Abstract
Thirty HIV-1 URF_01AE/ B′ complete or nearly full-length genome sequences sampled within Southeast Asia were obtained from the Los Alamos HIV Sequence Database. Phylogenetic and recombinant analyses revealed that three sequences indeed displayed the identical recombinant structure. Of note, the three subjects, harboring novel CRF01_AE/B recombinants, did not have apparent epidemiological linkage. They fulfilled the criteria for the designation of a new circulating recombinant form (CRF) and constituted the 52nd CRF identified in the worldwide HIV-1 pandemic. In this chimera, two short subtype B segments were inserted into a backbone of CRF_01AE. The breakpoints corresponded to HXB2 nucleotide positions 2930, 3251, 8521, and 9004 approximately. This CRF is the first one identified by neatening and analyzing the sequences already presented in the Los Alamos HIV Sequence Database. This indicates that we should pay attention not only to explicit subtype sequences but also to those classified as a unique recombinant form (URF) so far.
Mutation, coupled with recombination, has resulted in the extensive genetic diversity of HIV-1. The high genetic diversity of HIV-1 allows its classification into groups, subtypes, subsubtypes, and circulating recombinant forms (CRFs).1 Group M HIV-1 dominates the AIDS pandemic with at least nine subtypes (A to D, F to H, J, and K) and multiple intersubtype recombinants. Currently, a total of 51 CRFs have been represented in the Los Alamos HIV database web site (http://hiv-web.lanl.gov/CRFs/CRFs.html). Meanwhile multiple unique recombinant forms (URFs) have been reported in many regions around the world.
Southeast Asia was thought to be an epicenter of the HIV-1 epidemic in Asia. The AIDS epidemic in this region was initiated by two different subtype strains in two separated risk groups, with subtype B among injecting drug users (IDUs) and CRF01_AE among those heterosexually exposed, throughout the early phase.2,3 CRF01_AE can clearly cross over from the heterosexual epidemic to IDUs and by 1995 it also became the predominant subtype in IDUs.4 The cocirculation of the two different strains in this region led to the generation of more complex recombinant strains that emerged in almost every country in Southeast Asia.5–10 Furthermore, some of the recombinant strains have spread widely in populations and have become CRFs. Up to now, five CRFs, originated from CRF01_AE and subtype B′ lineages, have been reported in Southeast Asia: CRF15_01B11 and CRF34_01B12 from Thailand, CRF33_01B13 and CRF48_01B14 from Malaysia, and CRF51_01B15 from Singapore. The HIV-1 epidemic in this region has now become increasingly heterogeneous.
The extensive use of complete and nearly full-length genome sequencing of HIV-1 strains has provided a powerful and accurate approach to the molecular epidemiology of regional epidemics. Consequently, the number of complete or nearly full-length HIV-1 genome sequences was growing larger and larger. The majority of these can be classified as subtypes and CRFs definitely, while the minority remains as URFs. To define a CRF, at least three epidemiologically unlinked HIV-1 sequences with identical mosaic structures should be characterized, at least two of them in the nearly full-length genome (8 kb)1. For the present study, we identified a novel CRF_AE/B′ candidate, which emerged in Southeast Asia, using sequences obtained from the Los Alamos HIV Sequence Database (www.hiv.lanl.gov).
Thirty HIV-1 URF_01AE/ B′ complete or nearly full-length genome sequences sampled within Southeast Asia were obtained from the Los Alamos HIV Sequence Database. These sequences were generated by sequencing from plasma or peripheral blood mononuclear cell (PBMCs) samples collected from Thailand (n=19), Malaysia (n=8), Myanmar (n=2), and China (n=1). In addition to these 30 sequences, we also retrieved subtype reference sequences from the same database. All the sequences were incorporated into one sequence dataset using SynchAlign, software http://www.hiv.lanl.gov/content/sequence/SYNCH_ALIGNS/SynchAligns.html, and edited using BioEdit software. A phylogenetic tree was constructed by the neighbor-joining method based on Kimura's two-parameter distance matrix with 1000 bootstrap replicates using Mega 5.04. With respect to recombination analysis, RIP, http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html, software was applied at first; jpHMM software (http://jphmm.gobics.de/) was used to further scrutinize and define the recombination breakpoints subsequently. The origin of each segment was analyzed by subregion neighbor-joining tree analysis. Detailed information about the three subjects harboring CRF52_01B recombinants was collected from the studies13,16,17 in which the three subjects were reported. Furthermore, we retrieved the records for the three subjects from the Los Alamos HIV Sequence Database in order to supplement elements such as patient name and risk factor.
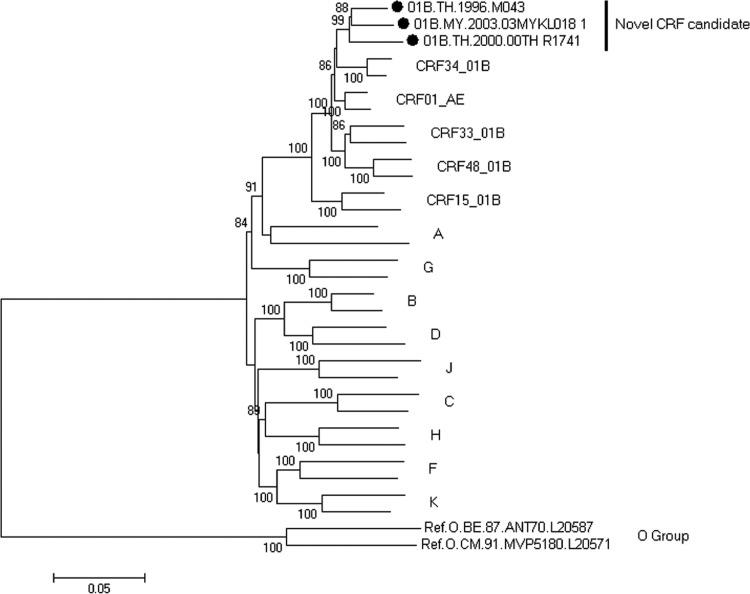
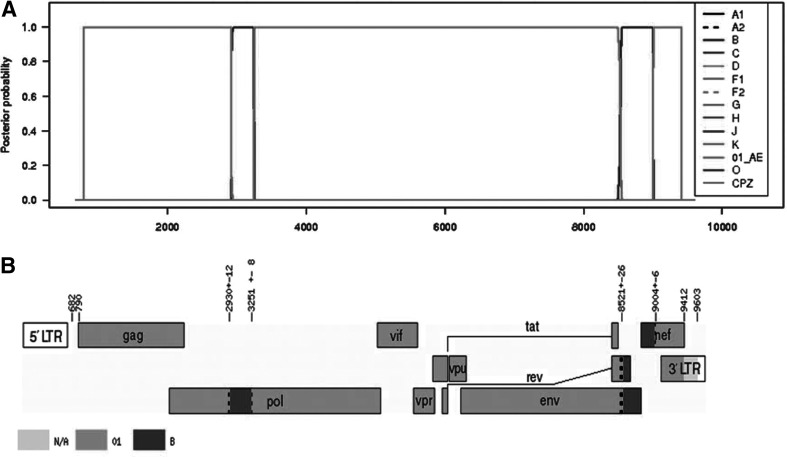
Using phylogenetic analysis, we found that three nearly full-length genome sequences from three patients formed a monophyletic branch supported by a high bootstrap value of 99%, located outside any HIV-1 subtypes and known CRFs in Southeast Asia (Fig. 1). Both RIP and jpHMM revealed that the three URFs nearly full-length genome sequences indeed displayed the identical recombinant structure composed of CRF_01AE and subtype B. In this chimera, two short subtype B segments were inserted into a backbone of CRF_01AE. The breakpoints corresponded to HXB2 nucleotide positions 2930, 3251, 8521, and 9004 approximately (Fig. 2).
FIG. 1.
Identification of a novel circulating recombinant form (CRF) candidate. The neighbor-joining phylogenetic tree is constructed based on the nucleotide sequence of nearly full-length genomes using MEGA 5.04. The tree is rooted with group O. All the reference strains were retrieved from the Los Alamos HIV Sequence Database. Values on the branches represent the percentage of 1000 bootstrap replicates and bootstrap values over 70% are shown in the tree. The novel CRF candidate is marked with a solid circle. The scale bar indicates 5% nucleotide sequence divergence.
FIG. 2.
Recombinant analysis of the novel identified CRF52_01B. (A) Posterior probabilities of the subtypes at each sequence position (original sequence positions) calculated by jpHMM. Inset: The different symbols represent different reference strains. (B) Genome map (based on HXB2 numbering). Numbers in the figure above denote intervals for recombination breakpoints based on HXB2 numbering. The white regions denote missing information due to input fragment sequences. Subtype N/A denotes missing information due to uninformative subtype models (subtype: N/A).
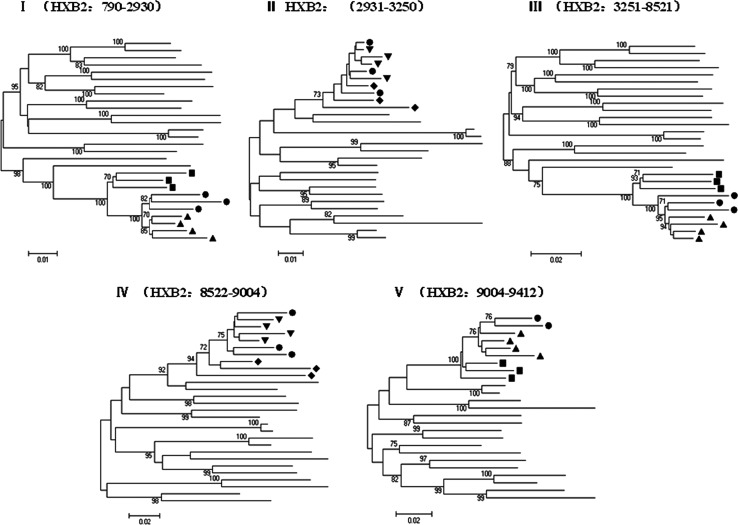
To confirm the subtype structure of CRF52_01B and to estimate likely parental lineages, we divided the HIV-1 genomes into five segments according to the breakpoints. Subregion phylogenetic analysis confirmed the four breakpoints identified using jpHMM. The genomic segments I, III, and V clustered with reference subtype CRF01_AE while the other two segments II and IV branched with reference subtype B (Fig. 3). As shown in Fig. 3, the subregion tree analysis demonstrated that CRF01_AE segments (I, III, and V) belonged to the cluster of Thailand CRF01_AE other than African CRF01_AE, showing that the three CRF01_AE segments indeed originated from CRF01_AE of Thailand origin. Similarly, subtype B segment IV analysis revealed that it originated from Thailand B rather than Western B. However, we could not determine the origin of the remaining subtype B segment II because the length was too short to obtain high bootstrap values.
FIG. 3.
Phylogenetic trees for genome segments of the novel identified CRF52_01B. The neighbor-joining tree is constructed using MEGA 5.04.Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported. Bootstraps of 70% and higher only are shown. The scale bars are shown at the bottom. •=CRF52_01B; ▴=Thailand CRF01_AE; ▪=African CRF01_AE;▾=Thailand B; □=Western B.
The profiles of the subtype structure of the three sequences were distinct from the other five CRFs originated from CRF01_AE and subtype B′ reported in Southeast Asia. Of note, the three nearly full-length genome sequences (01B.MY.2003.03MYKL018_1, 01B.TH.2000.00TH_R1741, and 01B.TH.1996.M043) were presented in three different studies13,16,17 from three different patients (Table 1) with no apparent epidemiological linkage. These data established that the three sequences show the same recombinant structure over the whole genome, fulfilling the criteria for designation of a new CRF. They constitute the 52nd CRF identified in the worldwide HIV-1 pandemic, herein called CRF52_01B.
Table 1.
Information on Subjects Harboring Novel CRF01_AE/B Recombinants in Southeast Asia
| Patient | Age | Gender | Country | Year of first positive HIV-1 test | Year of collection | Risk factor | Sequence name | Accession number |
|---|---|---|---|---|---|---|---|---|
| M043 | n/a | Female | Thailand | n/a | 1996 | Heterosexual | 01B.TH.1996.M043 | DQ354113 |
| MYKL018 | 32 | Male | Malaysia | 1997 | 2003 | Bisexual | 01B.MY.2003.03MYKL018_1 | DQ366664 |
| R1741 | 27 | Female | Thailand | 1999 | 2000 | Heterosexual | 01B.TH.2000.00TH_R1741 | AY945734 |
n/a, not available.
Detailed information about the three subjects harboring CRF52_01B recombinants is shown in Table 1. Unlike other CRFs identified worldwide, the most interesting element is that the three subjects belonged to two different countries, with two in Thailand and one in Malaysia, within Southeast Asia. However, the subjects were reported by year of first positive HIV-1 test, although that of subject M043 was not available. From Table 1, it is apparently that subject M043 was infected in or before 1996. The obviously wide geographic separation and early infection date of the three subjects suggested that CRF52_01B arose at least 15 years ago. To obtain accurate estimates regarding the time of origin and transmission direction of the novel CRF in Southeast Asia, more sequences with different locations and sampling times are needed in Bayesian coalescent analysis using BEAST software.18,19
All of the three subjects reported sexual exposure, with two being heterosexual and one bisexual. It has been reported that most women infected in Southeast Asia have been the monogamous wives or regular partners of higher risk men.20 Of interest, subject R1741 was one of the participants recruited from public family planning clinics in Thailand. Her only reported risk factor for HIV-1 infection was heterosexual exposure.16 Taking subject R1741 into account, she should have acquired the HIV-1 infection from her male sexual partner. It was suggested that the novel CRFs have spread into low-risk people early.
In this study, we identified a novel CRF (CRF52_01B) composed of CRF01_AE and subtype B in Southeast Asia by analyzing the 30 complete or nearly full-length genome sequences obtained from the Los Alamos HIV Sequence Database. CRF52_01B is the sixth identified CRF that has originated from CRF01_AE and subtype B lineages within Southeast Asia in the past several years. What is most interesting is that this CRF is the first one identified by neatening and analyzing the sequences already presented in the Los Alamos HIV Sequence Database. This emphasizes that we should pay attention not only to explicit subtype sequences but also to those classified as URFs so far. The prevalence and public significance of the novel CRF remain to be elucidated in the future.
Acknowledgments
This work was supported by the National Key S&T Special Projects on Major Infectious Diseases (Grants 2008ZX10001-004 and 2008ZX10001-012).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Robertson DL. Anderson JP. Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–57. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 2.Ou CY. Weniger BG. Luo CC, et al. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341(8854):1171–1174. doi: 10.1016/0140-6736(93)91001-3. [DOI] [PubMed] [Google Scholar]
- 3.Kalish ML. Baldwin A. Raktham S, et al. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: Implications for HIV vaccine trials. AIDS. 1995;9(8):851–858. doi: 10.1097/00002030-199508000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Subbarao S. Vanichseni S. Hu DJ, et al. Genetic characterization of incident HIV type 1 subtype E and B strains from a prospective cohort of injecting drug users in Bangkok, Thailand. AIDS Res Hum Retroviruses. 2000;16(8):699–707. doi: 10.1089/088922200308693. [DOI] [PubMed] [Google Scholar]
- 5.Tovanabutra S. Polonis V. De Souza M, et al. First CRF01_AE/B recombinant of HIV-1 is found in Thailand. AIDS. 2001;15(8):1063–1065. doi: 10.1097/00002030-200105250-00018. [DOI] [PubMed] [Google Scholar]
- 6.Yang R. Xia X. Kusagawa S. Zhang C. Ben K. Takebe Y. On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS. 2002;16(10):1401–1407. doi: 10.1097/00002030-200207050-00012. [DOI] [PubMed] [Google Scholar]
- 7.Ramos A. Nguyen L. Hu DJ, et al. New HIV type 1 CRF01_AE/B recombinants displaying unique distribution of breakpoints from incident infections among injecting drug users in Thailand. AIDS Res Hum Retroviruses. 2003;19(8):667–674. doi: 10.1089/088922203322280892. [DOI] [PubMed] [Google Scholar]
- 8.Takebe Y. Motomura K. Tatsumi M. Lwin HH. Zaw M. Kusagawa S. High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: Geographical hot spot of extensive recombination. AIDS. 2003;17(14):2077–2087. doi: 10.1097/00002030-200309260-00009. [DOI] [PubMed] [Google Scholar]
- 9.Tee KK. Pon CK. Kamarulzaman A. Ng KP. Emergence of HIV-1 CRF01_AE/B unique recombinant forms in Kuala Lumpur, Malaysia. AIDS. 2005;19(2):119–126. doi: 10.1097/00002030-200501280-00003. [DOI] [PubMed] [Google Scholar]
- 10.Wang B. Lau KA. Ong L-Y, et al. Complex patterns of the HIV-1 epidemic in Kuala Lumpur, Malaysia: Evidence for expansion of circulating recombinant form CRF33_01B and detection of multiple other recombinants. Virology. 2007;367(2):288–297. doi: 10.1016/j.virol.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Tovanabutra S. Watanaveeradej V. Viputtikul K, et al. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses. 2003;19(7):561–567. doi: 10.1089/088922203322230923. [DOI] [PubMed] [Google Scholar]
- 12.Tovanabutra S. Kijak GH. Beyrer C, et al. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses. 2007;23(6):829–833. doi: 10.1089/aid.2006.0300. [DOI] [PubMed] [Google Scholar]
- 13.Tee KK. Li X-J. Nohtomi K. Ng KP. Kamarulzaman A. Takebe Y. Identification of a novel circulating recombinant rorm (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr. 2006;43(5):523–529. doi: 10.1097/01.qai.0000242451.74779.a7. [DOI] [PubMed] [Google Scholar]
- 14.Li Y. Tee KK. Liao H, et al. Identification of a novel second-generation circulating recombinant form (CRF48_01B) in Malaysia: A descendant of the previously identified CRF33_01B. J Acquir Immune Defic Syndr. 2010;54(2):129–136. doi: 10.1097/QAI.0b013e3181d82ce5. [DOI] [PubMed] [Google Scholar]
- 15.Ng OT. Eyzaguirre LM. Carr JK, et al. Identification of new CRF51_01B in Singapore using full genome analysis of three HIV type 1 isolates. AIDS Res Hum Retroviruses. 2012 doi: 10.1089/aid.2011.0177. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanaveeradej V. Benenson MW. Souza MD, et al. Molecular epidemiology of HIV Type 1 in preparation for a phase III prime-boost vaccine trial in Thailand and a new approach to HIV Type 1 genotyping. AIDS Res Hum Retroviruses. 2006;22(8):801–807. doi: 10.1089/aid.2006.22.801. [DOI] [PubMed] [Google Scholar]
- 17.Kijak GH. Tovanabutra S. Sanders-Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v. Virology. 2007;358(1):178–191. doi: 10.1016/j.virol.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 18.Drummond A. Rambaut A. Shapiro B. Pybus O. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 19.Drummond A. Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7(1):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F. Kilmarx PH. Supawitkul S, et al. HIV-1 seroprevalence, risk factors, and preventive behaviors among women in Northern Thailand. J Acquir Immune Defic Syndr. 2000;25(4):353–359. doi: 10.1097/00042560-200012010-00010. [DOI] [PubMed] [Google Scholar]