Abstract
The speciation of iron in intact human Jurkat leukemic cells and their isolated mitochondria was assessed using biophysical methods. Large-scale cultures were grown in medium enriched with 57Fe citrate. Mitochondria were isolated anaerobically to prevent oxidation of iron centers. 5 K Mössbauer spectra of cells were dominated by a sextet due to ferritin. They also exhibited an intense central quadrupole doublet due to S = 0 [Fe4S4]2+ clusters and low-spin (LS) FeII heme centers. Spectra of isolated mitochondria were largely devoid of ferritin but contained the central doublet and features arising from what appear to be FeIII oxyhydroxide (phosphate) nanoparticles. Spectra from both cells and mitochondria contained a low-intensity doublet from non-heme high-spin (NHHS) FeII species. A portion of these species may constitute the ‘labile iron pool’ (LIP) proposed in cellular Fe trafficking. Such species might engage in Fenton chemistry to generate reactive oxygen species. EPR spectra of cells and mitochondria exhibited signals from reduced Fe/S clusters, and HS FeIII heme and non-heme species. The basal redox state of mitochondria within cells is reduced as monitored by heme redox states; this redox poise is unaltered during the anaerobic isolation of the organelle. Contributions from heme a, b and c centers were quantified using electronic absorption spectroscopy. Metal concentrations in cells and mitochondria were measured using ICP-MS. Results were collectively assessed to estimate the concentrations of various Fe-containing species in mitochondria and whole cells – the first “ironome” profile of a human cell.
Iron is an essential component of human cellular metabolism, due to its extensive redox, catalytic and substrate-binding abilities (1). A comprehensive molecular-level understanding of cellular iron metabolism will not only require understanding the properties of individual Fe-containing proteins, but a systems-level understanding of iron trafficking and regulation. Connecting these two levels, a major objective of our research program, is especially challenging.
Iron is imported into the cell via two major pathways. One involves transferrin, a protein that reversibly binds FeIII ions (2). The other transferrin-independent pathway involves a cell-surface ferrireductase that reduces FeIII to FeII prior to uptake (3, 4). In both pathways, FeII ions are eventually pumped into the cytosol where some incorporate into apo-proteins and others are trafficked into various cellular compartments.
Mitochondria are “traffic hubs”, as they reportedly account for 20-30% of cellular Fe (5). Cytosolic Fe enters mitochondria via mitoferrins (6) and perhaps by other unidentified transporters. Mitochondria may also import Fe by direct contact with transferrin-containing endosomes (7) and via the siderophore 2,5-dihydroxybenzoic acid (8). Most cellular Fe/S clusters and all heme prosthetic groups are biosynthesized in mitochondria. The final step of heme biosynthesis, inserting FeII into protoporphyrin IX, occurs in the mitochondria (9).
The Fe used to build Fe/S clusters in the mitochondria is transferred onto Fe/S scaffold proteins in the matrix of the organelle (10). These clusters are inserted into various recipient apo-proteins including respiratory complexes I – IV (11). RCI contains 2 [Fe2S2] clusters and 6 [Fe4S4] clusters (12). RCII, a.k.a. succinate dehydrogenase, contains a low-spin (LS) heme b as well as [Fe2S2], [Fe4S4] and [Fe3S4] clusters (13). RCIII, a.k.a. cytochrome bc1, contains a [Fe2S2] Rieske cluster, one heme c and two heme b centers (14). Cytochrome c contains a LS heme c center, while RCIV, a.k.a. cytochrome c oxidase, harbors a di-copper center (CuA), a heme a center, and an active-site heme a3 center interfaced to the CuB center (15).
Ferritin is a cytosolic protein complex that stores Fe as an insoluble magnetically-interacting ferrihydrite. This prevents Fe from participating in Fenton chemistry (FeII + H2O2 → FeIII OH− + ·OH) (16). Ferritin can store up to ~4500 iron atoms in its inner core, estimated to correspond to 60% to 88% of total cellular iron (17). The complex also mobilizes Fe under iron-depleted conditions.
Mössbauer spectra of ferritin at low temperatures (4.2 K) exhibit a broad sextet pattern similar to mononuclear high-spin (HS) FeIII complexes. At ~ 70 K and higher, the sextet collapses into a broad quadrupole doublet with Mössbauer parameters of a superparamagnetic FeIII species (18). Unlike mononuclear HS FeIII species, ferritin is EPR-silent at low temperatures and gives rise to a broad EPR signal in the g ~ 2 region as temperatures are raised above ~ 50 K (19).
A proportion of cellular iron has been hypothesized to be loosely coordinated and to function either as feedstock for the assembly of Fe centers or as trafficking species that can be imported into organelles. These forms of Fe are collectively referred to as the Labile Iron Pool (LIP). There is substantial, albeit circumstantial, evidence for an LIP in the cytosol and mitochondrial matrix (20). The mitochondrial LIP may be used for Fe-S cluster and heme biosynthesis, whereas the cytosolic LIP may report on the overall Fe status of the cell, be imported into various organelles as needed (21), and serve as feedstock for cytosolic Fe/S cluster assembly (22). Due to its weak coordination, the LIP may also participate in Fenton chemistry and thus generate ROS. ROS-promoted damage may be associated with aging and neurodegenerative diseases (21).
In this study, we evaluated, on the systems-level, the speciation and distribution of iron (i.e. the ironome) in whole human Jurkat cells and their mitochondria using a biophysical approach centered on Mössbauer spectroscopy but also involving EPR, UV-vis and ICP-MS. We were unable to resolve individual Fe-containing species, but could resolve different types or categories of Fe-associated species. Our results indicate that ferritin and mitochondrial Fe are indeed dominant players in cellular Fe metabolism. NHHS FeII species were detected at concentrations exceeding previous estimates, and we provide evidence for FeIII oxyhydroxo nanoparticles in both isolated mitochondria and whole cells. These results were integrated into a semi-quantitative model describing the distribution and speciation of iron in a human cell.
Experimental Procedures
Bioreactor and Cell Culture
T-REx Jurkat cells (Invitrogen, R72207) were grown in a custom-built 25 L bioreactor under an atmosphere of 75% N2, 20% O2, and 5% CO2 as established with a high-precision gas mixer (MG Industries). All gases were 99.9% pure (certified grade) and were delivered to the culture medium at a combined flow rate of ~ 3 mL/min. Temperature was maintained at 37° C by circulating water through the jacket of an all-glass bioreactor (Chemglass). The 12″ diameter stainless-steel lid plate had 4 evenly spaced 1″ ID ports with screw tops surrounding a central stir-motor connection. Two 1″ × 3″ and two 1″ × 30″ Teflon rods were inserted into the ports. The long rods were used to add media, harvest cells, and bubble gases through the culture. The short rods were used to deliver gases to the headspace and exhaust gases. A small hole was drilled into the short Teflon rod used to exhaust gases, through which a thin tube connected to a syringe was threaded. This tube was used to remove small samples of culture for cell count and to assess viability. A rounded-rectangular (3″ × 24″) Teflon paddlewheel was attached to the underside of the lid, and a stir-motor (Arrow Engineering Co., Inc model 350) rotated it at ~ 60 rpm. The bioreactor was wrapped with a black cloth during cell growth.
Starter cell cultures were grown in a CO2 incubator (Nuaire, Model NU-5500), starting from a 100 mL culture, which was scaled up to 1 L once growth was evident. When the 1 L culture reached a density of ~3 × 106 cells/mL (the maximum density), it was used to inoculate 2 L of medium in the bioreactor. Additional medium was added when cells reached maximum density. When the culture volume was < 10 L, gases were delivered to the headspace above the liquid; when the volume was > 10 L, gases were bubbled through the liquid. Cells in all culture volumes were grown in RPMI 1640 medium (Sigma Aldrich) supplemented with 5% Newborn Calf Serum (Invitrogen) and an antimycotic-antibiotic cocktail (Invitrogen) to give final concentrations of 100 units/L penicillin, 100 μg/L streptomycin, 0.25 μg/L amphotericin B, and 10 μM 56Fe or 57FeIII citrate. Pluronic F-68 (Sigma Aldrich) was added to a final concentration of 0.05% w/v to prevent hydrodynamic damage to the cells. Samples were removed daily and inspected by phase-contrast microscopy for viability and contamination, using 0.4% Trypan Blue solution. Cells were harvested when the cells reached maximum density. Cells were removed from the bioreactor using a peristaltic pump and centrifuged at 800×g for 10 min (Beckman Coulter Avanti J-26 XP centrifuge, JLA 8.1 rotor).
Whole cell Mössbauer and EPR sample preparation
EPR and Mössbauer samples of cells were prepared from 1 to 3 L of cultures that had reached maximum density. Cells were washed with phosphate-buffered saline (PBS, pH 7.4) containing 1 mM EGTA, followed by another wash with EGTA-free PBS buffer. Cells were then packed into 3 mL Mössbauer cups or 4 mm OD quartz EPR tubes by centrifugation at 800×g for 1 hr. The supernatant was removed and samples were frozen in liquid N2 aerobically.
Mitochondria Isolation
Pellets of freshly harvested cells (wet weight 50-60 g) from 24 L of culture were washed twice with PBS buffer (pH 7.4). Pelleted cells were re-suspended in ~ 500 mL of degassed Mitochondria Isolation Buffer (MIB: 225 mM D-mannitol, 75 mM sucrose, 5 mM HEPES, 1 mM EGTA and 1 mM PMSF, pH 7.4) to a cell density of ~2 × 107 cells/mL. From this point onwards, all manipulations were conducted anaerobically, mostly in an Ar-atmosphere MBraun Labmaster glove box containing less than 5 ppm O2 as monitored by a Teledyne O2 analyzer (Model 310). For centrifugation steps, samples were sealed in airtight centrifuge bottles inside the glove box before removing for centrifugation; afterwards, they were returned to the box before being opened for further manipulations. Cells were disrupted anaerobically by nitrogen cavitation at 800 psi for 30-40 min, using a disruption vessel (Model 4635, Parr Instruments). The cavitation extract was centrifuged at 800-times;g for 10 min and the pellet was discarded. The supernatant was centrifuged again at 9000×g for 30 min using a Sorvall Evolution centrifuge with SLA-1500 rotor. The resulting pellet, which contained crude mitochondria, was re-suspended in ~ 2 mL of MIB buffer, layered over a discontinuous gradient of 7.5 mL of 6% Percoll/3 mL of 17% Histodenz/3 mL of 35% Histodenz in MIB as described (23) and centrifuged at 45,000×g for 1 hr using a Beckman Coulter Optima L-90K ultracentrifuge with a SW 32 Ti swinging-bucket rotor. Mitochondria were collected from the 17-35% Histodenz interface and washed once with MIB. Mitochondria were then packed into Mössbauer cups by centrifugation in the ultracentrifuge (SW 32 Ti rotor) at 10,000×g for 1 hr. The supernatant was removed and the mitochondrial samples were frozen and stored in liquid N2.
Protein concentrations
Protein concentrations were determined using a BCA Protein Assay kit (Thermo Scientific Pierce Protein Research Products) as per manufacturer‘s instructions. Bovine serum albumin (BSA) was used to generate a standard curve (0-2 mg/mL). Absorbances were measured at 562 nm.
Biophysical Studies
EPR spectra of whole cells and isolated mitochondria were collected on an X-band EMX spectrometer (Bruker Biospin Corp., Billerica, MA) equipped with an Oxford Instruments ER900A cryostat. Spin quantifications were performed with SpinCount (http://www.chem.cmu.edu/groups/hendrich/facilities/index.html), using 1.00 mM CuSO4-EDTA as standard. Mössbauer spectra were acquired using a Model MS4 WRC spectrometer (SEE Co. Edina, MN) and a model LHe6T spectrometer (SEE Co., Edina MN). The latter instrument was equipped with a variable field superconducting magnet capable of generating 0 - 6 Tesla fields. Both instruments were calibrated using a spectrum of α-Fe foil collected at room temperature.
Packing Efficiencies
Jurkat cells were packed into an EPR tube by centrifugation at 800×g for 1 hr (Beckman Avanti-J26 XP centrifuge). Isolated mitochondria were similarly packed at 10000×g (Beckman Ultracentrifuge). The supernatant was discarded. Packed pellets of volume Vpellet consisted of the sample and interstitial buffer (Vpellet = Vsample + Vint). To determine packing efficiency, defined as 100·Vsample/Vpellet, the pellet was re-suspended in a known volume of buffer (Vbuffer1) containing 100 μM of a membrane-impermeable fluorescent Compound 5 (24). The sample was packed again for 1 hr. The supernatant, of volume Vsup1 and containing Compound 5 at concentration Csup1, was removed. Csup1 was determined using a fluorescence spectrometer (Koala 90080, ISS Inc). The conservation of matter requires that
where Vint1 is the volume of the interstitial buffer in the pellet. This equation was solved for Vint1 allowing the first packing efficiency (100·Vsample1/Vpellet1) to be determined.
The pellet was re-suspended in a known volume of buffer (Vbuffer2) lacking the fluorescent compound and the suspension was packed again. In this case, the conservation of matter requires that
Csup2, Vsup2 and Vpellet2 were measured as above, allowing Vint2 and a second packing efficiency (100·Vsample2/Vpellet2) to be calculated. The two packing efficiencies were averaged.
ICP–MS
Packed whole cells and isolated mitochondria from EPR tubes were diluted with a known volume of buffer (PBS buffer for whole cells, MIB for mitochondria). Suspensions were placed in 15 mL BD Falcon tubes and digested in concentrated trace-metal-grade nitric acid (final concentration 20-30%) for ~12 hrs. Samples were diluted with distilled and deionized water to a final acid concentration of 3%. The metal concentrations of digested samples were determined in both H2 reaction and He collision modes using ICP-MS (Agilent Technologies model 7700x). Values obtained from both modes were adjusted for dilution factors and packing efficiencies, and then averaged.
Electron Absorption Spectroscopy
Packed cell and mitochondrial samples from EPR tubes were diluted 3-fold with isolation buffer. Suspensions were placed in a custom 2 mm pathlength quartz UV-Vis cuvette (Precision cells), sealed with a septum, and removed from the glove box. Spectra were acquired on an Hitachi U3310 spectrometer with a Head-on photomultiplier tube, then simulated using OriginPro as described (25).
Western Blots
Forty μg of protein from cell extracts or mitochondria was loaded and separated on a 12% polyacrylamide gel (Bio-Rad) using SDS-containing running buffer and 100 V potential. Proteins were transferred to Immun-Blot PVDF membranes (Bio-Rad) overnight at 20 V. The membranes were incubated for 2 hrs using Blocker™ Casein solution (Thermo Scientific). Mouse monoclonal primary antibodies (Abcam) specific to human mitochondrial porin, human endoplasmic reticular protein PDI, human nuclear protein p84 were all diluted 1:1000 and mouse monoclonal antibody specific to human lysosomal protein LAMP1 was diluted 1:10,000 in Blocker™ casein solution. Membranes were incubated with primary antibody solutions for 1 hr, followed by another blocking step for 30 min using Blocker™ casein. Membranes were then incubated with goat anti-mouse HRP conjugated secondary antibody (Invitrogen) diluted 1:3000 in Blocker™ casein solution, followed by detection using the Thermo Scientific Enhanced Chemiluminescent Western Blotting Substrate. Images were obtained using the FujiFilm LAS-4000 mini imager and analyzed using ImageJ.
Electron Microscopy
Mitochondria were fixed in 3% (v/v) glutaraldehyde in MIB, washed 3 times with MIB, fixed in 1% (v/v) osmium tetroxide, infiltrated and embedded in epoxy resin by polymerization at 60° C overnight. Ultrathin sections were obtained using an Ultracut E microtome (Reichert-Jung) and post-stained on drops of 2% (w/v) uranyl acetate and 100 mM lead citrate as described (26). EM images were obtained on a JEOL 1200 EX Transmission Electron Microscope.
Results
Analytical Characterization
Fourteen batches of Jurkat cells were grown in medium containing ~ 6 WM endogenous 56Fe (as measured by ICP-MS) and supplemented with 10 μM 57FeIII citrate. The percent enrichment (ca. 75%) indicated that cells incorporated both sources of Fe. Mitochondria were isolated from 9 of these batches. Due to limited amounts of material, not every batch was characterized by every technique; characterizations performed on each batch are summarized in Tables S1 and S2.
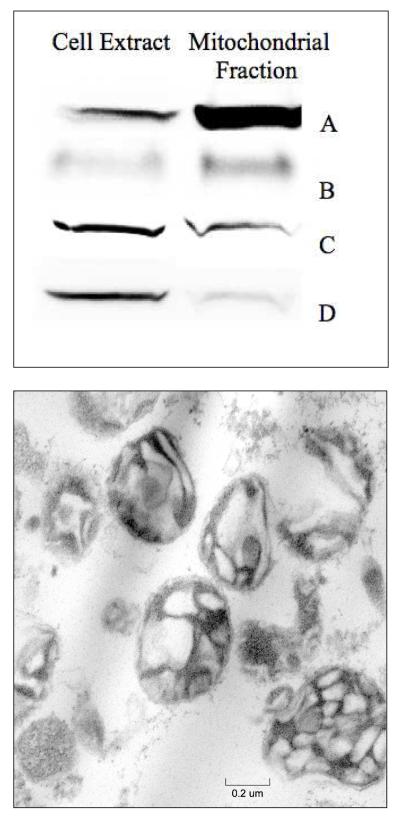
Purities of 6 batches of isolated mitochondria were evaluated by Western blots. Membrane integrity from 3 batches was assessed by EM (Table S2). Western analysis indicated a 10-fold increase in the mitochondrial porin protein in isolated mitochondria relative to the same mass of cell extract protein. Isolated mitochondria contained small levels of contaminating proteins, including marker proteins LAMP1 from lysosomes, p84 from nuclei, and PDI from ER (Fig. 1, top panel). EM images of isolated mitochondria (Fig. 1) generally showed intact organelles with sharp cristae, though some unidentified density was evident. In summary, EM and Western blot analyses suggest that the isolated mitochondria used in this study were 70% - 80% pure and generally intact.
Figure 1.
Characterization of isolated mitochondria. Top, western blot of isolated mitochondria; A, Porin (mitochondria); B, LAMP1 (lysosomes); C, Endoplasmic reticulum (PDI); and D, p84 (nuclei). Bottom, electron micrograph of isolated mitochondria, magnification = 40,000×.
We determined the absolute concentration of metal ions and protein in 5 batches each of mitochondria and whole cells (Table 1; individual determinations in Table S4). These values were obtained by dividing observed concentrations in packed mitochondria and cells by measured packing efficiencies (65% ± 10% and 81% ± 6%) respectively. See Table S3 for individual packing efficiency results.
Table 1.
Analytical properties of whole Jurkat cells and isolated mitochondria. Reported metal, UV-vis and EPR concentrations refer to packed cells and mitochondria after dividing by packing efficiencies. Values reported for isolated mitochondria were obtained by also dividing measured values by 0.75, to account for presumed metal-free impurities. The absolute uncertainty in percentages obtained by Mössbauer spectroscopy is ± 3%. Replicates n is given in the column to the right of the corresponding parameter. Concentrations of respiration-related mitochondrial proteins are estimated from the collective results of this study.
| Mitochondria | n | Whole Cells | n | |
|---|---|---|---|---|
| Protein (mg/mL) | 52 ± 12 | 4 | 62 ± 11 | 4 |
| [Fe] (μM) | 1120 ± 95 | 5 | 400 ± 70 | 5 |
| [Cu] (μM) | 115 ± 8 | 5 | 28 ± 4 | 5 |
| [Zn] (μM) | 167± 94 | 5 | 408 ± 135 | 5 |
| [Mn] (μM) | 14 ± 3 | 5 | 7.2 ± 0.6 | 5 |
| FeIII oxyhydroxy nanoparticles (%) | 37 | 3 | 18 | 2 |
| Ferritin-like (%) | 15 | 3 | 40 | 2 |
| Central Doublet (%) | 27 | 3 | 27 | 2 |
| Non-Heme HS FeII | 8 | 3 | 11 | 2 |
| HS FeII Hemes | 4 | 3 | 4 | 2 |
| g = 1.94 (μM) | 3.3 ± 0.6 | 4 | 0.3±0.1 | 4 |
| g = 1.90 (μM) | 3.3 ± 0.6 | 4 | 0.3±0.1 | 4 |
| g = 2.00 (μM) | 0.2 ±0.06 | 4 | ~ 0 | 4 |
| g = 4.3 (μM) | 6 ±0.6 | 4 | 1.5 ± 0.5 | 4 |
| g = 6.0 + (6.4, 5.4) | 0.5 ±0.3 | 4 | ~ 0 | 4 |
| Reduced [Heme a] (μM) | 37 ± 5 | 4 | 10 ± 2 | 4 |
| Reduced [Heme b] (μM) | 21 ± 4 | 4 | 5 ± 2 | 4 |
| Reduced [Heme c] (μM) | 75 ± 11 | 4 | 20 ± 3 | 4 |
| Cytochrome c oxidase (μM) | ~ 18 | n/a | ||
| Cytochrome c (μM) | ~ 70 | n/a | ||
| Cytochrome bc1 (μM) | ~ 4 | n/a | ||
| Succinate Dehydrogenase (μM) | ~ 4 | n/a | ||
| Respiratory Complex I (μM) | ~ 1 | n/a |
Biophysical characterization of mitochondria
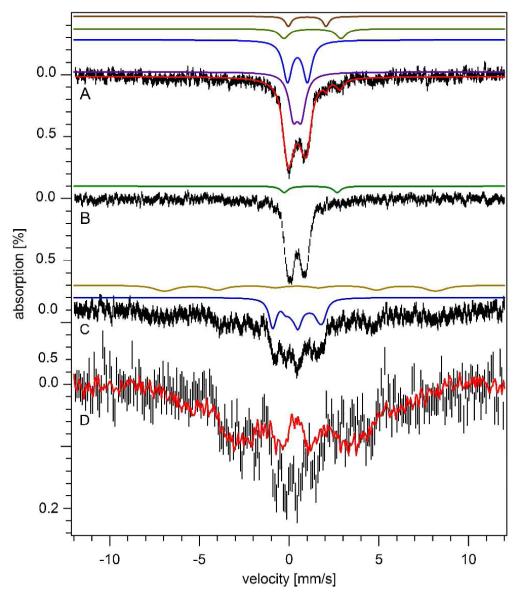
Three batches of isolated mitochondria were analyzed by Mössbauer spectroscopy (Table S2). Results are summarized in Table 1. Low-temperature low-field Mössbauer spectra exhibited 4 distinguishable species. The spectrum from batch M04 is shown in Fig. 2A while others are given in Fig. S1. The dominating features consisted of two overlapping quadrupole doublets in the center of the spectrum. These included the central doublet (CD) with parameters typical of S = 0 [Fe4S4]2+ clusters and LS ferrous heme centers (δ = 0.46 mm/s; ΔEQ = 1.2 mm/s), and a broad doublet with parameters (δ = 0.48 mm/s and ΔEQ = 0.57 mm/s) typical of FeIII (phosphate) oxyhydroxide nanoparticles. Such particles have been observed in certain genetic strains of yeast mitochondria (27, 28). The blue and purple lines in Fig. 2A simulate these two doublets.
Figure 2.
Mössbauer spectra of mitochondria isolated from Jurkat cells. A, 5 K and 0.05 T; the red line is a total simulation (see Table 1 for percentages); B, same as A but at 70 K, with simulation of NHHS FeII; C, same as A except at 6 T and 4.3 K. The brown line simulates the ferritin sextet while the blue line simulates the CD; D, same as C but after subtraction of sextet and CD simulations. The red line is a 6 T, 4.3 K spectrum of yeast mitochondria isolated from Aft1-1up cells (24). The applied magnetic field was parallel to the γ radiation in A and B, perpendicular in C and D.
The magnetic properties associated with the two doublets were investigated at high applied magnetic field (Fig. 2C). The solid blue line simulates the CD and confirms that it arises from diamagnetic [Fe4S4]2+ clusters and LS ferrous heme centers. The nanoparticle doublet broadened significantly at 6 T. The spectrum was compared to that of a yeast sample that was dominated by FeIII oxyhydroxide nanoparticles (24). Spectral features (Fig. 2D) were similar, suggesting that the second dominant doublet in the low-field spectrum of human mitochondria arose from a similar type of nanoparticle.
Low-temperature, low-field Mössbauer spectra of human mitochondria included 3 minor features. A quadrupole doublet was simulated (brown line, Fig. 2A) with parameters typical of HS FeII hemes (δ = 1.00 mm/s; ΔEQ = 2.00 mm/s). Another doublet was simulated using parameters (δ = 1.30 mm/s; ΔEQ = 3.00 mm/s) typical of non–heme HS FeII ions coordinated by O and N donors. The concentrations of heme centers in mitochondria were quantified by electronic absorption spectroscopy (Fig. 5A and Table 1).
Figure 5.
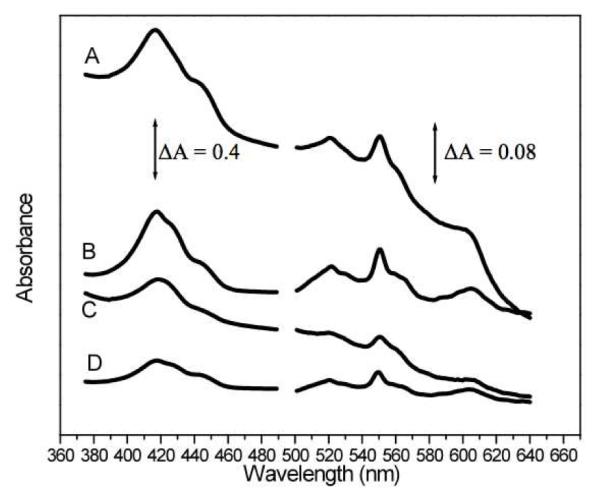
Electronic absorption spectra of mitochondrial (A) and cell (C) suspensions. Simulated spectra B and D were generated by combining individual spectra of isolated proteins containing the different types of hemes.
The 5 K 0.05 T spectrum of mitochondria also included broad features that were barely distinguishable from the baseline and were spread over the velocity range (Fig. 2A). At 70 K, these features collapsed into the center (Fig. 2B) as is typical of ferritin. At 6 T, the same features sharpened, revealing the same positions as the ferritin sextet in whole cell spectra (Fig. 2C). These features in mitochondria could arise from either contaminating ferritin, mitochondrial ferritin, or other ferritin-like material.
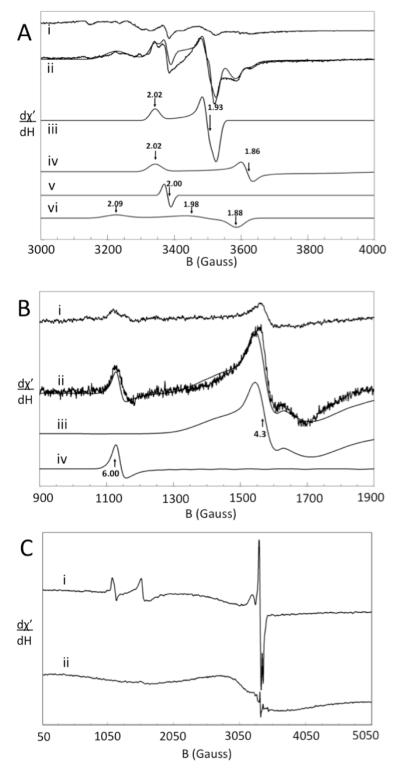
Low-temperature EPR of isolated mitochondria in the g = 2 region (Fig. 4A, ii) revealed a number of features (Fig. 4A). Spectra were decomposed into four signals with g –values indicated in Fig. 4A, iii – vi. Spin quantifications of most of these signals are given in Table 1. The signal with g = 1.98 (Fig. 4A, vi) may arise from Fe/S clusters but this is uncertain. The g = 2.00 signal originates from an organic radical, while the g = 1.94 signal probably arises from the [Fe2S2]+ cluster of succinate dehydrogenase (29). A very low-intensity signal at g = 2.15 was reproducibly present (Fig. 4A, i and ii), but its origin is unknown.
Figure 4.
X-band EPR spectra of Jurkat cells and mitochondria. A, high-field region of (i) cells and (ii) mitochondria (ave. of 5 and 3 scans, respectively). Simulations iii, iv, v, and vi were of the gave = 1.94, 1.90, 2.00, and 1.98 signals, respectively. The solid line overlaying ii is a combined simulation. Temperature, 8 K; frequency, 9.47 GHz; microwave power, 2.012 mW. B, low-field region of (i) cells and (ii) mitochondria (ave. of 5 and 3 scans, respectively). Simulations iii and iv are of the g = 4.3 and 6.0 features, respectively. The solid line overlaying ii is a combined simulation (same EPR conditions). C, wide-sweep spectra of mitochondria at 8 K (i) and 80 K (ii). Frequency, 9.46 GHz, power, 20.12 mW. In all spectra, modulation amplitude was 10 G, modulation frequency, 100 kHz, conversion time, 164 ms, and sweep time, 336 sec.
The low-field region (Fig. 4B) exhibited a g = 4.3 signal assigned to non-heme HS FeIII species with rhombic symmetry, as well as a signal at g = 6.0 which probably arises from the active site of cytochrome c oxidase in a mixed redox state, with heme a3 and CuBIII in the Fe and CuI states (30). At high temperatures, a broad EPR signal in the g = 2 region developed, with inverse Curie law dependence (Fig. 4C) as is characteristic of superparamagnetic FeIII oxyhydroxide nanoparticles.
Biophysical characterization of Jurkat cells
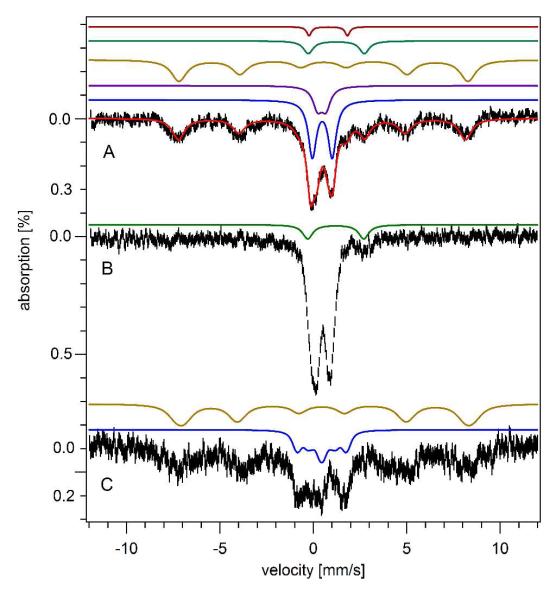
The dominant feature in the 5 K 0.05 T Mössbauer spectrum (Fig. 3A) of whole Jurkat cells was a broad sextet. This feature was simulated (brown line) with effective δ = 0.55 mm/s, ΔEQ = 0.25 mm/s, and Heff = 480 kG. This species was reminiscent of HS FeIII ions in ferritin (31). At 4.3 K and 6 T (Fig. 3C), magnetic hyperfine interactions were similar to those of ferritin (δ = 0.54 mm/s; ΔEQ = 0.20 mm/s; Heff = 450 kG; η = 1.00). At 70 K, the sextet collapsed into a doublet (Fig. 3B) (δ = 0.50 mm/s; ΔEQ = 0.75 mm/s), again typical of ferritin. We conclude that the sextet arises from ferritin.
Figure 3.
Mössbauer spectroscopy of Jurkat cells. A, 5 K and 0.05 T. Colored lines above the spectrum are individual simulations; the overlaid red line is a total simulation (see Table 1 for percentages). B, 70 K 0.05 T. The green line simulates the NHHS FeII doublet. C, 6 T and 4.3 K. The brown and blue lines simulate the sextet and CD respectively.
Other features of the 5 K, 0.05 T Jurkat cell Mössbauer spectrum were essentially identical to those in spectra of mitochondria, including a CD, a nanoparticle doublet, a quadrupole doublet from HS FeII hemes and a doublet arising from NHHS FeII species. Low-temperature EPR spectra of cells (Fig. 4, i in A and B) were similar to those of isolated mitochondria, including many features in the high field region and the g = 6.0 and 4.3 signals at low field. The signals were 5-7 fold less intense in cell spectra than in mitochondrial spectra. Concentrations of FeII heme centers in cells were quantified using electronic absorption spectroscopy (Fig. 5C and Table 1).
Discussion
We determined the absolute concentrations of iron and other transition metals in Jurkat cells and mitochondria isolated from these cells by determining packing efficiencies, and then using these values to correct measured concentration of metals in packed samples. There are few previous reports of absolute metal ion concentrations in mammalian cells as most are given as Fe:protein concentration ratios. Some reports are consistent with ours (32-34) while others range from being 10-fold higher (35) to nearly 50-fold lower (36). Assuming a cellular volume of 200 fL (37) we calculate from published data (35) ~ 5 mM Fe in lymphocytes, 12 times higher than we measured. Another determination in the same type of cells indicated 0.3-0.4 μmol Fe/g protein (36), 45-fold lower than we measured. Rat intestinal epithelial cells reportedly contained ~ 1.4 μmol Fe/g protein (5), ~10-fold lower than our measurements. Besides being outside the range of concentrations that we observed, these reported concentrations are inconsistent with the Mössbauer and EPR intensities observed here. For example, a 5 mM Fe sample would exhibit far greater % effect and spin intensities than what we observed; spectra would be unobtainable with samples that contained < 100 μM Fe. The spectroscopic intensities observed here require Fe concentrations in Jurkat cells and mitochondria within the uncertainties of the concentrations reported here.
Our results allow us to estimate the fraction of Jurkat cell volume due to mitochondria (Vmito/Vcell). Heme a is exclusively found in cytochrome c oxidase, a mitochondrial protein complex. The UV-Vis spectral absorption due to reduced heme a can be readily quantified. Comparing the intensity of this feature in cell vs. mitochondrial suspensions (Table 1) suggests that the fraction of cell volume due to mitochondria (Vmito/Vcell) is ≈ 0.27. Heme c may also be found exclusively in mitochondria (as cytochrome c and cytochrome bc1), and a similar analysis suggests the same ratio. After correcting for dilution, packing efficiencies, and an estimated 25% heme-free impurities, our results indicate Vmito/Vcell ≈ 0.20 ± 0.04. This agrees with previous electron microscopy studies which have measured this ratio in liver cells to be 0.18 (38).
The conservation of matter suggests that
where [Fe]other is the average [Fe] in all non-mitochondrial compartments of the cell excluding the ferritin contribution in the cytosol. By assuming Vmito/Vcell = 0.2 and values in Table 1 and Fig. 6, this relationship implies that [Fe]other ~ 70 μM. This concentration includes contributions from Fe in other organelles and cytosolic Fe other than ferritin.
Figure 6.
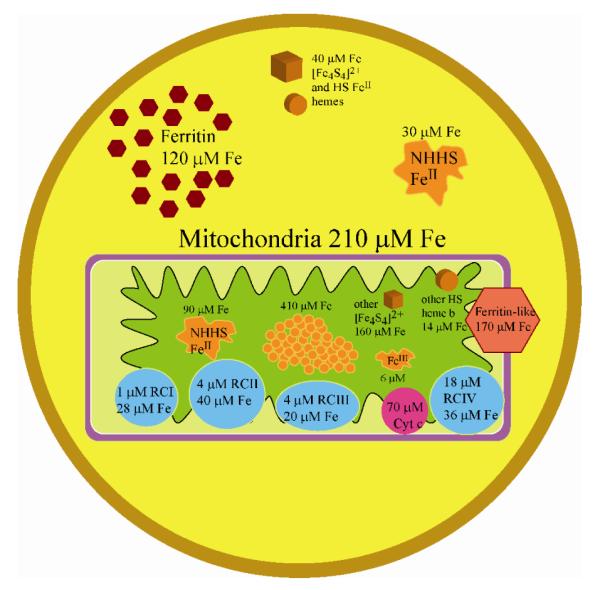
Ironome profile of Jurkat cells and mitochondria. See text for details.
Our results also allow us to estimate the concentrations of respiratory complexes in human mitochondria (Table 1). The concentration of cytochrome c oxidase is ~ 18 μM, half of the concentration of heme {a+a3}. This concentration (and others in Table 1 and below) refer to moles per mitochondrial volume; the concentration within the particular subcompartment of the mitochondria where RCIV is located (the IM) will be substantially higher. Heme a is LS and likely contributes to the CD, while heme a3 is HS and contributes to the HS FeII doublet.
Cytochrome c oxidase binds 3 Cu ions (15), suggesting that about half of mitochondrial Cu is associated with this enzyme. Most of the remainder may be associated with a Cu pool (39). Since no CuII EPR signals were apparent in mitochondrial spectra, most or all of the Cu ions in this pool would appear to be in the CuI state. Similar conservation-of-matter relationships suggest that the average concentrations of Cu, Mn, and Zn in non-mitochondrial locations within the cell are 6, 4, and 375 μM, respectively. Approximately 80%, 40% and 10% of the Cu, Mn, and Zn in the cell, respectively, are located in mitochondria.
About 3% of cytochrome c oxidase molecules are in the {FeIII heme a3 CuBI} state and exhibit the signal at g = 6.0. This signal was considered a “transient” catalytic intermediate arising from the one-electron reduction of the oxidized OH state (40, 41). However, our results demonstrate that it is stable and present reproducibly under non-turnover conditions in both cells and anaerobically isolated mitochondria. Establishing whether it functions in catalysis will require further study, but its presence indicates that the thermodynamic reduction potential of the heme a3 (FeIII/FeII) couple is less positive than that of the CuBII/CuBI couple under in vivo conditions. This conclusion is supported by a previous determination of E0‘ = 210 mV vs. SHE for heme a3 and 340 mV for CuB (42). We calculate a potential of ca. 170 mV for the solution in equilibrium with this site, under the (anaerobic) conditions of our experiments.
We estimate a concentration of ~ 4 μM for RCIII (cytochrome bc1) in mitochondria, based primarily on the spin concentration of the g = 1.90 signal which we assign to the Rieske Fe/S cluster. RCIII contains 1 and 2 equiv/mol of LS heme c1 and LS heme b, respectively (14), both of which would have contributed to the CD in our study.
We estimate a similar concentration for RCII, based on the spin concentration of the g = 1.94 signal which presumably arises from the RCII [Fe2S2]1+ cluster (25, 29). This enzyme also contains 1 LS Heme b, an [Fe3S4] cluster and an [Fe4S4] cluster (13). Collectively, the heme b contribution of both respiratory complexes is ~ 8 μM, suggesting that ~ 14 μM of heme b is due to other mitochondrial proteins such as catalase and cytochrome c peroxidase. The heme c contribution from cytochrome bc1 is minor, suggesting that the concentration of cytochrome c in mitochondria is ~ 70 μM.
Heme c centers are LS, as is heme a of cytochrome c oxidase and, we estimate, about half of the heme b centers. Collectively, this corresponds to ~ 100 μM or ca. 9% of mitochondrial Fe. This Fe would contribute to the CD. Since the CD represents ~ 27% of the Fe in the mitochondria, we conclude that ~ 17% (200 μM) is due to Fe in the form of [Fe4S4]2+ clusters. After subtracting ~ 4 μM [Fe4S4] for the cluster in RCII, ~ 46 μM remains for other [Fe4S4]-containing proteins in mitochondria.
The ratios of RCI:RCIII:RCIV have been measured in rat and bovine tissues to be ca. 1:3:8 (43). The ratio of RCIII:RCIV in our mitochondrial samples was 3:18, which suggests an RCI concentration between 0.5 and 1 μM. Since RCI contains 6 [Fe4S4] clusters and 2 [Fe2S2] clusters (12), ~ 6 μM of [Fe4S4] clusters due to RCI would contribute to the CD, leaving ~ 40 μM [Fe4S4] clusters for the other mitochondrial proteins. As with yeast mitochondria (25), the concentration of [Fe2S2]2+ clusters in human mitochondria appears to be low since a doublet due to this species was not detected.
Our calculations indicate that ca. 20% of the cell volume is due to mitochondria; this implies that ~ 55% of the Fe in the cell should be mitochondrial. Of this, ca. 27% (60 μM) is due to the CD. This corresponds to ~ 12% of the Fe in the cell spectrum. But the CD in the cell spectrum represents 27% of spectral intensity, corresponding to ~100 μM. The difference (100 – 60 = 40 μM) represents [Fe4S4]2+ clusters and/or LS FeII hemes in the cell that are not in mitochondria.
Cells contain ~ 30 μM of HS FeII species in the cytosol. A portion of these are probably associated with the LIP, as this has been suggested to be mostly in the FeII state (44). The size of the LIP has been estimated to be between 0.5 – 10 μM in mammalian cells (45, 46), and our results are consistent with this. However, our results cannot exclude the possibility that the concentration of the LIP in human cells is 3 – 60 fold higher than these previous estimates.
FeIII oxyhydroxide nanoparticles are present in both mitochondria and whole cells, at concentrations of ~ 410 and 70 μM, respectively. Similar nanoparticles have been observed in mitochondria and vacuoles of yeast cells (24, 25) where they are associated with phosphate or polyphosphate groups. At high fields, the Mössbauer spectra of the nanoparticles in yeast exhibit a wide distribution of hyperfine fields (47). In contrast, the magnetically interacting FeIII ions in ferritin afford a narrow distribution of fields (48, 49). The nanoparticles observed in Jurkat cells and mitochondria also exhibited a wide distribution of hyperfine fields, suggesting that they, like their yeast counterparts, have phosphate groups associated. These particles might be independent of ferritin or found in partially loaded ferritin; we are currently working to distinguish these possibilities.
Our results also provide insight into the redox status of the mitochondria within Jurkat cells, at least when packed into Mössbauer/EPR tubes. In contrast, mitochondria were isolated anaerobically. Almost all heme centers (and the same group of Fe/S clusters) in both anaerobically isolated mitochondria and in whole cells (grown in air, then packed into cuvettes) were reduced. The only exception was a small portion of heme a3 in cytochrome c oxidase which was partially oxidized in both types of samples.
The distribution of iron within Jurkat cells and mitochondria are summarized in Fig. 6. A surprisingly large portion of the Fe in mitochondria is present as FeIII oxyhydroxide (phosphate) nanoparticles, which might be in equilibrium with the nonheme HS FeII and FeIII species in the organelle. Similar species in yeast mitochondria may function as feedstock for Fe/S cluster and heme biosynthesis (24, 25). Respiratory complexes (including cytochrome c) account for ~17% of mitochondrial Fe. Another 14% is present as “other” [Fe4S4]2+ clusters and heme b centers. About 15% of mitochondrial Fe is ferritin-like; this may be a contaminant of cytosolic ferritin or ferritin-like material within the mitochondria (mitochondrial ferritin?). Approximately 20% of the volume of Jurkat cells is occupied by mitochondria, yet this fraction accounts for about 55% of the total Fe. Another 40% of cellular Fe is stored in the cytosol as ferritin and the remaining 5% is present (outside of the mitochondria) as Fe/S containing proteins, nonheme HS FeII and FeIII species. Thus, the vast majority of Fe in human cells is either stored (as ferritin) or used (within mitochondria). The bulk of the latter Fe is used to synthesize Fe/S clusters and heme centers that are primarily installed in respiratory complexes, which are used, in turn, to generate cellular energy. Only ~ 5% of cellular Fe is used for all other Fe-associated processes. This does not imply a lesser functional importance for these other process; we simply lack the resolution and sensitivity required to further characterize the Fe species associated with these processes.
The ironomes of yeast and human cells are remarkably similar. From the perspective of this study, the ironome of human mitochondria appears midway between that of fermenting and respiring yeast mitochondria. The redox poise of the organelle and the relative proportion of Fe-containing species contained therein are similar. Clearly, yeast provides an excellent model of human cells for studying Fe metabolism.
We find it intriguing that human mitochondria contain oxyhydroxide (phosphate) nanoparticles similar to those found in yeast. We suspect that they are formed independent of ferritin, in which case they would probably not be under the genetic control of the cell. The significant levels of NHHS FeII in human cells are also potentially significant, in that this type of Fe can generate ROS through Fenton chemistry. We plan to investigate how these nanoparticle and nonheme HS FeII species change under different growth and genetic conditions. We hope to establish how the formation of these species can be controlled, which may improve our ability to understand the molecular basis of Fe-associated diseases.
Supplementary Material
Acknowledgements
Compound 5 was a generous gift from Kevin Burgess (Texas A&M University). We thank Ann Ellis and Rick Littleton at the Microscopy and Imaging Center at Texas A&M University for help in collecting images. This study was supported by the National Institutes of Health (GM084266) and the Robert A. Welch Foundation (A1170).
Funding Statement: The National Institutes of Health (GM084266) and the Robert A. Welch Foundation (A1170) sponsored this study.
Abbreviations
- LIP
labile iron pool
- ICP-MS
inductively coupled plasma mass spectrometry
- ROS
reactive oxygen species
- EPR
electron paramagnetic resonance
- HS
high-spin
- LS
low spin
- CD
central doublet
- NHHS
nonheme high-spin
- RCI – RCIV
mitochondrial respiratory complexes I – IV
Footnotes
Supporting Information Available. A summary of whole cell batches (Table S1) and mitochondrial batches (Table S2) prepared and analyzed, packing efficiency results of whole cells and mitochondria (Table S3), transition metal concentrations from ICP-MS of whole cells and mitochondria (Table S4), additional Mössbauer spectra (Figure S1) and EM images of isolated mitochondria (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Anderson GJ, Vulpe CD. Mammalian iron transport. Cell. Mol. Life Sci. 2009;66:3241–3261. doi: 10.1007/s00018-009-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huebers H, Josephson B, Huebers E, Csiba E, Finch C. Uptake and release of iron from human transferrin. Proc. Natl. Acad. Sci. 1981;78:2572–2576. doi: 10.1073/pnas.78.4.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hider RC. Nature of nontransferrin-bound iron. European Journal of Clinical Investigation. 2002;32:50–54. doi: 10.1046/j.1365-2362.2002.0320s1050.x. [DOI] [PubMed] [Google Scholar]
- 4.Jordan I, Kaplan J. The mammalian transferrin-independent iron transport system may involve a surface ferrireductase activity. Biochem. J. 1994;302:875–879. doi: 10.1042/bj3020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richmond VS, Worwood M, Jacobs A. The Iron Content of Intestinal Epithelial Cells and Its Subcellular Distribution: Studies on Normal, Iron-Overloaded and Iron-Deficient Rats. British Journal of Haematology. 1972;23:605–614. doi: 10.1111/j.1365-2141.1972.tb07095.x. [DOI] [PubMed] [Google Scholar]
- 6.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw B. Mitoferrin is essential for erythroid iron assimilation. Nature Letters. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 7.Sheftel AD, Zhang A-S, Brown C, Shirihai OS, Ponka P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 2007;110:125–132. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 8.Devireddy LR, Hart DO, Goetz D, Green MR. A Mammalian Siderophore Synthesized by an Enzyme with a Bacterial Homologue Involved in Enterobactin Production. Cell. 2010;141:1006–1017. doi: 10.1016/j.cell.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochimica et Biophysica Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Pandolfo M, Pastore A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J Neurol. 2009;256:9–17. doi: 10.1007/s00415-009-1003-2. [DOI] [PubMed] [Google Scholar]
- 11.Ye H, Rouault TA. Human Iron-Sulfur Cluster Assembly, Cellular Iron Homeostasis, and Disease. Biochemistry. 2010;49:4945–4956. doi: 10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine Complex I is a complex of 45 different subunits. The Journal of Biological Chemistry. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 13.Cecchini G. Function and structure of Complex II of the respiratory chain. Annual Reviews. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 14.Crofts AR. The cytochrome bc1 complex: function in the context of structure. Annu. Rev. Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 15.Tsukihara T,H,A, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 16.Theil EC. Ferritin: Structure, Gene Regulation, and Cellular Function in Animals, Plants and Microorganisms. Ann. Rev. Biochem. 1987;56:289–315. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- 17.Dubiel SM, Zablotna-Rypien B, Mackey JB, Williams JM. Magnetic properties of human liver and brain ferritin. Eur Biophys J. 1999;28:263–267. doi: 10.1007/s002490050208. [DOI] [PubMed] [Google Scholar]
- 18.Papaefthymiou GC. The Mössbauer and magnetic properties of ferritin cores. Biochimica et Biophysica Acta. 2010;1800:886–897. doi: 10.1016/j.bbagen.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Baader SL, Bill E, Trautwein AX, Bruchelt G, Matzanke BF. Mobilization of iron from cellular ferritin by ascorbic acid in neuroblastoma SK-N-SH cells: an EPR study. FEBS Letters. 1996;381:131–134. doi: 10.1016/0014-5793(96)00098-1. [DOI] [PubMed] [Google Scholar]
- 20.Glickstein H, El RB, Shvartsman M, Cabantchik ZI. Intracellular labile iron pools as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood. 2005;106:3242–3250. doi: 10.1182/blood-2005-02-0460. [DOI] [PubMed] [Google Scholar]
- 21.Kruszewski M. Labile iron pool: the main determinant of cellular response to oxidative stress. Mutation Research. 2003;531:81–92. doi: 10.1016/j.mrfmmm.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Lill R, Mühlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. TRENDS in Biochemical Sciences. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Wiley SE, Rardin MJ, Dixon JE. Localization and function of the 2Fe-2S outer mitochondrial membrane protein MitoNEET. Methods in Enzymology. 2009;456:233–246. doi: 10.1016/S0076-6879(08)04413-3. [DOI] [PubMed] [Google Scholar]
- 24.Cockrell AL, Holmes-Hampton GP, McCormick SP, Chakrabarti M, Lindahl PA. Mössbauer and EPR Study of Iron in Vacuoles from Fermenting Saccharomyces cerevisiae. Biochemistry. 2011;50:10275–10283. doi: 10.1021/bi2014954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales JG, Holmes-Hampton G, Miao R, Guo Y, Münck E, Lindahl PA. Biophysical Characterization of Iron in Mitochondria Isolated from Respiring and Fermenting Yeast. Biochemistry. 2010;49:5436–5444. doi: 10.1021/bi100558z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis EA. Poststaining grids for Transmission Electron Microscopy. Methods in Molecular Biology. 369:97–106. doi: 10.1007/978-1-59745-294-6_6. [DOI] [PubMed] [Google Scholar]
- 27.Miao R, Martinho M, Morales JG, Kim H, Ellis A, Lill R, Hendrich MP, Münck E, Lindahl PA. EPR and Mössbauer Spectroscopy of Intact Mitochondria Isolated from Yah1p-Depleted Saccharomyces cerevisiae. Biochemistry. 2008;47:9888–9899. doi: 10.1021/bi801047q. [DOI] [PubMed] [Google Scholar]
- 28.Miao R, Kim H, Koppolu MK, Ellis A, Scott RA, Lindahl PA. Biophysical Characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae. Biochemistry. 2009;48:9556–9568. doi: 10.1021/bi901110n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudder BN, Morales JG, Stubna A, Münck E, Hendrich MP, Lindahl PA. Electron paramagnetic resonance and Mössbauer spectroscopy of intact mitochondria from respiring Saccharomyces cerevisiae. J Biol Inorg Chem. 2007;12:1029–1053. doi: 10.1007/s00775-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 30.Beinert H. EPR Spectroscopy of components of mitochondrial electron-transfer system. Methods in Enzymology. 1978;LIV:133–150. doi: 10.1016/s0076-6879(78)54014-7. [DOI] [PubMed] [Google Scholar]
- 31.Hunt C, Pankhurst QA, Dickson DPE. Applied field Mössbauer studies of the iron storage proteins ferritin and haemosiderin. Hyperfine Interactions. 1994;91:821–826. [Google Scholar]
- 32.Tangerås A, Flatmark T, Bäckström D, Ehrenberg A. Mitochondrial iron not bound in heme and iron-sulfur centers: estimation, compartmentation and redox state. Biochimica et Biophysica Acta. 1980;589:162–175. doi: 10.1016/0005-2728(80)90035-3. [DOI] [PubMed] [Google Scholar]
- 33.Tangerås A. Iron content and degree of lipid peroxidation in liver mitochondria isolated from iron-loaded rats. Biochimica et Biophysica Acta. 1983;757:59–68. [PubMed] [Google Scholar]
- 34.Kim Y-M, Chung H-T, Simmons RL, Billiar TR. Cellular Non-Heme Iron Content Is a Determinant of Nitric Oxide-mediated Apoptosis, Necrosis, and Caspase Inhibition. The Journal of Biological Chemistry. 2000;275:10954–10961. doi: 10.1074/jbc.275.15.10954. [DOI] [PubMed] [Google Scholar]
- 35.Ekmekcioglu C, Prohaska C, Pomazal K, Steffan I, Schernthaner G, Marktl W. Concentrations of seven trace elements in different hematological matrices in patients with type 2 diabetes as compared to healthy controls. Biological Trace Element Research. 2001;79:205–219. doi: 10.1385/BTER:79:3:205. [DOI] [PubMed] [Google Scholar]
- 36.Carpentieri U, Myers J, Thorpe L, Daeschner CW, III, Haggard ME. Copper, Zinc, and Iron in Normal and Leukemic Lymphocytes from Children. Cancer Research. 1986;46:981–984. [PubMed] [Google Scholar]
- 37.Chapman EH, Kurec AS, Davey FR. Cell volumes of normal and malignant mononuclear cells. J Clin Pathol. 1981;34:1083–1090. doi: 10.1136/jcp.34.10.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weibel ER, Stäubli W, Gnägi HR, Hess FA. Correlated Morphometric and Biochemical Studies on the Liver Cell. The Journal of Cell Biology. 1969;42:68–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leary SC, Winge DR, Cobine PA. “Pulling the plug” on cellular copper: The role of mitochondria in copper export. Biochimica et Biophysica Acta. 2009;1793:146–153. doi: 10.1016/j.bbamcr.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jancura D, Berka V, Antalik M, Bagelova J, Gennis RB, Palmer G, Fabian M. Spectral and Kinetic Equivalence of Oxidized Cytochrome c Oxidase as Isolated and “Activated” by Reoxidation. The Journal of Biological Chemistry. 2006;281:30319–30325. doi: 10.1074/jbc.M605955200. [DOI] [PubMed] [Google Scholar]
- 41.Kaila VRI, Verkhovsky MI, Wikström M. Proton-Coupled Electron Transfer in Cytochrome Oxidase. Chem. Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 42.Lanne B, Väangård T. Redox Titrations of Cytochrome c Oxidase: An Analysis of a Multi-Electron System. Biochimica et Biophysica Acta. 1978;501:449–457. doi: 10.1016/0005-2728(78)90112-3. [DOI] [PubMed] [Google Scholar]
- 43.Lenaz G, Genova ML. Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject. Antioxidants & Redox Signaling. 2010;12:961–1008. doi: 10.1089/ars.2009.2704. [DOI] [PubMed] [Google Scholar]
- 44.Epsztejn S, Kakhlon O, Glickstein H, Breuer W, Cabantchik ZI. Fluorescence Analysis of the Labile Iron Pool of Mammalian Cells. Analytical Biochemistry. 1997;248:31–40. doi: 10.1006/abio.1997.2126. [DOI] [PubMed] [Google Scholar]
- 45.Petrat F, Groot HD, Rauen U. Subcellular distribution of chelatable iron: a lase scanning microscopic study in isolated hepatocytes and liver endothelial cells. Biochem. J. 2001;356:61–69. doi: 10.1042/0264-6021:3560061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gackowski D, Kruszewski M, Banaszkiewicz Z, Jawien A, Olinski R. Lymphocyte labile iron pool, plasma iron, transferrin saturation and ferritin levels in colon cancer patients. Acta Biochimica Polonica. 2002;49:269–272. [PubMed] [Google Scholar]
- 47.Seguin A, Sutak R, Bulteau A-L, Garcia-Serres R, Oddou J-L, Lefevre S, Santos R, Dancis A, Camadro J-M, Latour J-M, Lesuisse E. Evidence that yeast frataxin is not an iron storage protein in vivo. Biochimica et Biophysica Acta. 2010;1802:531–538. doi: 10.1016/j.bbadis.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Bauminger ER, Cohen SG, Dickson DPE, Levy A, Ofer S, Yariv J. Mössbauer spectroscopy of Escherichia coli and its iron-storage protein. Biochimica et Biophysica Acta. 1979;623:237–242. doi: 10.1016/0005-2795(80)90252-4. [DOI] [PubMed] [Google Scholar]
- 49.St. Pierre TG, Bell SH, Dickson DPE, Mann S, Webb J, Moore GR, Williams RJP. Mössbauer spectroscopic studies of the cores of human, limpet and bacterial ferritins. Biochimica et Biophysica Acta. 1985;870:127–134. doi: 10.1016/0167-4838(86)90015-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.