PS II would function to vector molecular oxygen and, by extension, ROS away from the Mn4CaO5 cluster towards the surface of the photosystem. Murray and Barber (18) used the CAVER Program (19) to examine the 3.5 Å crystal structure of T. elongatus (10); later examining similar PS II crystals perfused with xenon (20). Gabdulkhakov et al. (21) used nobel gas and dimethyl sulfoxide co-crystallization studies in combination with CAVER to examine the 2.9 Å structure of T. elongatus (12), and Ho and Styring calculated solvent-accessible surfaces for the 3.0 Å T. elongatus structure of Loll et al. (11). We hypothesized that amino acid residues in contact with such oxygen channels in the interior of PS II would be significantly more susceptible to oxidative amino acid modification than residues not exposed to molecular oxygen and/or ROS. Consequently, the identification of such oxidatively modified residues in the interior of PS II should serve to complement and extend the largely computational studies mentioned above.
Materials and Methods
PS II membranes were isolated from market spinach by the method of Berthold et al. (22, 23) modified by Ghanotakis and Babcock (22, 23). After isolation, the PS II membranes were suspended at 2 mg chl/ml in 50 mM Mes-NaOH, pH 6.0, 300 mM sucrose, 15 mM NaCl buffer and frozen at −80°C. The proteins in the samples were resolved on a 12.5–20% acrylamide gradient by LiDS-PAGE using a non-oxidizing gel system (24). This was required, as standard PAGE is known to introduce numerous protein oxidation artifacts (24, 25). In the non-oxidizing system the gels are polymerized with riboflavin (in the presence of diphenyliodonium chloride + toluensulfinate) followed by exposure to UV light. The upper reservoir contains thioglycolate. Preliminary experiments indicated that proteins resolved in this system exhibited extremely low levels of artifactual protein oxidation when compared to proteins resolved under standard PAGE conditions (Fig. S1). After electrophoresis, the gels were stained with Coomassie Blue, destained, and protein bands containing CP47, CP43, D1, and D2 were excised. These were then processed for trypsin digestion using standard protocols. In some cases, the tryptic peptides were processed using a C18 ZipTip® prior to mass analysis.
Chromatography was performed using a Finnigan Surveyor MS pump and a Finnigan Micro AS autosampler. The column used was a Water’s X-Bridge C18 3.5µm 2.1 × 100 mm with a flow rate of 200µl/min. The mobile phases consist of a 95:5 water:acetonitrile with 0.1% formic acid aqueous phase and a 95:5 acetonitrile:water with 0.1% formic acid organic phase. The gradient was as follows: The organic phase composition was 10% for the first 5 min, ramped to 20% for the next 10 min (total time 15 min), ramped to 50% for the next 25 min (total time 40 min), ramped to 80% in the next 35 min (total time 75 min), held at 80% for 10 min followed by a quick ramp to 10% in 5 min and a 10 min hold to equilibrate the column (total time 100 min).
Mass spectrometry was performed on a Thermo Scientific LTQ-FT™, a hybrid instrument consisting of a linear ion trap and a Fourier transform ion cyclotron resonance mass spectrometer. The experiments used the standard electrospray source operating with a source voltage of 5 kV and a capillary temperature of 275°C. Sheath and auxiliary gas flows were 18 and 5 respectively. A typical scan sequence involved a positive ion FT-ICR scan at 100K resolution (100K at m/z 400). During the FT-ICR acquisition, six MS2 scans were acquired by the linear ion trap determining the parent ions from the six largest ions observed from a preview of the FT-ICR scan. The CID scans are acquired with an isolation width of 2 and a normalized collision energy of 35 (both Thermo Scientific machine settings). After acquiring the tandem mass spectra twice, the ion is placed into an exclusion list for 30 sec. Charge state screening is enabled with monoisotopic precursor selection.
Two biological replicate experiments were performed. Identification and analysis of peptides containing oxidative modifications were performed using the MassMatrix Program ver. 1.3.1 (26, 27) which had been programmed to include the possible oxidative modifications (28, 29). A FASTA library containing the spinach PS II protein sequences was searched as was a decoy library that contained these same proteins but with reversed amino acid sequences. No hits to the decoy library were observed. For the determination of the quality of the peptide calls within MassMatrix, max(pp1, pp2) was ≥ 8.5 and ppTag ≥ 5.0 (26, 27). These parameters yield a p value of ≤ 0.0001; only oxidized peptides which exhibited these extremely low p value were considered. Given the high quality of the data used in this study, the union of the replicate data sets was examined. The identified oxidatively modified residues were mapped onto the T. vulcanus PS II structure (13) of the D1, D2, CP43 and CP47 proteins using PYMOL (31).
Results
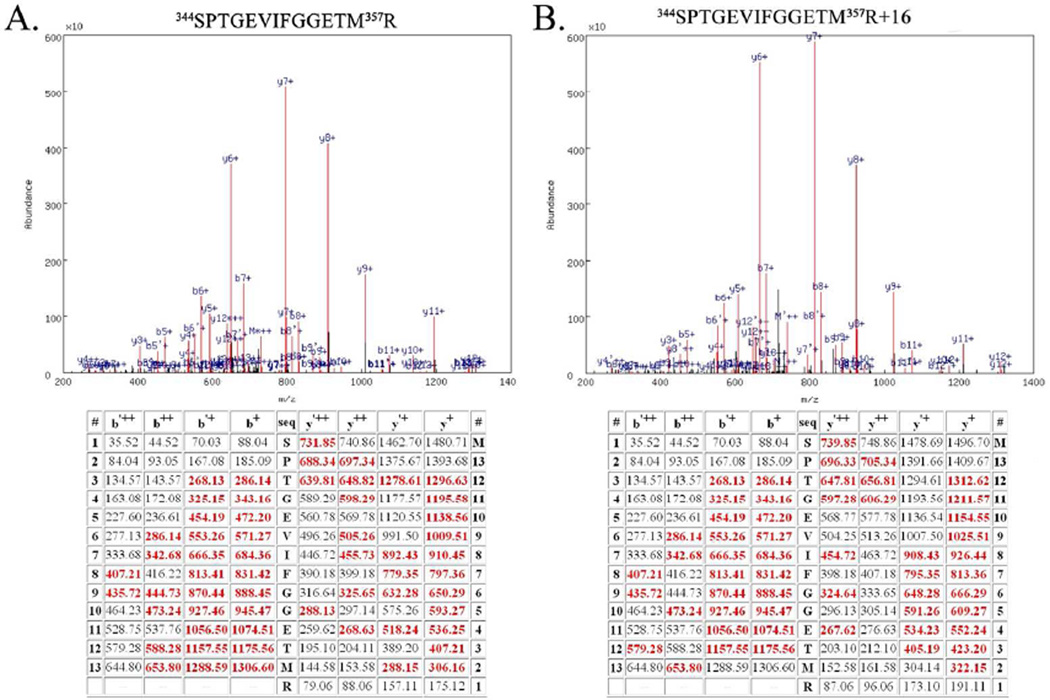
Figure 1 illustrates the quality of the mass spectrometry data used for the identification of oxidized amino acid residues in the D1, D2 CP47 and CP43 proteins. Using a p value ≤ 0.0001 assured extremely high quality peptide identifications. In this context it should be noted that in MASCOT searches (30), peptide identifications are typically performed with p values ≤ 0.05 (www.matrixscience.com). The direct consequence of our using such low p values is that in all cases nearly complete y- and b-ion series for the oxidatively modified peptides were observed. (Fig. 1 and Figs. S2–S5)
Figure 1.
Example Mass Spectrometry Data from the Unmodified Peptide 344SPTGEVIFGGETM357R and the Oxidatively Modified Peptide 344SPTGEVIFGGETM357R+16. A. Top, spectrum of the CID dissociation of the unmodified peptide 344SPTGEVIFGGETM357R. Various identified ions are labeled. Bottom, table of all predicted masses for the y- and b- ions generated from this peptide sequence. Ions identified in the CID spectrum (above) are shown in red. The b’++, b’+ y’++ and y’+ ions are generated by the neutral loss of water. B. Top, spectrum of the CID dissociation of the modified peptide344SPTGEVIFGGETM357R+16. Various identified ions are labeled. Bottom, table of all predicted masses for the y- and b- ions generated from this peptide sequence. Ions identified in the CID spectrum are shown in red. The b’++, b’+ y’++ and y’+ ions are generated by the neutral loss of water. Note the additional mass of 16 amu (in comparison with Fig. 1A) arising from modification of 357R. This modification is evident in all identified y-ions species but is absent in all identified b-ions. Both of these spectra are from Biological Replicate #1.
A total of seventy-three oxidatively modified amino acid residues were identified in the D1, D2, CP43, and CP47 proteins of spinach PS II. This represents ≈ 4 % of the residues found in these four proteins. These are summarized in Table 1 along with the type of oxidative modifications observed and the residue location (surface, buried but in contact with cavity/channel, or buried and not in contact with cavity or channel) within the T. vulcanus crystal structure. In some instances more than one type of oxidative modification was observed at a particular amino acid residue position. It is important to note that these oxidized residues are not uniformly modified on all PS II complexes. In most instances the oxidation of a particular residue or groups of residues is expected to be quite rare. The actual degree of modification of any particular residue is quite difficult to quantitate and lies well beyond the scope of this communication. Indeed, such quantification is problematic since oxidative modification of many amino acid residues yields derivatives that are more polar than the parent residue. Consequently, peptides containing such modifications are more polar and more readily resolved by reversed-phase HPLC, making quantitative comparison to the unmodified, more hydrophobic, parent peptide difficult. It should also be noted that, in general, mass spectrometry coverage of intrinsic membrane proteins is challenging. The overall sequence coverage observed in this study for the proteins examined was: D1, 24%; D2, 27%; CP47, 41%; and CP43, 26%, which is quite comparable to that observed for these proteins by other investigators (for instance Nakamura et al. (32) observed; D1, 21%; D2, 22%; CP47, 23%; and CP43, 32%). However, the coverage of the residues located in the extrinsic loops of these proteins was significantly higher: D1, 35%; D2, 43%; CP47, 55%; and CP43, 43%. These are the domains of principal interest in this study, as the oxygen-evolving complex and proposed oxygen channels are all embedded in the lumenally exposed extrinsic loop regions of these proteins. Within this context it should be noted that any oxidative modifications to amino acid residues lying in the transmembrane helixes of these proteins would be very difficult to identify and would probably escape detection due to their poor resolution during reversed-phase chromatography.
Table 1.
Oxidatively Modified Residues in the D1, D2, CP43, and CP47 Proteins. Individual residues are listed along with the modifications observed. In some instances, different modifications were observed for the same residue on different peptides. For a complete list of oxidative modification types, the amino acids targeted, and mass modifications searched for in this study, see (29). Key: aa, aldehyde addition (−32 amu); ca, carbonyl addition (+14 amu) cysh, cysteine hydroxyl (−16 amu); gad, Glu/Asp decarboxylation (−30 amu); go, general oxidation (+16 amu); stcb, serine/threonine carbonyl (−2 amu).
| Protein Modified Residues | |
|---|---|
| D1 |
2Ta + stcb or go, 3Aa + go, 5La + go, 6Ea + gad, 17F + go, 18C + cysh, 130Eb + go, 133Lb + go, 135Fb + go, 239F + go, 241Q + ca, 242E + gad |
| D2 |
13K + go, 14D + gad, 15L + ca, 16F + go, 19M + go, 20D + ca, 21D + gad, 238P + ca, 239T + go, 242E + gad, 247M + go, 345E + gad, 347L + go, 349R + ca |
| CP43 |
27D + go, 28Q + ca, 29E + gad, 195D + gad, 196V + ca, 354Ec + gad, 355Tc + go, 356Mc + aa or go, 357Rc + ca or go, 367E + gad, 368P + go, 369L + ca, 375L + ca, 376D + gad, 379M + go, 460D + go, 461R + ca, 462D + gad, 469M + go |
| CP47 |
58Q + go, 60M + go, 327A + go, 329S + go, 330M + go or aa, 358R + go, 359M + go or aa or ca, 360P + go or ca, 361T + go, 363Fc + go, 364Ec + gad, 365Tc + go, 374D + gad, 376I + go, 377V + ca, 378R + go, 393E + gad, 394Q + ca, 398T + go,423R +ca, 424Ac + go, 425Q + ca, 428E + gad, 484P + ca, 485D + gad, 486L + go or ca, 487D + gad, 488V + go, 489Q + ca |
Not resolved in the Thermosynechococcus vulcanus structure (13)
Buried residues not adjacent to apparent cavities/channels
Buried residues adjacent to cavities/channels partially occupied by structural waters
The D1, D2, CP47 and CP43 proteins are highly homologous between higher plants and cyanobacteria (>85% similarity). Consequently, it is possible to map the modified residues present on these intrinsic proteins from spinach directly to the homologous residues in the T. vulcanus crystal structure. Indeed, 90% of the observed oxidatively modified residues in these spinach proteins are conserved (or conservatively replaced) in the corresponding T. vulcanus sequences. These results are shown in Fig. 2. As expected, the vast majority (81%) of the observed modifications are located on the surface of the PS II complex, which is exposed to the oxygenated bulk solvent. Additionally, four oxidized residues at the N-terminus of the D1 (2T, 3A, 5L, and 6E) are also almost certainly surface-exposed; however, this could not be verified by direct comparison to the T. vulcanus structure, as these residues are not resolved. Consequently, they were not included in the above calculations and are not shown in Fig. 2. Eight residues were observed to be modified which, while not located on the surface of the complex, are adjacent to cavities/channels within the complex, while three residues are buried and are not adjacent to such cavities/channels (Table 1).
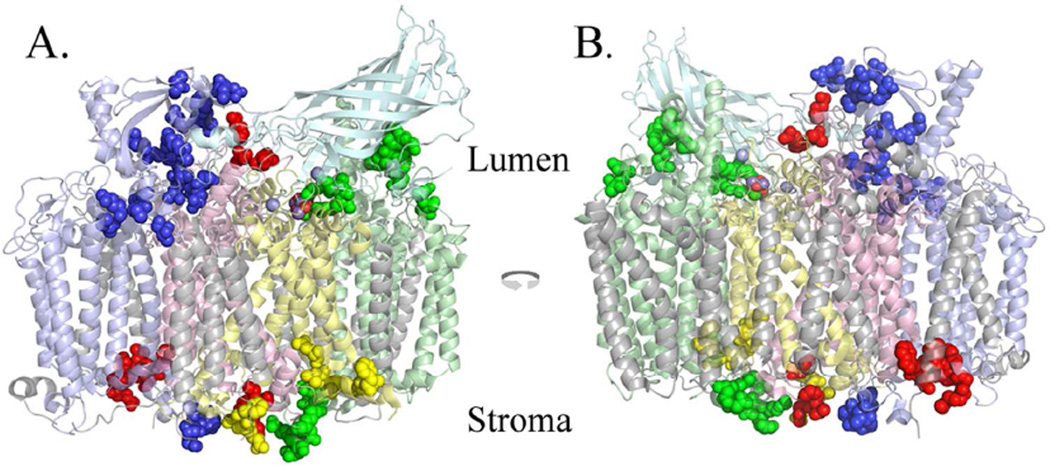
Figure 2.
Overview of the Oxidized Residues Identified in the D1, D2, CP43 and CP47 Proteins. Oxidized residues are shown as spheres superimposed on monomer I of the Thermosynechococcus vulcanus structure. Since the PsbU and PsbV subunits are not present in higher plant PS II, these chains are not shown. A., the view from outside Monomer I, looking towards the dimeric complex within the plane of the membrane. B., the view from Monomer II looking towards its interface with Monomer I within the plane of the membrane. Color key: CP47, pale blue; CP43, pale green; D1, pale yellow; D2, pale red; PsbO, pale cyan; all other chains, grey. The modified residues are shown in darker shades of these colors. Chlorides are shown as light blue spheres, and the manganese and oxygen of the Mn4O5Ca cluster are shown as purple and red spheres, respectively. It should be noted that many surface residues are modified on the PsbO subunit but these are not shown in this illustration. Figs. 1 and 2 were produced using PYMOL (31).
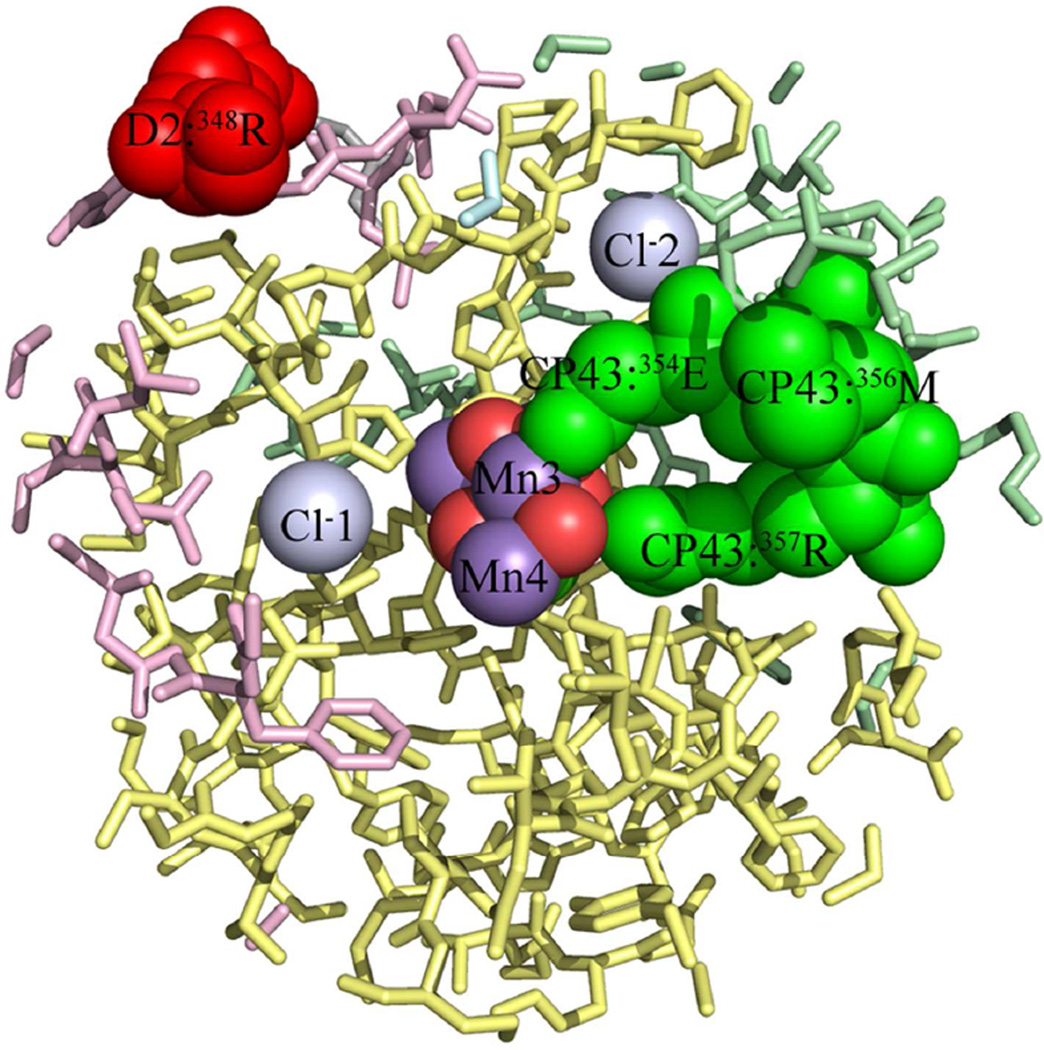
Fig. 3 illustrates all of the modified residues identified which are located within 15 Å of the metal cluster. The residues 354E, 355T, 356M and 357R of CP43 and residue 349R of the D2 protein are oxidatively modified (Fig. 3). This latter residue is exposed on the surface of the complex. The FT-ICR spectra identifying the oxidative modification of 354E, 355T, 356M and 357R of CP43 residues and the analogous unmodified peptides from both biological replicates are shown in Fig. 1 and Figs. S2–S5. In all cases, extremely high quality data were obtained with excellent b- and y- ion coverage. It should be noted that 354E is an inner sphere bidentate ligand to the Mn4O5Ca cluster, bridging Mn2 and Mn3. 357R is a second sphere ligand to the metal cluster and is hydrogen bonded to O2 and O4 (13). All four of these modified residues are in contact with cavities/channels within the photosystem. 354E is in contact with a channel partially occupied with structurally resolved water molecules (W538, W539, W397, W393, etc.) and is also in contact with a cavity containing Cl−2 and W498, W506, W547, W548, and W943. 355T is also in contact with this cavity. 356M is in contact with a channel partially occupied by the structurally resolved water molecules (W369, W546, W402, etc.). 357R is in contact with the same channel, with the closest structurally resolved water molecules being W1015, W1016, and W550. This channel leads to the surface of the complex.
Figure 3.
Oxidized Residues in the Vicinity of the Mn4O5Ca Cluster Active Site of PS II. A 15 Å sphere centered on Mn3 of the Mn4O5Ca cluster is shown. The CP43 residues 354E, 355T, 356M, and 357R are all buried residues which are in contact with cavities/channels on the interior of the complex. Please note that the D2 residue 349R of spinach is the equivalent of 348R of T. vulcanus and is a surface residue within the cyanobacterial structure. In this view 355T is not visible in this view. Color key: CP47, pale blue; CP43, pale green; D1, pale yellow; D2, pale red; PsbO, pale cyan; all other chains, grey. The modified residues are shown in darker shades of these colors. The oxidized residues are labeled, as are Cl−1 and Cl−2 and Mn3 and Mn4. These are shown as spheres. Some foreground residues, principally of the D1 chain, have been removed for clarity.
Discussion
The oxidative modification of these four CP43 residues (354E, 355T, 356M and, 357R) appears to indicate that they have been modified by ROS generated within the PS II complex. The close proximity of these residues to the Mn4O5Ca cluster supports the hypothesis that these residues are associated with an O2/ROS exit pathway leading from the metal cluster to the surface of the complex. In the earlier computational studies 354E, 356M and 357R were identified as being associated with a number of proposed channels. All three residues were located in the “narrow” channel (33) (also termed the “D, E, F” channel (21)) which was hypothesized to be a proton exit pathway. 356M and 357R were also proposed to be associated with the “back” (33), “(i)” (18) or “A1, A2” (21) channel. Ho and Styring (33) and Gabdulkhakov et al. (21) hypothesized that this channel was involved in vectoring water towards the Mn4O5Ca cluster, while Murray and Barber (18) suggested that this was an O2 exit pathway. Finally, 354E was also suggested to be located in the “large” (33), “(ii)” (18) or “B1,B2” (21) channel. Ho and Styring (33) and Gabdulkhakov et al. (21) suggested that this channel was involved in vectoring O2 away from the Mn4O5Ca cluster, while Murray and Barber (18) hypothesized that this was a water/proton exit pathway. 355T was not identified in any of these prior studies as being associated with any channel. It should be noted that several limitations, in large measure, exist with all of these largely computational studies. First, both the CAVER program (19), used by Murray and Barber (18) and Gabdulkhakov et al. (21), and calculations based on solvent accessibility used by Ho and Styring (33), examine static crystal structures. This is also true of the nobel gas (20, 21) and dimethyl sulfoxide (21) co-crystallization studies which have been reported. Large protein complexes exhibit substantial movement on the nsec time scales (34). Such movement in PS II could substantially alter the overall shape and dimensions of the identified channels within the photosystem. For substrate water, this problem has been recognized and addressed computationally by molecular dynamic simulations of water movement within the photosystem (34, 35); however, at this time, the movement of O2 has not been examined by these methods. Second, the majority of these studies have been performed on PS II structures of moderate resolution (≥ 2.9 Å). At these resolutions the accurate modeling of the location of the amino acid side chains is problematic. The accurate positioning of residue side chains could have important consequences with respect to the continuity and radius of proposed channels. At this time, no similar studies based on the high resolution (1.9 Å) PS II structure of Umena et al. (13) have been reported with respect to the identification of O2 channels. As noted above, nobel gas derivitization has been used in an attempt to identify oxygen egress channels in PS II. These gasses bind to hydrophobic domains hypothesized to be associated with O2 transport within the photosystem (20, 21). However, it should also be noted that in other systems hydrophilic O2 binding domains have been identified which do not interact with nobel gasses but which do interact with chloride and water (36, 37). Finally, all of these studies have been performed on crystal structures of PS II complexes which, while uniformly reported to be in S1, are clearly in an unknown, more highly reduced S-state (38). Even if these crystal structures were to accurately represent the protein architecture of the S1 state, an implicit assumption has been made that no conformational changes occur during the various S-state transitions, particularly the S3 → [S4] → S0 state transition associated with O2 formation, release, and water binding which could affect the location, shape and dimensions of possible O2/ROS exit pathways. Numerous recent studies have indicated that protein conformational changes do occur during S-state cycling (39–43), although the specific residues involved and the effects on possible channels within the photosystem have not been determined.
As noted above, oxidative modification of amino acid residues requires that they be exposed to ROS. PS II, particularly when under stress, can produce a variety of ROS at a variety of sites (15). Several studies have identified the production of ROS, particularly OH•, by the donor side of PS II. OH• is produced at or near the Mn4CaO5 cluster under moderately high temperature (44, 45) conditions. Additionally, it has been documented that the Mn4O5Ca cluster can directly absorb near-UV radiation (46–48), leading to damage to the manganese cluster. This damage could also lead to the partial oxidation of water, H2O2 production, and the generation of OH•. Both of these conditions could be experienced by the field-grown spinach used in these studies. It has also recently been shown that loss of the chloride associated with the active site under these conditions leads to the production of H2O2 (49). The loss of chloride also leads to the loss of manganese associated with the Mn4O5Ca cluster (50). This manganese has been hypothesized to react with the H2O2 in a classical Fenton reaction to produce OH• (15). It must be stressed, however, that at this time we cannot discriminate between these or other mechanisms that produce the ROS responsible for the oxidative modifications that we observe. Using mass spectrometry, it is also very difficult, and in most cases impossible, to distinguish hydroxide ion radical modifications of amino acid side chains from those produced by singlet oxygen, superoxide or other oxidative species (51, 52).
Our results indicate that the residues 354E, 355T, 356M and 357R of CP43 are oxidatively modified in spinach PS II and, consequently, these residues have been exposed to oxygen/ROS. These residues probably represent only a subset of the total oxidatively modified residues in the photosystem. For instance, oxidatively modified residues in the transmembrane domains of these proteins would generally be difficult to detect given the hydrophobic nature of the tryptic peptides derived from these helical domains. We hypothesize that 354E, 355T, 356M and 357R of CP43 are adjacent to molecular oxygen/ROS egress channel(s) which vector these products away from the Mn4O5Ca cluster towards the surface of the PS II complex. One possibility is that 356M and 357R which are in contact with the so called “back” channel (33) (also known as the “(i)” (18) or “A1,A2” (21) channels) that leads to surface of the PS II is an oxygen/ROS exit pathway as was originally suggested by Murray and Barber (18). 354E and 355T, however, do not appear to be in contact with this “back” channel in these analyses of the static crystal structures (18, 21, 33). Dynamic motions of PS II may modulate the dimensions and/or the precise location of the “back” channel, allowing 354E and 355T to become oxidatively modified. It is additionally possible, however, that the “narrow” channel (33) (also termed the “D, E, F” channel (21)) that is adjacent to 354E, 356M and 357R is an oxygen/ROS egress pathway. While in static structures this channel appears too narrow to allow the egress of oxygen/ROS, dynamic molecular motions or S-state-dependant conformational changes may facilitate this process. Alternatively, all of the oxidized residues (354E, 355T, 356M and, 357R) which we have identified in the vicinity of the Mn4O5Ca cluster may be associated with additional exit pathways for oxygen/ROS which have not been identified computationally. The differentiation between these and other hypotheses is currently underway.
Finally, it has not escaped our attention that another group of oxidized residues which we have identified (the D1 residues 130E, 133L, and 135F) may be involved in an ROS egress pathway operating on the reducing side of the photosystem. Oxidative modifications of amino acid residues in this vicinity have been previously observed, although the identities of the modified residues were not determined (53). These residues are in close proximity to PheoD1 which, under stress conditions, has the potential (Em = −610 mV, pH 7.0) of being a source of reducing-side ROS (15). This possibility will be examined in a separate communication.
Supplementary Material
Acknowledgments
Funding Statement:
Funding was provided by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE-FG02-98ER20310 to T.M.B and L.K.F., which supported the protein chemistry aspects and data analysis for this manuscript and the National Institutes of Health Grants RR019900 and GM58843 to P.A.L., which supported the mass spectrometry experiments of this manuscript.
Abbreviations
- amu
atomic mass units
- CID
collision-induced dissociation
- FT-ICR
Fourier transform-ion cyclotron resonance
- LiDS
lithium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- PS II
Photosystem II
- ROS
reactive oxygen species
- Sn
oxidation states (n=1,2,3,4) of the Mn4O5Ca cluster
Footnotes
Supporting Information Available
One figure showing the efficacy of the non-oxidizing gel system in suppressing artifactual electrophoresis-induced protein oxidation. Additionally, four figures showing the MS2 of the unmodified and modified peptides containing residues 354E, 355T, 356M and 357R of CP43 and their associated tables of the identified ions in these spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bricker TM, Burnap RL. The extrinsic proteins of Photosystem II. In: Wydrzynski T, Satoh K, editors. Photosystem II: The Water/Plastoquinone Oxido-Reductase of Photosynthesis. Dordrecht: Springer; 2005. pp. 95–120. [Google Scholar]
- 2.Nelson N, Yocum CF. Structure and function of Photosystems I and II. Ann Rev Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 3.Bricker TM, Roose JL, Fagerlund RD, Frankel LK, Eaton – Rye JJ. The extrinsic proteins of Photosystem II. Biochim Biophys Acta. 2012;1817:121–142. doi: 10.1016/j.bbabio.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Yi X, Hargett SR, Frankel LK, Bricker TM. The PsbP protein, but not the PsbQ protein, is required for normal thylakoid membrane architecture in Arabidopsis thaliana. FEBS Lett. 2009;583:2142–2147. doi: 10.1016/j.febslet.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 5.Yi X, Liu H, Hargett SR, Frankel LK, Bricker TM. The PsbP protein is required for Photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J Biol Chem. 2007;34:24833–24841. doi: 10.1074/jbc.M705011200. [DOI] [PubMed] [Google Scholar]
- 6.Yi X, McChargue M, Laborde SM, Frankel LK, Bricker TM. The manganese-stabilizing protein is required for Photosystem II assembly/stability and photoautotrophy in higher plants. J Biol Chem. 2005;280:16170–16174. doi: 10.1074/jbc.M501550200. [DOI] [PubMed] [Google Scholar]
- 7.Yi X, Hargett SR, Frankel LK, Bricker TM. The PsbQ protein is required in Arabidopsis for Photosystem II assembly/stability and photoautotrophy under low light conditions. J Biol Chem. 2006;281:26260–26267. doi: 10.1074/jbc.M603582200. [DOI] [PubMed] [Google Scholar]
- 8.Zouni A, Witt H-T, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Crystal structure of Photosystem II from Synechococcus elongatus at 3.8Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya N, Shen J-R. Crystal structure of oxygen-evolving Photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc Natl Sci Acad (USA) 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 11.Loll B, Kern N, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of Photosystem II. Nature. 2006;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 12.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-Å resolution and the roles of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 13.Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving Photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 14.Bricker TM, Ghanotakis DF. Introduction to oxygen evolution and the oxygen-evolving complex. In: Ort DR, Yocum CF, editors. Oxygenic Photosynthesis: The Light Reactions. Dordrecht: Kluwer Academic Publishers; 1996. pp. 113–136. [Google Scholar]
- 15.Pospisil P. Production of reactive oxygen species by Photosystem II. Biochim Biophys Acta. 2009;1787:1151–1160. doi: 10.1016/j.bbabio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Anderson LB, Maderia M, Ouellette AJA, Putnam-Evans C, Higgins L, Krick T, MacCoss MJ, Lim H, Yates JR, III, Barry BA. Posttranslational modifications in the CP43 subunit of Photosystem I. Proc Natl Acad Sci (USA) 2002;99:14676–14681. doi: 10.1073/pnas.232591599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreaden TM, Chen J, Rexroth S, Barry BA. N-Formylkynurenine as a marker of high light stress in photosynthesis. J Biol Chem. 2011;286:22632–22641. doi: 10.1074/jbc.M110.212928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray JW, Barber J. Structural characteristics of channels and pathways in Photosystem II including the identification of an oxygen channel. J Struct Biol. 2007;159:228–237. doi: 10.1016/j.jsb.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Petřek M, Otyepka M, Banáš P, Košinová P, Koča J. CAVER: A new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinform. 2006;7:315–315. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray JW, Maghlaoui K, Kargul J, Sugiura M, Barber J. Analysis of xenon binding to Photosystem II by x-ray crystallography. Photosyn Res. 2008;98:523–527. doi: 10.1007/s11120-008-9366-2. [DOI] [PubMed] [Google Scholar]
- 21.Gabdulkhakov A, Guskov A, Broser AM, Kern J, M h F, Saenger W, Zouni A. Probing the accessibility of the Mn4Ca cluster in Photosystem II; channels calculation, noble gas derivatization, and co-crystallization with DMSO. Structure. 2009;17:1223–1234. doi: 10.1016/j.str.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Berthold DA, Babcock GT, Yocum CF. A highly resolved oxygen-evolving Photosystem II preparation from spinach thylakoid membranes. FEBS Lett. 1981;134:231–234. [Google Scholar]
- 23.Ghanotakis DF, Babcock GT. Hydroxylamine as an inhibitor between Z and P680 in Photosystem II. FEBS Lett. 1983;153:231–234. [Google Scholar]
- 24.Rabilloud T, Vincon M, Garin J. Micropreparative one- and two-dimensional electrophoresis: Improvement with new photopolymerization systems. Electrophoresis. 1995;16:1414–1422. doi: 10.1002/elps.11501601234. [DOI] [PubMed] [Google Scholar]
- 25.Sun G, Anderson VE. Prevention of artifactual protein oxidation generated during sodium dodecyl sulfate-gel electrophoresis. Electrophoresis. 2004;25:959–965. doi: 10.1002/elps.200305800. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinform. 2007;8:133–137. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectroemtry to probe the structure of macromolecular complexes. Ann Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 29.Renzone G, Salzano AM, Arena S, D'Ambrosio C, Scaloni A. Mass spectrometry-based approaches for structural studies on protein complexes at low-resolution. Curr Proteom. 2007;4:1–16. [Google Scholar]
- 30.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence database using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Software. The PyMOL Molecular Graphics System, Version 1.4. Schrödinger, LLC; [Google Scholar]
- 32.Nakamura T, Dohmae N, Takio K. Characterization of a digested protein complex with quantitative aspects: An approach based on accurate mass chromatographic analysis with Fourier transform-ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:2558–2566. doi: 10.1002/pmic.200300812. [DOI] [PubMed] [Google Scholar]
- 33.Ho FM, Styring S. Access channels and methanol binding site to the CaMn4 cluster in photosystem II based on solvent accessibility simulations, with implications for substrate water access. Biochim Biophys Acta. 2008;1777:140–153. doi: 10.1016/j.bbabio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Vassiliev S, Comte P, Mahoob A, Bruce D. Tracking the flow of water through PS II using molecular dynamics and streamline tracing. Biochemistry. 2010;49:1873–1881. doi: 10.1021/bi901900s. [DOI] [PubMed] [Google Scholar]
- 35.Vassiliev S, Zaraiskaya T, Bruce D. Exploring the energetics of water permeation in Photosystem II by multiple steered molecular dynamics simulations. Biochim Biophys Acta. 2012;1817:1671–1678. doi: 10.1016/j.bbabio.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Colloc’h N, Gabison L, Monard G, Altarsha M, Chiadmi M, Marassio G, Santos JS-dO, El Hajji M, Castro B, Abraini JH, Prange T. Oxygen Pressurized X-Ray Crystallography: Probing the Dioxygen Binding site in cofactorless urate oxidase and implications for its catalytic mechanism. Biophys J. 2008;95:2415–2422. doi: 10.1529/biophysj.107.122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kommoju P-R, Chen Z-W, Bruckner RC, Mathews FS, Schuman Jorns MS. Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate. Biochemistry. 2011;50:5521–5534. doi: 10.1021/bi200388g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano J, Kern J, Irrgang KD, Latimer MJ, Bergmann U, Glatzel P, Pushkar Y, Biesiadka J, Loll B, Sauer K, Messinger J, Zouni A, Yachandra VK. X-ray damage to the Mn4Ca complex in single crystals of Photosystem II: a case study for metalloprotein crystallography. Proc Natl Acad Sci (USA) 2005;102:12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noguchi T, Sugiura M. Flash-induced Fourier transform infrared detection of the structural changes during the S-state cycle of the oxygen-evolving complex in Photosystem II. Biochemistry. 2001;40:1497–1502. doi: 10.1021/bi0023807. [DOI] [PubMed] [Google Scholar]
- 40.Halverson KM, Barry BA. Evidence for spontainious structural changes in a dark-adapted state of Photosystem II. Biophys J. 2003;85:2581–2588. doi: 10.1016/s0006-3495(03)74680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper IB, Barry BA. Pertubations at the chloride site during the photosynthetic oxygen-evolving cycle. Photosyn Res. 2007;92:345–356. doi: 10.1007/s11120-007-9147-3. [DOI] [PubMed] [Google Scholar]
- 42.Noguchi T. FTIR detection of water reactions in the oxygen-evolving centere of Photosystem II. Philos Trans R Soc Lond B Biol. 2007;363:1189–1195. doi: 10.1098/rstb.2007.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klauss A, Sikora T, Süss B, Dau H. Fast structural changes (200–900 ns) may prepare the photosynthetic manganese complex for oxidation by Tyr-Z. Biochim Biophys Acta. 2012;1817:1196–1207. doi: 10.1016/j.bbabio.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 44.Pospíšil P, Šnyrychová S, Nauš J. Dark production of reactive oxygen species in photosystem II membrane particles at elevated temperature: EPR spin-trapping study. Biochim Biophys Acta. 2007;1767:854–859. doi: 10.1016/j.bbabio.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita A, Nijo N, Pospíšil P, Morita N, Takenaka D, Aminaka R, Yamamoto Y. Quality control of Photosystem II: reactive oxygen species are responsible for the damage to Photosystem II under moderate heat stress. J Biol Chem. 2008;283:28380–28391. doi: 10.1074/jbc.M710465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hakala M, Tuominen I, Keränen M, Tyystjärvi T, Tyystjärvi E. Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of Photosystem II. Biochim Biophys Acta. 2005;1706:68–80. doi: 10.1016/j.bbabio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Ohnishi N, Allakhverdiev SI, Takahashi S, Higashi S, Watanabe M, Nishiyama Y, Murata N. Two-step mechanism of photodamage to Photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry. 2005;44:8494–8499. doi: 10.1021/bi047518q. [DOI] [PubMed] [Google Scholar]
- 48.Tyystjarvi E. Photoinhibition of Photosystem II and photodamage of the oxygen evolving manganese cluster. Coord Chem Rev. 2008;252:361–376. [Google Scholar]
- 49.Yadav DK, Pospíšil P. Role of chloride ion in hydroxyl radical production in photosystem II under heat stress: Electron paramagnetic resonance spin-trapping study. J Bioenerg Biomembr. 2012 doi: 10.1007/s10863-012-9433-4. [DOI] [PubMed] [Google Scholar]
- 50.Pospíšil P, Haumann M, Dittmer J, Sole VA, Dau H. Stepwise transition of the tetra-manganese complex of photosystem II to a binuclear Mn2(μ-O)2 complex in response to a temperature jump: a time-resolved structural investigation employing X-ray absorption spectroscopy. Biophys J. 2003;84:1370–1386. doi: 10.1016/s0006-3495(03)74952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schey KL, Finley EL. Identification of peptide oxidation by tandem mass spectrometry. Acc Chem Res. 2000;33:299–306. doi: 10.1021/ar9800744. [DOI] [PubMed] [Google Scholar]
- 52.Schöneich C, Sharov VS. Mass spectrometry of protein modifications by reactive oxygen and nitrogen species. Free Rad Biol Med. 2006;41:1507–1520. doi: 10.1016/j.freeradbiomed.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Sharma J, Panico M, Shipton CA, Nilsson F, Morris HR, Barber J. Primary structure characterization of the photosystem II D1 and D2 subnits. J Biol Chem. 1997;272:33158–33166. doi: 10.1074/jbc.272.52.33158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.