Abstract
Vaginal bacterial communities play an important role in human health and have been shown to influence HIV infection. Pigtailed macaques (Macaca nemestrina) are used as an animal model of HIV vaginal infection of women. Since the bacterial microbiota could influence retrovirus infection of pigtailed macaques, the genital microbiota in 10 cycling macaques was determined by pyrosequencing. The microbiota of all macaques was polymicrobial with a median of 13 distinct genera. Strikingly, the genera Sneathia and Fusobacterium, both in the phylum Fusobacteria, accounted for 18.9% and 13.3% of sequences while the next most frequent were Prevotella (5.6%), Porphyromonas (4.1%), Atopobium (3.6%), and Parvimonas (2.6%). Sequences corresponding to Lactobacillus comprised only 2.2% of sequences on average and were essentially all L. amylovorus. Longitudinal sampling of the 10 macaques over an 8-week period, which spanned at least one full ovulatory cycle, showed a generally stable presence of the major types of bacteria with some exceptions. These studies show that the microbiota of the pigtailed macaques is substantially dissimilar to that found in most healthy humans, where the genital microbiota is usually dominated by Lactobacillus sp. The polymicrobial makeup of the macaque bacterial populations, the paucity of lactobacilli, and the specific types of bacteria present suggest that the pigtailed macaque microbiota could influence vaginal retrovirus infection.
Introduction
Vaginal infection of pigtailed macaques (Macaca nemestrina) with SIV or SHIV is used as a primate model of HIV sexual transmission in women as well as for testing microbicides and other treatments for prevention of mucosal HIV infection.1–4 The bacterial microbiota in the lower genital tract of pigtailed macaques could have an impact on vaginal infection with SIV or SHIV since in humans, epidemiological studies indicate that women with a genital microbiota that has the characteristics of bacterial vaginosis have a significantly increased susceptibility to sexual transmission of HIV.5–7 Bacterial vaginosis (BV) is a condition in women in which the predominant genital bacterial types are a polymicrobial and variable mixture of bacteria including Gardnerella vaginalis, Prevotella, bacteria in the Order Clostridiales, and other anaerobes.8 This spectrum of bacteria is quite different from that commonly found in women without BV, where the predominant type of bacteria is in most cases Lactobacillus species.
Several previous studies of pigtailed macaque genital microbiota have been performed, but those studies used cultivation to characterize the bacteria.9–11 Cultivation biases are well known,12 and recent studies that used molecular techniques to identify genital bacteria indicate that many of the bacteria that are present in the genital tract of women are not cultivable or are difficult to cultivate.13,14 For example, bacteria from the genus Sneathia, in the phylum Fusobacteria, cannot be cultured using current methods, but make up a substantial proportion of the microbiota in some women with BV.15,16 Furthermore, the type of culture medium used for isolation of genital bacteria can bias the types of cultivatable bacteria found.17
To avoid possible culture bias, a recent study used Multitag Pyrosequencing (MTPS) of the 16S ribosomal RNA (rRNA) gene to characterize the genital microbiota of rhesus macaques (Macaca mulatta).18 That study found the rhesus vaginal microbiome had relatively low levels of Lactobacillus and had a relatively polymicrobial microbiota. Some of the genera of bacteria present in the rhesus macaques included Peptoniphilus, Sneathia, Porphyromonas, Mobiluncus, Atopobacter, Dialister, Thioreductor, Prevotella, and Streptococcus.
The goal of the present study was to use MTPS of the 16S gene to determine the types of bacteria that constitute the genital microbiota of cycling pigtailed macaques. The microbiota was sampled and characterized six to eight times over an 8-week period to determine its stability. We hypothesized that the genital microbiota of pigtailed macaques would be more highly dominated by Lactobacillus than was the microbiota of rhesus macaques since pigtailed macaques and humans have the reproductive similarity of having cycles that take place throughout the year whereas rhesus macaques do not.1
Materials and Methods
Animals and specimen collection
All animal studies were reviewed and approved by the Centers for Disease Control and Prevention (CDC)'s Institutional Animal Care and Use Committee. The pigtail macaques were purchased from the breeding colony of Yerkes National Primate Research Center (n=2) and from New Iberia Research Center (n=8). Upon arrival at CDC, macaques were given complete physical examinations, blood work (CBC chemistries), and an evaluation of the female reproductive tract including colposcopic examination for signs of inflammation. All were deemed to be normal, healthy, cycling mature females 10–16 years of age at sample collection, and weighing 6–11 kg. All macaques tested negative for simian immunodeficiency virus (SIV), simian retroviruses, and simian T cell leukemia viruses. Animals were not Depo-provera treated or synchronized for menstrual cycle.
Ten mature, cycling pigtailed macaques were anesthetized using standard doses of ketamine once per week for 12 weeks. Progesterone and estrogen were measured in blood each week to confirm cycling (data not shown). Cervical vaginal lavage (CVL) samples were collected on weeks 4 through 12 by infusing 8 ml of phosphate-buffered saline into the vaginal vault via a sterile 10-ml syringe attached to a sterile gastric feeding tube, followed by immediate recovery and freezing at −80°C.
Pyrosequencing and identification of bacteria
Bacteria in CVL were pelleted by centrifugation and DNA was isolated using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH). The Multitag Pyrosequencing has been described previously and used 12 bar-coded primer sets each containing the 27F and 355R 16S rRNA gene primers.18 The Bayesian Classifier provided by the Ribosomal Database II Project (RDP 10) was used to identify bacteria using forward reads only. An average of 2137 sequences were obtained for each sample (min=797, max=4394). Total numbers of bacteria in samples were estimated using real time PCR with primers F340 and R514.19 To identify the species of Lactobacillus, Lactobacillus 16S rRNA gene sequences were assembled with reference sequences into phylogenetic trees with a 96% overlap identity and 80% confidence threshold using Geneious Pro 4.6.1 software (Auckland, New Zealand). Reference sequences were AF257097 (L. crispatus), M58820 (L. gasseri), AF243176 (L. jensenii), AF243177 (L. vaginalis), and AY526083 (L. iners).
Results
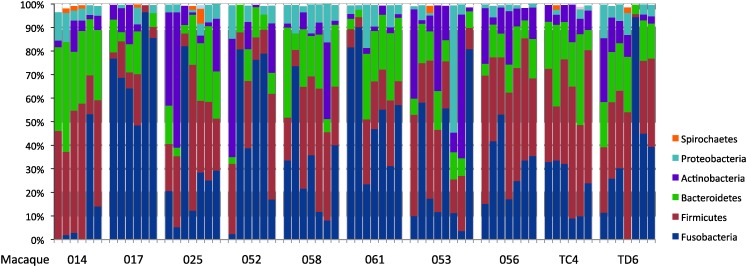
Lower genital tract bacterial microbiota were identified by MTPS of the 16S rRNA gene in 67 samples collected from 10 pigtailed macaques sampled approximately weekly over an 8-week period. At the phylum level, samples contained on average 36% Fusobacteria sequences, 29.6% Firmicutes sequences, 17.2% Bacteroidetes sequences, 10.1% Actinobacteria sequences, and 5.3% Proteobacteria sequences (Fig. 1). Together those five phyla comprised >97% of the sequences in all samples except for one sample that had 6% of its sequences from the phylum Spirochaetes (macaque #025, week 10).
FIG. 1.
Phyla of bacteria found in macaques. Each bar represents the relative proportions of 16S sequences corresponding to bacterial phyla in the macaque genital tract at one time point. Time points are displayed chronologically from left to right. For example, the six bars for macaque 014 are weeks 4, 5, 8, 9, 11, and 12. All macaques were sampled at weeks 4, 8, 9, 11, and 12.
Firmicutes sequences were detected at all time points in all 10 macaques (Fig. 1). Bacteriodetes sequences were present at all time points in all macaques except for one time point in macaque 017. Fusobacteria sequences were found in all samples except two time points from macaque 014 and one time point from macaque TD6. Actinobacteria were present at all time points from four macaques (025, 053, 056, and TC4), but undetected at one or two time points in the other six. Proteobacteria were present at all sampling times in only two macaques (014 and 058). Thus, the phyla Firmicutes, Bacteriodetes, and Fusobacteria were relatively consistent in their presence throughout the 8-week period in all 10 macaques while the presence of Actinobacteria and Proteobacteria was variable.
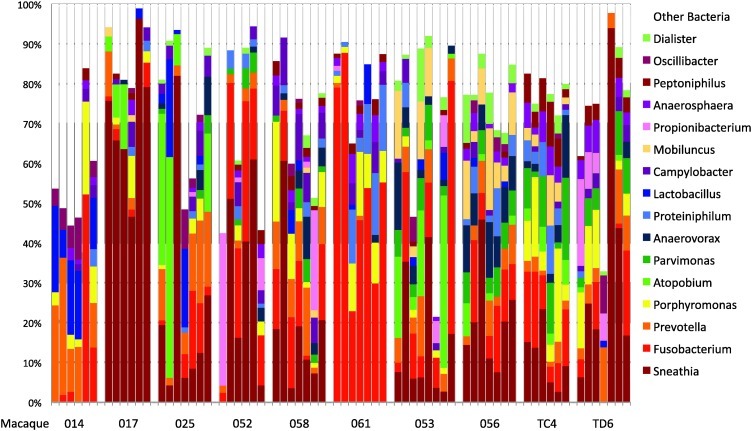
The genera present in the samples were also determined and analysis was limited to genera found at ≥1% of the total community in each sample with the premise that the most abundant bacteria contribute most significantly to the functionality of the microbial community. Sequences corresponding to 100 different genera of bacteria were found in at least one of the 67 samples at ≥1% of the sequences from each macaque. The 20 most predominant of the genera are shown in Table 1 and in Fig. 2. Strikingly, the genera Sneathia and Fusobacterium, both in the phylum Fusobacteria, accounted for an average of 18.9% and 13.3% of sequences in all the samples (Table 1). The next most frequent sequences were those corresponding to Prevotella, Porphyromonas, Atopobium, and Parvimonas (5.6%, 4.1%, 3.6%, and 2.6%, respectively).
Table 1.
Predominant Genera in Macaque Samples
| |
|
Present in (%)a |
|
|---|---|---|---|
| Taxa | Percent of sequencesb | Samples | Macaques |
| Sneathia | 18.9 | 75 | 90 |
| Fusobacterium | 13.3 | 82 | 100 |
| Prevotella | 5.6 | 64 | 100 |
| Porphyromonas | 4.1 | 61 | 90 |
| Atopobium | 3.6 | 34 | 70 |
| Parvimonas | 2.6 | 52 | 70 |
| Anaerovorax | 2.3 | 48 | 90 |
| Proteiniphilum | 2.2 | 49 | 100 |
| Lactobacillus | 2.2 | 31 | 80 |
| Campylobacter | 2.1 | 58 | 100 |
| Mobiluncus | 1.9 | 30 | 50 |
| Propionibacerium | 1.8 | 19 | 60 |
| Anaerosphaera | 1.6 | 54 | 100 |
| Peptoniphilus | 1.3 | 46 | 90 |
| Dialister | 1.1 | 40 | 70 |
| Oscillibacter | 1.0 | 28 | 80 |
| Butyricicoccus | 0.9 | 33 | 70 |
| Corynebacerium | 0.9 | 24 | 70 |
| Streptococcus | 0.8 | 25 | 90 |
| Soehngenia | 0.8 | 37 | 90 |
| Other | 31.0 | ||
Present at ≥1% of sequences in a sample, or in any one sample from a macaque.
Average number of sequences in the 67 samples from 10 macaques.
FIG. 2.
Genera of bacteria found in macaques. Each bar represents the relative proportions of 16S sequences indentifying bacterial genera in the macaque genital tract at one time point. Time points are displayed the same as in Fig. 1. Only the 16 most predominant genera are displayed for clarity.
All macaques had a bacterial microbiota that was polymicrobial with a median of 13 genera found in the 67 samples (Table 2). Only two samples had as few as two genera while the highest number of genera in a sample was 24. Macaque 017 had the lowest number of genera (median of 6) over the 8-week period while 058 had the highest (median of 17) (Fig. 2). While this difference was significant using the Mann–Whitney test (p=0.014) there were no significant differences between any of the macaques using a multiple group comparison (Kruskal–Wallis), indicating that overall, the number of genera in macaques over this time period was not a stable parameter.
Table 2.
Number of Genera in Macaque Genital Samples
| |
Number of genera (≥1% of sequences) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Week | 014 | 017 | 025 | 052 | 058 | 061 | 053 | 056 | TC4 | TD6 |
| 4 | 10 | 6 | 12 | 18 | 10 | 9 | 14 | 16 | 15 | 21 |
| 5 | 13 | 7 | 5 | 11 | 14 | |||||
| 7 | 9 | 6 | 8 | 24 | 9 | 18 | 12 | |||
| 8 | 14 | 4 | 4 | 19 | 13 | 13 | 11 | 15 | 14 | 16 |
| 9 | 22 | 14 | 16 | 7 | 17 | 13 | 9 | 14 | 15 | 20 |
| 10 | 18 | 23 | 7 | 17 | 2 | |||||
| 11 | 9 | 2 | 15 | 7 | 21 | 13 | 15 | 16 | 17 | 11 |
| 12 | 15 | 6 | 10 | 17 | 18 | 7 | 5 | 13 | 16 | 13 |
A blank indicates that a sample was not obtained.
Since Sneathia was the genus with the highest average number of sequences in this group of animals it was analyzed in further detail. Sneathia levels were relatively stable in five of the macaques over the 8-week period (Fig. 2). For example, macaque 017 consistently had relatively high levels of Sneathia sequences (48–96%) at all time points while macaques 056 and TC4 had lower Sneathia levels (5–25% of sequences) of Sneathia at all time points (except for week 7 of animal 056). Animals 014 and 061 did not have any Sneathia sequences over the 8 weeks. In contrast, the other animals had more variation in the levels of Sneathia with two animals (052 and TD6) varying over the sampling period from no detectable Sneathia to relatively high levels. Total copy numbers of Sneathia 16S rRNA were also analyzed (see Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/aid). Total copy numbers of Sneathia were significantly higher in animal 017 than in 053 and TD6 (p<0.05, Dunn's Multiple Comparisons Test, excluding 014 and 061 from the analysis), but all other comparisons were not significant. Taken together these data indicate that the relative abundance of bacteria from the genus Sneathia was relatively stable in most of the macaques.
Sequences corresponding to Lactobacillus comprised only 2.2% of all sequences and were found in 31% of all samples (Table 3). Only five of the macaques had Lactobacillus sequences at >2% of total sequences at one or more sampling times. Animal 014 had the highest levels of Lactobacillus on average, although levels dipped below 1% of sequences on week 11. The Lactobacillus sequences from all the animals corresponded to L. amylovorus (see Supplementary Fig. S1).
Table 3.
Proportion of Sequences Corresponding to Lactobacillus
| |
% of sequences corresponding to Lactobacillus |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Week | 014 | 017 | 025 | 052 | 058 | 061 | 053 | 056 | TC4 | TD6 |
| 4 | 22 | <1 | 2 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| 5 | 7 | <1 | <1 | <1 | 2 | |||||
| 7 | 2 | 27 | <1 | 2 | <1 | <1 | <1 | |||
| 8 | 19 | <1 | 1 | <1 | 6 | 1 | 3 | <1 | <1 | <1 |
| 9 | 17 | <1 | 20 | <1 | 3 | <1 | <1 | <1 | <1 | 2 |
| 10 | <1 | <1 | 10 | <1 | <1 | |||||
| 11 | <1 | <1 | <1 | <1 | <1 | <1 | 7 | 1 | <1 | <1 |
| 12 | 13 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
A blank indicates that a sample was not obtained at that time point.
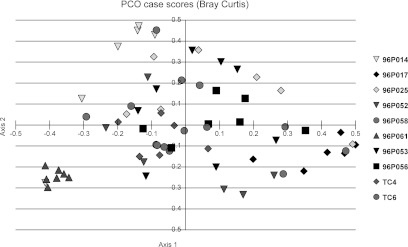
To further assess the stability of the microbiota in the macaques over the 8-week period, principle coordinates analysis (PCO) was performed. PCO analysis of the genera in the samples showed clustering of the time points from four animals (Fig. 3). The most notable clustering was in animal 061 with all six time points closely arranged followed by 014 and 017 with four time points closely arranged. Animal TC4 also had some clustering of the data points. The other six animals' points were not closely clustered indicating less stability by this measure.
FIG. 3.
Principal coordinate analysis of macaque microbiota. Each macaque is represented by one type of symbol and each point is a separate sampling time. For example, the six points representing the six sampling times for macaque 061 are closely clustered indicating the relative relatedness of the bacterial microbiota over time in this animal.
Analysis of Shannon diversity scores (see Supplementary Fig. S2) showed a large variability in diversity over the 8-week period for all animals except macaque TC4. TC4 had the highest median Shannon score (2.37) while macaque 017 had the lowest (0.84). The Shannon scores of macaques TC4 and 017 over the 8-week period were significantly different (p<0.05, Dunn's Multiple Comparisons Test) but all other comparisons were not significant. These data indicate that the diversity of the genital microbiota in most of the macaques was variable over the 8-week period.
Discussion
This study found that the genital microbiota in this group of pigtailed macaques over an 8-week period was quite different from a Lactobacillus-dominated type of microbiota commonly found in women.13,20–23 There were several characteristics of the macaque microbiota that were substantially different from a healthy microbiota in women.
First, levels of lactobacillus were relatively low. Thus, only two macaques had a level of Lactobacillus sequences>10% with the highest level reaching 27% of sequences at one time point. In contrast, women with a Lactobacillus-dominated genital microbiota typically have >75% Lactobacillus sequences.13–16,22
Second, the macaque genital microbiota was relatively polymicrobial, which is a feature not usually found in healthy microbiota in women, but that is common in BV.13–16 Thus, the number of genera found in the macaques ranged from a median of 6 to a median of 17. In contrast, women without BV typically have one to four genera that constitute ≥98% of the microbiota.13,14,16 Interestingly, many of the genera that were found at high levels in the pigtailed macaques are equivalent to those present in women with BV; Sneathia, Prevotella, Porphyromonas, and Atopobium sequences were found in the macaques frequently and are also often found in women with BV.13, 5 However, there were also several notable differences between the genital microbiota in the pigtailed macaques and BV. First, Gardnerella vaginalis is present at relatively high levels in many women with BV (average of approximately 5% of sequences), but was found in only three samples from two macaques and at relatively low levels (1.2%, 2.1%, and 6.5% of the sequences in those samples). Second, sequences from two genera, Fusobacterium and Parvimonas, were found frequently in the macaques but are not frequent in BV.
Third, there were no major differences in proinflammatory cytokine–chemokine levels in macaque vaginal secretions (data not shown) that correlated with differences in the microbiota, as is observed when comparing humans with and without bacterial vaginosis.24 However, this lack of notable differences in cytokine–chemokine levels between pigtailed macaques could be due in part to the fact that none of the macaques had microbiota that corresponded to what is regarded as healthy microbiota in humans.
A remarkable finding in this study was the very high levels of bacteria from the phylum Fusobacteria with the two genera Sneathia and Fusobacterium together accounting for more than a third of the sequences. It is interesting to compare this to a study that determined the genital microbiota of rhesus macaques.18 In that study, Sneathia was found at >10% of sequences in 8 out of 11 of the rhesus macaques. In contrast, Fusobacterium was found at >10% of sequences in only one. Thus, while Sneathia was highly represented in both rhesus and pigtailed macaques, Fusobacterium was not. Another interesting comparison between the rhesus and pigtailed studies was that Lactobacillus was found at relatively low levels in both types of macaque when compared with humans, but was identified as L. amylovorus in the pigtailed macaques and L. johnsonii in the rhesus macaques. Yu et al. 25 reported that L. johnsonii, L. amylovorus, and L. acidophilus were all present in a group of Chinese rhesus macaques, but L. johnsonii was most prevalent. This is in contrast to humans where the most prevalent species are L. iners, L. crispatus, L. jensenii, and L. gasseri.22,26
It is of interest to compare the vaginal bacterial microbiota of pigtailed macaques with the gut microbiota at the phylum level. While the gut microbiota of pigtailed macaques has not been analyzed with culture-independent techniques, the gut microbiota of rhesus macaques and humans have been determined and are both composed mostly of the phyla Bacteroidetes and Firmicutes with little representation of Fusobacteria and Actinobacteria.27–29 Relatively high proportions of Fusobacteria and Actinobacteria are a common characteristic of the genital microbiota of pigtailed macaques (Fig. 1), rhesus macaques,18 and humans with BV.13,20 This microbiota pattern is substantially different from gut microbiota, indicating that the genital microbiota samples do not merely reflect contamination with feces.
This study has the limitations that pigtailed macaques from only one primate center were analyzed and that Nugent scores and vaginal pH were not determined. Also, vaginal cultures were not obtained, which could have been used to compare cultivatable and noncultivatable microbiota. It is possible that different housing conditions or diets found at other primate centers could impact the makeup of the genital microbiota. However, all 10 macaques over the entire assessment period of 8 weeks showed a genital microbiota that was dissimilar to healthy microbiota in humans but had some similarities to BV in humans. Because in humans, women who lack a healthy genital microbiota have increased susceptibility to sexual transmission of HIV, these results suggest that the genital microbiota of pigtailed macaques in captivity could augment infection with SIV or SHIV. However, the relationship between pigtailed macaque microbiota, inflammation, and virus susceptibility needs further clarification.
Supplementary Material
Acknowledgments
We thank Drs. Ron Otten and Nat Promadej-Lanier for macaque samples. This work was supported by NIH Grants U19 AI076981 and P30 AI082151, by the James B. Pendleton Charitable Trust, and by the Interagency Agreement Y1-AI-0681-02 between CDC and NIH. The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Cole AM. Patton DL. Rohan LC, et al. The formulated microbicide RC-101 was safe, antivirally active following intravaginal application in pigtailed macaques. PLoS One. 5(11):e15111. doi: 10.1371/journal.pone.0015111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang Y. Tian B. Saifuddin M, et al. RT-SHIV, an infectious CCR5-tropic chimeric virus suitable for evaluating HIV reverse transcriptase inhibitors in macaque models. AIDS Res Ther. 2009;6:23. doi: 10.1186/1742-6405-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Promadej-Lanier N. Srinivasan P. Curtis K, et al. Systemic and mucosal immunological responses during repeated mucosal SHIV(162P3) challenges prior to and following infection in pigtailed macaques. Virology. 2008;375(2):492–503. doi: 10.1016/j.virol.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 4.Kent SJ. Dale CJ. Ranasinghe C, et al. Mucosally-administered human-simian immunodeficiency virus DNA and fowlpoxvirus-based recombinant vaccines reduce acute phase viral replication in macaques following vaginal challenge with CCR5-tropic SHIVSF162P3. Vaccine. 2005;23(42):5009–5021. doi: 10.1016/j.vaccine.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Atashili J. Poole C. Ndumbe PM. Adimora AA. Smith JS. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. AIDS. 2008;22(12):1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaul R. Nagelkerke NJ. Kimani J, et al. Prevalent herpes simplex virus type 2 infection is associated with altered vaginal flora and an increased susceptibility to multiple sexually transmitted infections. J Infect Dis. 2007;196(11):1692–1697. doi: 10.1086/522006. [DOI] [PubMed] [Google Scholar]
- 7.Cherpes TL. Meyn LA. Krohn MA. Lurie JG. Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37(3):319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- 8.Cherpes TL. Hillier SL. Meyn LA. Busch JL. Krohn MA. A delicate balance: Risk factors for acquisition of bacterial vaginosis include sexual activity, absence of hydrogen peroxide-producing lactobacilli, black race, and positive herpes simplex virus type 2 serology. Sex Transm Dis. 2008;35(1):78–83. doi: 10.1097/OLQ.0b013e318156a5d0. [DOI] [PubMed] [Google Scholar]
- 9.Patton DL. Sweeney YC. Cummings PK. Meyn L. Rabe LK. Hillier SL. Safety and efficacy evaluations for vaginal and rectal use of BufferGel in the macaque model. Sex Transm Dis. 2004;31(5):290–296. doi: 10.1097/01.olq.0000124614.91448.d4. [DOI] [PubMed] [Google Scholar]
- 10.Patton DL. Sweeney YT. Balkus JE, et al. Preclinical safety assessments of UC781 anti-human immunodeficiency virus topical microbicide formulations. Antimicrob Agents Chemother. 2007;51(5):1608–1615. doi: 10.1128/AAC.00984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton DL. Sweeney YC. Rabe LK. Hillier SL. The vaginal microflora of pig-tailed macaques and the effects of chlorhexidine and benzalkonium on this ecosystem. Sex Transm Dis. 1996;23(6):489–493. doi: 10.1097/00007435-199611000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Amann RI. Ludwig W. Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredricks DN. Fiedler TL. Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353(18):1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 14.Hyman RW. Fukushima M. Diamond L. Kumm J. Giudice LC. Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102(22):7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravel J. Gajer P. Abdo Z, et al. Microbes, Health Sackler Colloquium: Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spear GT. Sikaroodi M. Zariffard MR. Landay AL. French AL. Gillevet PM. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198:1131–1140. doi: 10.1086/591942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falsen E. Pascual C. Sjoden B. Ohlen M. Collins MD. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: Description of Lactobacillus iners sp. nov. Int J Syst Bacteriol. 1999;49(Pt 1):217–221. doi: 10.1099/00207713-49-1-217. [DOI] [PubMed] [Google Scholar]
- 18.Spear GT. Gilbert D. Sikaroodi M, et al. Identification of rhesus macaque genital microbiota by 16S pyrosequencing shows similarities to human bacterial vaginosis: Implications for use as an animal model for HIV vaginal infection. AIDS Res Hum Retroviruses. 2010;26(2):193–200. doi: 10.1089/aid.2009.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barman M. Unold D. Shifley K, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76(3):907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hummelen R. Fernandes AD. Macklaim JM, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010;5(8):e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepin J. Deslandes S. Giroux G, et al. The complex vaginal flora of West African women with bacterial vaginosis. PLoS One. 2011;6(9):e25082. doi: 10.1371/journal.pone.0025082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spear GT. Gilbert D. Landay AL, et al. Pyrosequencing of the genital microbiotas of HIV-seropositive and -seronegative women reveals Lactobacillus iners as the predominant Lactobacillus species. Appl Environ Microbiol. 2011;77(1):378–381. doi: 10.1128/AEM.00973-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling Z. Liu X. Chen X, et al. Diversity of cervicovaginal microbiota associated with female lower genital tract infections. Microb Ecol. 2011;61(3):704–714. doi: 10.1007/s00248-011-9813-z. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell CM. Balkus J. Agnew KJ, et al. Bacterial vaginosis, not HIV, is primarily responsible for increased vaginal concentrations of proinflammatory cytokines. AIDS Res Hum Retroviruses. 2008;24(5):667–671. doi: 10.1089/aid.2007.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu RR. Cheng AT. Lagenaur LA, et al. A Chinese rhesus macaque (Macaca mulatta) model for vaginal Lactobacillus colonization and live microbicide development. J Med Primatol. 2009;38(2):125–136. doi: 10.1111/j.1600-0684.2008.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonio MA. Hawes SE. Hillier SL. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180(6):1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 27.Dethlefsen L. Relman DA. Microbes and Health Sackler Colloquium: Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna P. Hoffmann C. Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4(2):e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckburg PB. Bik EM. Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.