Abstract
After screening a large number of clinical samples of HIV-1 subtype C in India, a subset of viral strains containing sequence insertions upstream of the viral enhancer has been identified. The sequence insertions contained binding sites for at least two different transcription factors NF-κB and RBEIII, importantly, in a mutually exclusive fashion. Furthermore, while some of the viral strains contained insertions of κB-like sites, a few others contained dual insertions of the RBEIII and κB sites together but only one of the two was intact. NF-κB acquisition appears to be the most common phenotype unique for subtype C with nearly half of the variant strains containing such insertions. Given that subtype C already contains three functional NF-κB sites in the viral enhancer, acquisition of a fourth NF-κB motif in some variant viral strains is intriguing. Further investigation is warranted to examine the significance of the sequence insertions for the replicative fitness of the variant viral strains.
Of the various genetic subtypes of HIV-1, the subtype C strains are responsible for more than half of the global infections.1 It is not understood if the diverse genetic subtypes of HIV-1 differ from one another with respect to the biological and pathogenic properties, however, such experimental evidence has been gradually accumulating in the recent past.2 The differences in the biological properties of the diverse viral subtypes perhaps could be ascribed to the subtype-specific molecular variations. Such subtype-specific molecular variations have been mapped to the promoter sequences, cis-regulatory elements, and regulatory, structural, and accessory proteins.
The promoter sequences of the viral subtypes are genetically diverse among which the subtype C long terminal repeat (C-LTR) is the most divergent viral promoter.3 Several transcription factor binding sites (TFBS) in the LTR demonstrate subtype-associated differences including in the NF-κB, NF-AT, USF, and other regulatory elements such as the TATA box, and the TAR region.4 Of these variations in the HIV-1 LTR, subtype-specific patterns within the enhancer element, exclusively consisting of the NF-κB motifs, are important given the profound impact NF-κB has on gene expression from the viral promoter. The enhancer in most of the viral subtypes, including the prototype subtype B, consists of two identical and canonical NF-κB motifs with the exception of subtypes C and A/E. While in the subtype A/E, the upstream NF-κB site has been replaced by a GABP binding motif,5 the genetic variation within the subtype C enhancer is more complex. Although a large proportion of the C-LTRs contains three NF-κB sites, a significant minority of the LTRs contains a fourth motif that is either a canonical NF-κB site or a κB-like site.6 Additionally, C-LTRs with only two NF-κB sites have also been reported. Thus, the subtype C viral enhancer is characterized by a significant magnitude of variation in terms of the number of NF-κB sites and sequence differences within these motifs.
In approximately 38% of subtype B viral isolates, immediately upstream of the viral enhancer, insertion of unique sequences of 15 to 34 bp length has been reported previously adding an additional level of complexity to the genetic diversity of this important regulatory sequence.7 Insertion of these sequences, commonly known as the most frequent naturally occurring length polymorphism (MFNLP), predominantly generates an RBEIII motif. In subtype B, the RBEIII site binds a transcription factor complex of variable composition, RBF-2, that functions as a strong transcription repressor.8 Of note, the MFNLP insertions in the subtype B LTR (B-LTR) are strictly restricted to duplicating the RBEIII site only as a compensatory mechanism when the original site is inactivated by the natural variation.9 Importantly, MFNLP sequence insertions in subtype C LTR have not been reported previously, although a few sequences deposited to the databases contained such elements.
In a previous analysis, we found that approximately 6% (34/608) of the primary viral isolates of India contained the presence of length polymorphism in the viral enhancer.10 With the exception of a replication-competent subtype C molecular clone D24,11 the nature of the viral enhancer in these subtype C viral strains has not been determined. Here, we report the molecular nature of the sequence insertions in 25 of the 34 of these viral isolates that indicated length polymorphism in the LTR. The clinical profile of all the 25 subjects has been summarized (Table 1).
Table 1.
Clinical Profile of the Study Subjects and the Sequence Details
| |
|
|
|
|
LTR (MFNLP) |
GenBank accession No. |
|||
|---|---|---|---|---|---|---|---|---|---|
| S. No. | Clinical code | Gender | Age (Years) | CD4 (cells/mm3) | F-κB | κB like | RBEIII | LTR | env |
| 1 | BL42/02 | M | 35 | 412 | + | − | − | HQ202921 | JQ250636 |
| 2 | D24a | M | 39 | — | + | − | − | EF1786131 | EF469243 |
| 3 | 02/529 | F | 31 | 573 | + | − | − | HQ202939 | JQ250624 |
| 4 | S38 | M | 41 | — | + | − | − | HQ202922 | JQ250642 |
| 5 | KR15 | F | 42 | 234 | + | − | − | HQ202924 | JQ250639 |
| 6 | 516 | M | 34 | 345 | + | − | − | JQ268626 | JQ250634 |
| 7 | A35/01 | F | 36 | 512 | + | − | − | JQ268631 | JQ250635 |
| 8 | 2/498 | M | 33 | 342 | + | − | − | JQ268636 | JQ250622 |
| 9 | 2/566 | M | 26 | 421 | + | − | − | JQ268639 | JQ250627 |
| 10 | 2/567 | F | 29 | 344 | + | − | − | JQ268643 | JQ250628 |
| 11 | 2/481 | M | 26 | 253 | + | − | − | JQ268650 | JQ250621 |
| 12 | 2/532 | F | 28 | 227 | + | − | − | HQ202946 | JQ250625 |
| 13 | HY6 | M | 35 | — | − | + | − | HQ202925 | JQ250638 |
| 14 | 2/502 | F | 30 | 787 | − | + | − | JQ268653 | JQ250623 |
| 15 | KR22 | M | 40 | 80 | − | + | − | HQ202936 | JQ250640 |
| 16 | KR6 | M | 38 | — | − | + | − | HQ202934 | JQ250641 |
| 17 | 02/6 | M | 23 | 218 | − | − | + | HQ202930 | JQ250630 |
| 18 | 2/570 | F | 32 | 323 | − | − | + | HQ202937 | JQ250629 |
| 19 | 02/9 | F | 29 | 566 | − | − | + | HQ202938 | JQ250632 |
| 20 | SEVA9 | M | 34 | — | − | − | + | HQ202933 | JQ250644 |
| 21 | S45 | F | 27 | — | − | − | + | EU74705 | JQ250643 |
| 22 | HY49 | M | 27 | — | − | − | + | HQ202935 | JQ250637 |
| 23 | 02/7 | F | 25 | 50 | − | + | + | HQ202931 | JQ250631 |
| 24 | 3/BL61 | M | 38 | — | − | + | + | HQ202923 | JQ250633 |
| 25 | 02/538 | F | 36 | 61 | − | + | + | HQ202932 | JQ250626 |
D24 is an infectious molecular clone of subtype C containing four NF-κB sites in the LTR and has been reported previously with the following GenBank accession No. EF469243. There is no separate GenBank entry for the env sequence of the D24 molecular clone, hence the accession number of the full length molecular clone was considered for the env sequence.
LTR, long terminal repeat; MFNLP, most frequent naturally occurring length polymorphism.
The full-length LTR was amplified from each of the clinical samples in a nested polymerase chain reaction (PCR) using genomic DNA extracted from stored peripheral blood mononuclear cells (PBMCs). Nested PCR was performed using the outer primer pair N558 (5′ TGGAAGGGTTAATTTACTCTAAGGAAAGGAAAGAGATCCTTG 3′) and N424 (5′ GACACCAARGAAGCYTTAGAYAARATAGAG 3′) and an inner primer pair N698 (5′ ATGACGACGCGTTGGAAGGGTTAATTTACTCYMAGAAAAGRCAAGA 3′) and N854 (5′ GAATTCCTGCTAGAGATTTTCCACACTACCAAAAG 3′). Primers N1007 (FP 5′ TTAAGCTACAAGGCAAGGC 3′) and N1009 (RP 5′ GTTGTTCTCGGTGGGGCTTGG 3′) that anneal to the flanking vector backbone sequences were used for sequencing the cloned LTR inserts. Despite our best efforts, we could not amplify the LTR from nine of the clinical samples. The amplified LTR product was cloned into a promoter-less expression vector, upstream of a dual reporter cassette consisting of Luciferase-IRES-GFP that simultaneously expresses two different reporter genes—secreted luciferase and GFP.12 Multiple plasmid clones that transiently expressed GFP in HEK293 cells, three to five per each clinical sample, were identified. The sequence of the full-length LTR from each of the plasmid clones was determined in both the orientations. The LTR clone with the highest magnitude of homology to the consensus sequence of the replicate clones was identified from multisequence alignment, considered as representative of the viral isolates and used in subsequent analyses. Furthermore, the V3-V5 env sequence (7001–7667, HxB2 coordinates) from each of the 25 viral isolates was determined using the HIV-1 env Subtyping Kit (NIH AIDS Research and Reference Reagent Program). The full-length LTR and the V3-V5 env sequences of a representative clone for each of the viruses have been deposited in GenBank (Table 1).
As a standard measure of quality control, each individual sequence was subjected to the BLAST analysis against an in-house laboratory sequence database and the GenBank database. The sequence tools REGA and RIP3.0 available online were used for HIV-1 subtyping (http://dbpartners.stanford.edu/RegaSubtyping/) and viral recombination analysis (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html), respectively. Phylogenetic analysis was performed using the reference sequences of HIV-1 group M available from the Los Alamos database (http://www.hiv.lanl.gov/). The sequences were aligned using the Clustal W algorithm of the BioEdit software. The sequences were gap-stripped and edited manually for optimal alignment. The evolutionary history was inferred using the neighbor-joining algorithm of the MEGA4.1 software. The evolutionary distances were computed using the maximum composite likelihood method and expressed as the rate of base pair substitution per site. The percentage of replicate trees where the associated taxa clustered together in the bootstrap test of 1000 replicates was determined.
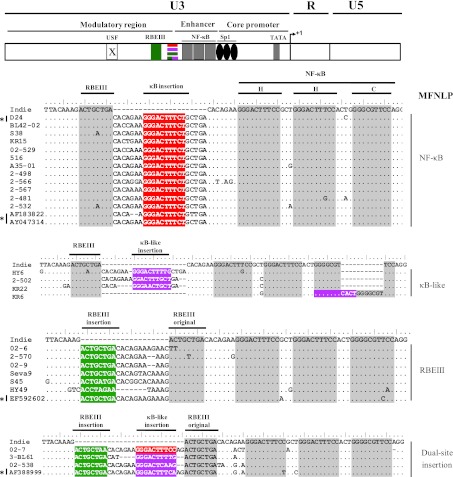
The multiple sequence alignment of the LTR sequences from the 25 viral strains with that of a subtype C reference molecular clone Indie-C1 and a search for the various transcription factor binding sites within this region identified that the major regulatory elements within the core promoter were conserved among all the viral strains (Fig. 1). The conserved regulatory elements include the TAR structure, the TATA box, the three Sp1 sites, the three canonical NF-κB sites, and the RBEIII motif. In a typical C-LTR, an RBEIII site (5′-ACTGCTGA-3′) is found immediately upstream of the viral enhancer consisting of the three NF-κB sites. In 24 of the 25 viral strains examined, we found sequence insertions between the RBEIII site and the viral enhancer.
FIG. 1.
The profiles of the sequence insertion in the HIV-1 subtype C LTR. The top panel depicts a schematic representation of a typical HIV-1 LTR showing important transcription factor binding sites including the RBEIII, NF-κB, Sp1, and TATA box. The USF-1 site is inactive in subtype C, indicated using a crossed box. The arrow indicates the transcriptional start site. Multiple sequence alignment of the viral enhancer and the upstream region derived from 25 Indian and 4 African viral strains has been presented with the Indie-C1 sequence at the top for comparison. The viral strains have been classified into four categories based on the nature of the transcription factor binding sites in the inserted sequences. The inserted transcription factor binding sites NF-κB, κB like, and RBEIII have been highlighted in red, pink, and green background colors, respectively, while the original sites are shaded gray. The asterisk represents sequences previously published. With the exception of the Indian D24, the rest of the published sequences belong to Africa. Dots and dashes represent sequence identity and deletions, respectively.
The nature of the sequence insertion in subtype C demonstrated many similarities and several important differences from that of subtype B. First, the location of the sequence insertion is identical in both of the viral subtypes, between the RBEIII site and the viral enhancer. In only one viral strain (KR6) a κB-like sequence was inserted between the viral enhancer and the Sp1 sites.
Second, the most important of all, unlike in subtype B where only the RBEIII site is inserted through the MFNLP, the nature of sequence insertion in subtype C appears to be broader. At least three different types of sequences were found inserted in the C-LTR consisting of the NF-κB (12/25), κB-like (4/25), or RBEIII (6/25) motifs. Additionally, three other viral strains contained a dual-site insertion (see below). Thus, the MFNLP in the C-LTR predominantly generated an NF-κB binding site in nearly half of the viral strains examined. Of note, the viral enhancer in these viral strains is expected to contain four functional NF-κB binding sites. In the κB-like sequences, typically the left half of the motif was found intact whereas the right half contained sequences that do not conform to NF-κB binding. Insertion of an NF-κB or a κB-like site into the viral promoter through MFNLP has not been reported previously in subtype B.
Third, the insertion of the NF-κB and RBEIII sites appears to be mutually exclusive. Three of the 25 viral strains contained sequence insertions that apparently contained binding motifs for two different transcription factors, however, only one of the two motifs appears to be intact. Two of these strains (3-BL61 and 02-538) contained a canonical RBEIII site but the coinserted κB-like site contains noncanonical base substitutions in the 3′ half of the motif. The single viral strain 02-7 contains the insertion of a canonical NF-κB site, however, this motif is associated with an RBEIII site that contains a single residue variation. It therefore appears that the MFNLP insertions in subtype C could acquire an NF-κB site or an RBEIII site but not both of them together. The mutually exclusive nature of these two sites is consistent with a large number of additional viral isolates we examined subsequently (Bachu et al., unpublished observations).
Fourth, in all of the 12 viral strains that contained an NF-κB site insertion, the inserted κB-site differed from the canonical NF-κB motif at position 10 with a “C to T” variation (5′- GGGACTTTCT-3′). This observation is consistent with many other subtype C viral strains that we examined subsequently (data not shown) or sequences available in the databanks (Fig. 1).
Fifth, as previously shown with subtype B,7 the duplication of the RBEIII site in subtype C was associated with an altered original RBEIII site, but in only two strains (02-6 and 2-570). We found two additional types of variants that were not reported in subtype B viruses. In two different strains (02-9 and Seva 9), RBEIII duplication appeared while the original site was intact; thus these strains contain two functional RBEIII sites. Additionally, the duplicated RBEIII element contained mutations that could make the site nonfunctional (S45 and KR22).
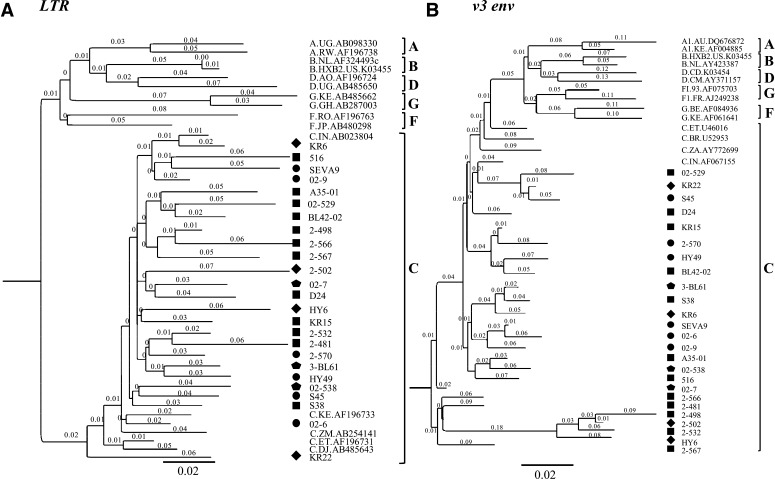
Lastly, while the inserted NF-κB site and the flanking sequences were highly conserved among the viral strains, the duplicated RBEIII site was characterized by a large sequence variation within and in the flanking sequences. Additionally, unlike the NF-κB insertion, which contained a fixed number of residues consisting of 21 bp, the length of the RBEIII insertion varied considerably among the viral isolates ranging from 12 to 20 bp. A phylogenetic analysis of the 25 viral isolates in the LTR and V3 of envelope (Fig. 2A and B, respectively) confirmed the subtype C origin of these strains as they tightly clustered with the reference subtype C sequences. In addition to the 25 Indian viral strains described here, four other subtype C viruses containing the MFNLP insertions were also examined and all of these viral strains conform to the observations delineated above (Fig. 1).
FIG. 2.
Phylogenetic analysis of the LTR and V3 env sequences. Phylogenetic analysis of (A) the LTR and (B) V3 env sequences derived from the 25 Indian viral isolates containing the promoter insertions. The reference sequences have been downloaded from the Los Alamos HIV sequence database. After gap-stripping of the sequences, 321 and 57 positions were used in the analysis of the LTR and V3 env regions, respectively. The trees are mid-point rooted. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The individual sequences have been highlighted with different shapes based on the nature of the sequence insertion. Square, diamond, circle and pentagon shapes represent sequence insertions containing the NF-κB, κB-like, RBEIII and RBEIII+κB-like motifs, respectively.
To our knowledge, the present work represents the first major analysis of sequence insertion in the subtype C viral promoter derived from a large number of primary viral isolates. The results are implicated in the viral evolution of HIV-1. The major subtypes of HIV-1 are believed to have evolved into the distinct genetic forms as they are known today in the late 1950s or early 1960s.13 The subsequent years witnessed the emergence of a large number of diverse recombinant forms although only a small minority of these strains appear to have survival competence. Of the several HIV-1 major subtypes and the plethora of the recombinant forms, viral strains belonging to subtype C dominate the global epidemics and cause half of the global infections.14
The experimental and epidemiological observations seem to collectively suggest that the subtype C strains enjoy higher levels of replication fitness as compared to other genetic subtypes of HIV-1, although such a proposition is highly controversial.15 Importantly, subtype C appears to dominate the “founder” viral strains where its introduction followed that of the other viral subtypes. The domination of subtype C is clearly manifested against several other subtypes in The Democratic Republic of Congo, Tanzania, and South Africa; against subtype B in the South American continent especially in southern Brazil; and against subtypes B and AE in China. Additionally, since the time of its origin, subtype C has expanded at a faster rate as compared to any other viral subtype.16 For instance, the incidence of the subtype C strains in China increased from 5.1% in 1992 to nearly 90% in 2002.15 Additionally, the apparent genetic stability of the subtype C strains in India appears to be quite unusual. Although other HIV-1 subtypes and several unique recombinant forms have been reported from India previously,17 the subtype C strains in this country appear to remain genetically stable without undergoing a major event of genetic recombination with other subtypes unlike in China or southern Brazil. None of the 51 CRFs known today has been reported from India or South Africa or other African countries where introduction of subtype C preceded that of other subtypes. These observations collectively seem to suggest that subtype C has attained a relative level of evolutionary stability so that any type of genetic recombination is not a viable option. Based on this, the results presented here acquire importance with the implication that the subtype C viral strains have been undergoing another round of adoptive evolutionary modification, but in the viral promoter.
The most significant finding of the present analysis is the acquisition of an additional NF-κB binding site in the C-LTR. Most of the major subtypes of HIV-1 contain two genetically identical NF-κB sites that constitute the viral enhancer. In contrast, a typical subtype C LTR is endowed with three canonical NF-κB binding sites.18 Stronger transcription from the subtype C LTR, as compared to the subtype B LTR, has been ascribed to the presence of the higher number of NF-κB binding sites,12,19 and this quality in part is believed to underlie the global prevalence of the subtype C strains. Given that the C-LTR is already endowed with three canonical κB sites, it is surprising that a few variant viral strains acquired an additional κB site.
NF-κB plays an important role in regulating the viral gene expression from the viral promoter and controlling viral latency. NF-κB, especially in the absence of Tat, mediates initiation of the viral transcription, remodels the nucleosomes especially nuc-0 and nuc-1 that flank the transcription start site, and recruits histone acetyltransferases such as p300 to the promoter. NF-κB can also enhance transcription elongation although this effect may not be as profound as its influence at the other levels. Importantly, NF-κB function is also critical in the presence of Tat in regulating gene expression from the viral promoter. Collectively, NF-κB plays a critical role in regulating the basal level transcription from the viral promoter in the absence of Tat and the overall gene expression in its presence. Acquisition of an additional κB site therefore must confer a significant replicative competence on the variant 4-κB viral strains of subtype C.
However, a higher magnitude of gene expression could also lead to increased immune activation and elevated immune surveillance thus essentially offsetting the advantages gained by a higher transmission rate. Furthermore, the increased number of NF-κB sites in the viral promoter could be detrimental to the establishment and maintenance of the viral latency given the profound influence this transcription factor has on transcription silencing. It therefore remains to be determined if the novel viral strains of HIV-1 subtype C would gain replication fitness in the context of the natural infection.
The present study also identified additional types of sequence insertions in the subtype C LTR including that of an RBEIII site. Unlike that of NF-κB, the RBEIII site insertion is a feature common to all the major subtypes of HIV-1 and extensively examined in subtype B.7 In the context of subtype B, the RBEIII site binds a transcription factor complex RBF-2 that largely functions as a strong transcription repressor.8 Interestingly, some of the new subtype C viral strains contain two genetically intact RBEIII sites (02-9, Seva9, and EF592602, Fig. 1) unlike in subtype B where RBEIII duplication was demonstrated only as a compensatory mechanism for the loss of the original site.9 Of note, the results also demonstrated the mutually exclusive nature of the NF-κB and RBEIII site insertions in the subtype C LTR. Although NF-κB is largely known to function as a transcription enhancer and RBEIII as a transcription suppressor, these factors could modulate viral gene expression in quite the opposite way depending on the physiological context. For instance, NF-κB binding sites in the LTR could maintain viral latency by recruiting the p50 homodimer thus essentially suppressing gene expression from the viral promoter.20 Likewise, RBF-2 could activate transactivation from the viral promoter in T cells21 or in combination with NF-κB.22 The mutually exclusive nature of the NF-κB and RBEIII sites in C-LTR therefore suggests that the duplication of these sites essentially serves a redundant function possibly common for both of these transcription factors.
In summary, the viral promoter of subtype C appears to be presently undergoing a major evolutionary adoptive modification. At least two major promoter variant viral strains of HIV-1 subtype C that acquired an additional NF-κB site or an additional RBEIII motif have been identified. Whether either one or both of these viral variants would gain higher replicative competence and establish successful viral epidemics like the precedent 3-κB site containing subtype C viral strains needs further evaluation.
Acknowledgments
M.B. is a recipient of the Council of Science and Industrial Research fellowship from the Government of India. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HMA HIV-1 Env Subtyping Kit from Dr. James Mullins. This work was supported by grants to T.K.K. and U.R. from the Department of BioTechnology, Government of India (grant BT/01CEIB/10/III/01) and intramural funds from JNCASR.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Geretti AM. HIV-1 subtypes: Epidemiology and significance for HIV management. Curr Opin Infect Dis. 2006;19(1):1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 2.Ranga U. Shankarappa R. Siddappa NB, et al. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol. 2004;78(5):2586–2590. doi: 10.1128/JVI.78.5.2586-2590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montano MA. Novitsky VA. Blackard JT, et al. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71(11):8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez dA. Soriano V. Alcamil J. Holguin A. New findings on transcription regulation across different HIV-1 subtypes. AIDS Rev. 2006;8(1):9–16. [PubMed] [Google Scholar]
- 5.Verhoef K. Sanders RW. Fontaine V. Kitajima S. Berkhout B. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-kappaB enhancer element into a GABP binding site. J Virol. 1999;73(2):1331–1340. doi: 10.1128/jvi.73.2.1331-1340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt GM. Johnson D. Tiemesse CT. Characterisation of the long terminal repeat regions of South African human immunodeficiency virus type 1 isolates. Virus Genes. 2001;23(1):27–34. doi: 10.1023/a:1011171027134. [DOI] [PubMed] [Google Scholar]
- 7.Estable MC. In search of a function for the most frequent naturally-occurring length polymorphism (MFNLP) of the HIV-1 LTR: retaining functional coupling, of Nef and RBF-2, at RBEIII? Int J Biol Sci. 2007;3(5):318–327. doi: 10.7150/ijbs.3.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadowski I. Lourenco P. Malcolm T. Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency. Curr HIV Res. 2008;6(4):286–295. doi: 10.2174/157016208785132563. [DOI] [PubMed] [Google Scholar]
- 9.Estable MC. Bell B. Merzouki A, et al. Human immunodeficiency virus type 1 long terminal repeat variants from 42 patients representing all stages of infection display a wide range of sequence polymorphism and transcription activity. J Virol. 1996;70(6):4053–4062. doi: 10.1128/jvi.70.6.4053-4062.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddappa NB. Dash PK. Mahadevan A, et al. Identification of unique B/C recombinant strains of HIV-1 in the southern state of Karnataka, India. AIDS. 2005;19(13):1426–1429. doi: 10.1097/01.aids.0000180795.49016.89. [DOI] [PubMed] [Google Scholar]
- 11.Dash PK. Siddappa NB. Mangaiarkarasi A, et al. Exceptional molecular and coreceptor-requirement properties of molecular clones isolated from an human immunodeficiency virus type-1 subtype-C infection. Retrovirology. 2008;5(1):25. doi: 10.1186/1742-4690-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddappa NB. Kashi VP. Venkatramanan M, et al. Gene expression analysis from human immunodeficiency virus type-1 subtype C promoter and construction of bicistronic reporter vectors. AIDS Res Hum Retroviruses. 2007;23(10):1268–1278. doi: 10.1089/aid.2006.0305. [DOI] [PubMed] [Google Scholar]
- 13.Rousseau CM. Learn GH. Bhattacharya T, et al. Extensive intra-subtype recombination in South African HIV-1 subtype C infections. J Virol. 2007;81(9):4492–4500. doi: 10.1128/JVI.02050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arien KK. Vanham G. Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol. 2007;5(2):141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tebit DM. Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11(1):45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 17.Ranga U. Banerjea A. Chakrabarti S. Mitra D. HIV/AIDS research in India–past, present and future. Current Sci. 2010;98(3):335–345. [Google Scholar]
- 18.Naghavi MH. Salminen MO. Sonnerborg A. Vahlne A. DNA sequence of the long terminal repeat of human immunodeficiency virus type 1 subtype A through G. AIDS Res Hum Retroviruses. 1999;15(5):485–488. doi: 10.1089/088922299311240. [DOI] [PubMed] [Google Scholar]
- 19.Jeeninga RE. Hoogenkamp M. Armand-Ugon M, et al. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74(8):3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams SA. Chen LF. Kwon H, et al. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25(1):139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koken SE. van Wamel JL. Geelen JL. Berkhout B. Functional analysis of the ACTGCTGA sequence motif in the human immunodeficiency virus type-1 long terminal repeat promoter. J Biomed Sci. 1994;1(2):83–92. doi: 10.1007/BF02257981. [DOI] [PubMed] [Google Scholar]
- 22.Naghavi MH. Estable MC. Schwartz S. Roeder RG. Vahlne A. Upstream stimulating factor affects human immunodeficiency virus type 1 (HIV-1) long terminal repeat-directed transcription in a cell-specific manner, independently of the HIV-1 subtype and the core-negative regulatory element. J Gen Virol. 2001;82(Pt 3):547–559. doi: 10.1099/0022-1317-82-3-547. [DOI] [PubMed] [Google Scholar]