ABSTRACT
In sub-Saharan Africa, cryptococcal meningitis (CM) continues to be a predominant cause of AIDS-related mortality. Understanding virulence and improving clinical treatments remain important. To characterize the role of the fungal strain genotype in clinical disease, we analyzed 140 Cryptococcus isolates from 111 Ugandans with AIDS and CM. Isolates consisted of 107 nonredundant Cryptococcus neoformans var. grubii strains and 8 C. neoformans var. grubii/neoformans hybrid strains. Multilocus sequence typing (MLST) was used to characterize genotypes, yielding 15 sequence types and 4 clonal clusters. The largest clonal cluster consisted of 74 isolates. The results of Burst and phylogenetic analysis suggested that the C. neoformans var. grubii strains could be separated into three nonredundant evolutionary groups (Burst group 1 to group 3). Patient mortality was differentially associated with the different evolutionary groups (P = 0.04), with the highest mortality observed among Burst group 1, Burst group 2, and hybrid strains. Compared to Burst group 3 strains, Burst group 1 strains were associated with higher mortality (P = 0.02), exhibited increased capsule shedding (P = 0.02), and elicited a more pronounced Th2 response during ex vivo cytokine release assays with strain-specific capsule stimulation (P = 0.02). The results of these analyses suggest that cryptococcal strain variation can be an important determinant of human immune responses and mortality.
IMPORTANCE
Cryptococcus neoformans is a common life-threatening human fungal pathogen that is responsible for an estimated 1 million cases of meningitis in HIV-infected patients annually. Virulence factors that are important in human disease have been identified, yet the impacts of the fungal strain genotype on virulence and outcomes of human infection remain poorly understood. Using an analysis of strain variation based on in vitro assays and clinical data from Ugandans living with AIDS and cryptococcal infection, we report that strain genotype predicts the type of immune response and mortality risk. These studies suggest that knowledge of the strain genotype during human infections could be used to predict disease outcomes and lead to improved treatment approaches aimed at targeting the specific combination of pathogen virulence and host response.
Introduction
More than 2.7 million new HIV infections and 1.8 million AIDS-related deaths occurred in 2010, despite the roll-out of antiretroviral therapy (1). Based on community cohorts, 13 to 44% of HIV/AIDS deaths are due to Cryptococcus infections (2). Consequently, cryptococcal meningitis (CM) may be the fourth leading cause of death due to infectious disease in Africa with estimates of a half million deaths annually (2). Even with access to antiretroviral therapy (ART), the 6-month survival rate remains approximately 40% (2–4). Efforts to improve the survival of HIV/AIDS patients in sub-Saharan Africa must focus on the development of effective treatments for opportunistic infections such as CM.
One possible approach to improving the clinical outcome is to customize the treatment based on specific pathogen virulence and host response. A healthy immune response to Cryptococcus depends on coordinated interactions between antigen-presenting cells (APCs) and effector T cells to generate a type 1 helper T-cell (Th1) response that stimulates classical activation of macrophages and destruction of internalized cryptococcal cells (5–9). Gamma interferon (IFN-γ) induces protective Th1 responses, and human studies have shown that IFN-γ in the cerebrospinal fluid (CSF) is associated with increased fungal clearance (10). In contrast, Th2-biased responses to Cryptococcus are not protective and result in disseminated, uncontrolled infections in mice (11, 12). Interleukin 10 (IL-10) produced by Th2 and regulatory T cells inhibits the synthesis of proinflammatory cytokines like IFN-γ and is generated in response to purified cryptococcal capsule (13, 14). Finally, IL-4 is a powerful inducer of Th2 development, suppresses Th1 development, and compromises the ability of IFN-γ to drive protective Th1 responses (13). Therefore, patients exhibiting Th2-biased immune responses to cryptococcal infections could be at higher risk for poor clinical outcomes.
Numerous relationships between pathogen genotypes and virulence have been demonstrated in bacterial and viral pathogens (15–18) as well as between Cryptococcus gattii genotypes and virulence in the outbreak on Vancouver Island, British Columbia, Canada (19, 20), suggesting that cryptococcal strain variation could play a role in determining the clinical outcome. However, clinical outcomes in cryptococcosis are multifactorial—both the pathogen and the host contribute to disease. Epidemiological characteristics of Cryptococcus neoformans, virulence-associated phenotypic traits, and host immune responses have been studied as individual factors influencing the outcome of cryptococcosis, but none of these studies have found associations between cryptococcal strain variation and human disease (21, 22). We propose that exogenous (pathogen) and endogenous (host) factors contribute concurrently to the pathophysiology of cryptococcosis. Consequently, an integrated investigation of the cryptococcal strain and the host immune response could provide valuable insight into cryptococcal pathogenesis and potentially lead to custom therapies that target the host-pathogen interface. Thus, we hypothesize that differences in strain genotype, as determined by multilocus sequence typing (MLST), and strain phenotype could be responsible for disease severity and clinical outcome in patients. To test these hypotheses, we analyzed cryptococcal strains and clinical data from a cohort of ART-naive Ugandans with AIDS and CM. We report that the genotype of the infecting strain is associated with the type of immune response and clinical outcome.
RESULTS
Cohort demographic characteristics.
A prospective cohort of 199 HIV-infected, ART-naive Ugandans with their first episode of CM was recruited at Mulago Hospital, Kampala, Uganda, over a 26-month enrollment period between 2006 and 2009 in order to study immune reconstitution inflammatory syndrome and other clinical outcomes after ART initiation (3). Of these patients, 111 patients had culture-positive CSF samples stored for subsequent genotypic analysis. Detailed clinical and demographic information from 98 of these patients were used for comparison of strain genotype to clinical outcome (Table 1).
TABLE 1 .
Patient demographic characteristics and clinical parameters
| Demographic characteristic or clinical parameter |
Mean ± SD (n)a |
P valuec | ||||
|---|---|---|---|---|---|---|
| Total subjectsb |
Patients in each burst group |
|||||
| Burst 1 (78 isolates) |
Burst 2 (18 isolates) |
Burst 3 (10 isolates) |
Hybrid (6 isolates) |
|||
| Age (yr) | 35 ± 8 (63) | 35 ± 9 (43) | 37 ± 8 (9) | 35 ± 4 (9) | 33 ± 4 (3) | 0.95 |
| Sex (no. of males/no. of females) | 51/32 (83) | 38/20 (58) | 7/7 (14) | 7/3 (10) | 1/2 (3) | 0.45 |
| Death (%)d | 78 (98) | 80 (71) | 80 (15) | 38 (8) | 100 (6) | 0.05 |
| CD4 count (no. of cells/μl) | 26 ± 24 (39) | 25 ± 23 (29) | 29 ± 31 (5) | 25 ± 28 (6) | NA | 0.94 |
| Viral load (log10 no. copies/μl) | 5.1 ± 0.7 (34) | 5.0 ± 0.8 (26 ) | 5.1 ± 0.5 (3) | 5.3 ± 0.4 (6) | NA | 0.72 |
| CSF opening pressure (mm H2O) | 339 ± 160 (56) | 364 ± 174 (36) | 268 ± 117 (11) | 360 ± 134 (7) | 225 ± 64 (2) | 0.25 |
| No. of CFU (log10 CFU/ml) | 4.3 ± 1.1 (64) | 4.4 ± 1.0 (45) | 4.0 ± 1.5 (10) | 4.0 ± 0.9 (8) | 4.2 ± 0.1 (3) | 0.53 |
| CRAG titer (log2 titer) | 11.7 ± 2.1 (56) | 11.8 ± 1.9 (43) | 10.8 ± 3.3 (8) | 12.1 ± 0.8 (5) | 11.3 (1) | 0.61 |
Values indicated in the table are means ± SD for all demographic characteristics and clinical parameters except Sex, which indicates males/females, and Death, which shows the percentage of patients that died. n indicates the number of patients. NA, not available.
Complete data were not available for all patients. Total subjects indicates the mean ± SD of patients, indicated by n, for which data were available.
P values were calculated by one-way ANOVA or Fisher’s exact test as appropriate.
Pairwise Fisher’s exact test comparisons showed that patients infected with Burst group 3 isolates had significantly lower mortality than patients infected with Burst group 1 isolates (P = 0.02) or the hybrid isolates (P= 0.03).
Cryptococcal strain genotypes.
Cryptococcal strains were colony purified from a total of 140 CSF specimens from the 111 patients with their first episode of CM. Genotypes of the Ugandan clinical (UgCl) strains were determined by MLST according to a consensus scheme that included eight loci: CAP59, GPD1, IGS1, LAC1, PLB1, SOD1, URA5, and TEF1 (23).
Eight isolates could not be fully genotyped, as some loci were recalcitrant to PCR amplification upon sequencing with either legacy (24) or consensus (23) MLST primers at >1 MLST locus. The loci from these nongenotyped strains showed the highest similarity to Cryptococcus neoformans var. neoformans (n = 4) or var. grubii (n = 4) (data not shown). None of the strains were Cryptococcus gattii based on sequencing or CGB agar screening. PCR serotyping showed combinations of both C. neoformans var. grubii and C. neoformans var. neoformans gene alleles in the strains (data not shown). Taken together, these data suggest the eight strains are C. neoformans var. grubii/neoformans “hybrids.”
Colony purification allowed us to analyze the greatest number of clinical isolates from the broadest spectrum of patients, but coinfection of patients with multiple strains could confound this approach (25). Thus, multiple isolates were collected from 24 patients. In 20 of the 24 patients, no differences in MLST alleles between isolate pairs were observed; however, multiple genotypically distinct isolates were identified in 4 patients (17%), suggesting coinfection with ≥2 strains. Subsequent analyses included single isolates from clonally infected patients as well as both isolates from patients infected with two genotypes.
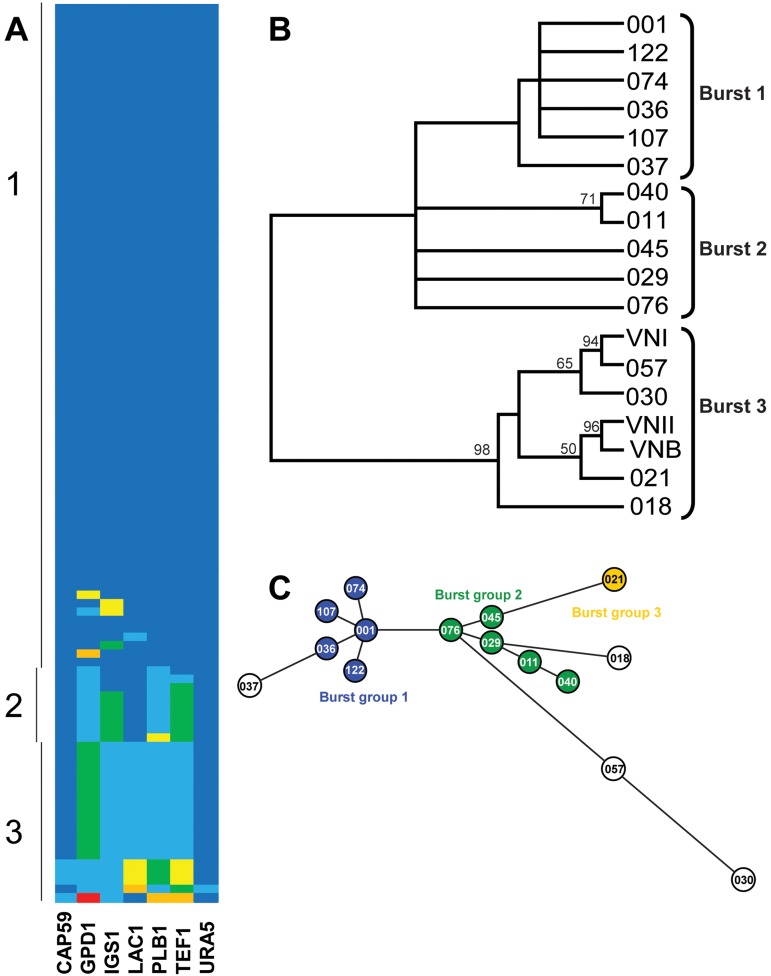
Sequences for each MLST locus were compared and reduced to nonredundant clonal sets, and a maximum parsimony analysis of the sequences at individual loci was performed (see Fig. S1 in the supplemental material). Only one strain showed a polymorphism at the URA5 locus. The SOD1 locus was completely clonal in our population. Locus allele identifiers for each strain were combined to produce a strain sequence type (ST) that serves as a representation of the strain genotype. A total of 15 sequence types were identified (Table 2). The sequence types consisted of 4 clonal clusters and 11 singleton sequence types (Fig. 1A). The largest clonal cluster (UgCl001) contained 74 isolates. The other clonal clusters contained 14, 5, and 3 isolates (represented in Table 2 by UgCl021, UgCl011, and UgCl057, respectively).
TABLE 2.
Ugandan clinical isolate sequences and sequence types
| Burst | UgCl isolateb |
Nucleotidea at the indicated position of the concatenated sequences in the UgCl isolate: |
STc |
|||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAP59 |
GPD1 |
IGS1 |
LAC1 |
PLB1 |
TEF1 |
URA5 | ||||||||||||||||||||||||||||||||||
| 458 | 527 | 563 | 576 | 683 | 778 | 915 | 956 | 1220 | 1303 | 1324 | 1388 | 1401 | 1459 | 1481 | 1482 | 1487 | 1548 | 1552 | 1688 | 1706 | 1708 | 1725 | 1826 | 1972 | 2014 | 2132 | 2273 | 2331 | 2355 | 2357 | 2422 | 2820 | 2832 | 3266 | 3430 | 3451 | 3933 | |||
| 1 | 001 | T | T | C | T | C | G | T | C | T | G | G | A | C | T | X | X | T | G | A | G | T | G | A | T | G | A | T | X | T | C | A | G | C | C | T | G | T | T | 93 |
| 107 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 91 | |
| 036 | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 94 | |
| 037 | . | C | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | 39 | |
| 074 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 95 | |
| 122 | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 92 | |
| 2 | 076 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | 31 |
| 029 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | C | . | . | . | 31 | ||
| 011 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | G | . | . | . | . | . | C | . | . | . | 77 | ||
| 040 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | A | G | . | . | . | . | . | C | . | . | . | 78 | |
| 045 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | C | A | . | . | 31 | |
| 3 | 018 | . | C | . | . | . | . | . | . | A | C | . | G | T | C | G | T | C | A | G | A | . | A | . | C | . | . | . | G | . | . | . | . | . | C | . | . | C | 4 | |
| 021 | . | C | . | . | . | . | A | . | A | C | . | G | T | C | G | T | C | A | G | A | . | A | . | . | . | . | C | . | G | . | . | . | . | . | C | A | . | . | 5 | |
| 057 | C | C | . | . | . | . | . | . | A | C | . | G | T | C | G | T | C | A | G | A | . | A | . | . | C | G | . | . | . | . | . | T | . | T | C | A | . | . | 63 | |
| 030 | C | C | T | A | . | A | . | . | A | C | . | G | T | C | G | T | C | A | G | A | . | A | . | . | . | . | . | . | G | T | C | . | T | T | C | A | A | . | 69 | |
The numbers are the locations of polymorphism relative to the start site of concatenated MLST loci. A dot indicates that the nucleotide was identical to that in UgCl001.
Clonal clusters indicated in bold type.
Sequence type (ST) designation based on BioloMICS.net database.
FIG 1 .
Multilocus sequence typing of Ugandan clinical isolates. (A) Pseudocolor representation of the allelic structure of the UgCl strain population. Alleles at the seven loci are indicated by different colors. Each row indicates a single strain. Vertical lines on the left represent Burst groups. (B) Consensus tree from 1,000 bootstrapped replicates of a maximum parsimony analysis of the concatenated sequences of all informative loci. Bootstrap values over 50% are shown. (C) Analytical separation of UgCl strains into nonredundant evolutionary groups based on Burst analysis of the sequence types. The colors in the circles indicate the 3 Burst groups. Outlier strains are not shaded.
A single representative strain for each sequence type was used to analyze evolutionary relationships between the UgCl strains and further stratify the isolates into distinct groups. Phylogenetic analyses, including maximum parsimony, maximum likelihood, and bootstrap analyses, were performed on the concatenated sequences of MLST loci for the 15 identified sequence types. Maximum parsimony analysis was performed with inclusion of reference strain sequences for the major C. neoformans var. grubii VNI, VNII, and VNB clades to place the resulting tree in the context of previously established evolutionary relationships (Fig. 1B). Maximum parsimony and maximum likelihood analyses produced comparable results (not shown). To further stratify the isolates, an MLST-specific Burst clustering analysis was performed (26). The Burst clustering hypothesized a division of the 15 sequence types into three nonredundant Burst groups and four more distant singleton strains (Fig. 1C). The more highly stratified three Burst groups were used in subsequent clinical and phenotypic analyses. For the 4 patients with multiple infections, 2 patients had infections with strains in the same Burst group, and thus the demographic and clinical comparisons were not repeated. Two of the patients had infections with strains in different Burst groups. In these instances, the clinical and geographic data from the two patients were included in the analysis of both respective Burst groups.
Patient clinical outcome.
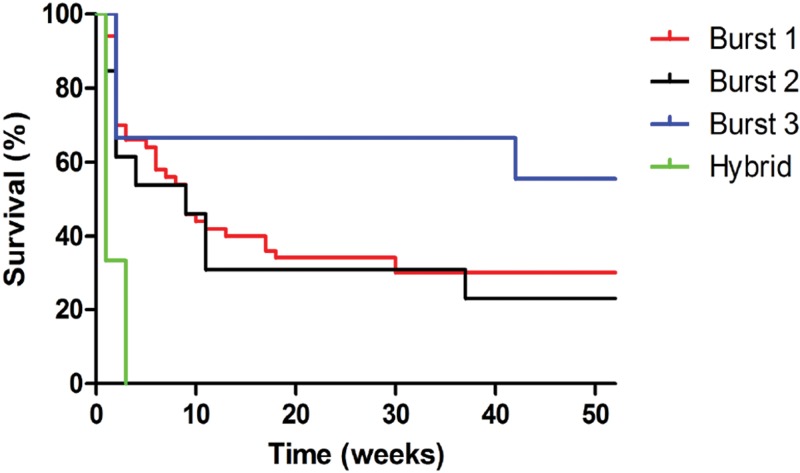
Having identified three distinct Burst groups and a hybrid group, we next asked whether cryptococcal genotype could predict disease severity. Patient 1-year clinical outcome data were available for 98 of the 111 culture-positive patients (Table 1). The overall 1-year mortality rate was 78%. Patients infected with Burst group 1 and 2 strains had higher mortality than Burst group 3 strains (79% [68/86] versus 38% [3/8] [P = 0.05 by the chi-square test]). Survival curves showed that patients infected with Burst group 3 strains lived longer than patients infected with the other genotypes (Fig. 2, P = 0.007 by the log rank test). None of the six patients infected with hybrid strains survived the infection. No other clinical or demographic parameter monitored, including CD4 count and HIV-1 viral load, was associated with genotype.
FIG 2 .
One-year survival of culture-positive patients presenting at Mulago Hospital in Kampala, Uganda, based on strain genotype. Patients infected with Burst group 3 strains lived longer than patients infected with other genotypes (P = 0.007 by log rank test). No patients infected with hybrid strains survived the infection.
Environmental exposure to specific cryptococcal strains in particular geographic regions is a possible explanation for the association between isolate genotype and patient death. The majority of patients lived in parishes within the Kampala district in Uganda, but no geographic clusters of patients infected with similar genotypes was observed (see Fig. S2 in the supplemental material).
Cryptococcal strain phenotypes.
At least three major virulence factors are important for cryptococcal pathogenesis: mating type, melanin, and polysaccharide capsule (21, 22). Mating type determination and in vitro assays of melanin and capsule production were performed to identify any associations with genotype that could explain the differences in lethality.
The mating type of the strain was determined by PCR using STE20 primers specific for mating type and serotype. The vast majority of the C. neoformans var. grubii strains were mating type α (αA); only two were mating type a (aA). Because only 2 out of 132 C. neoformans var. grubii strains were mating type a, sufficient variation was not present to allow for comparison of mating type to genotype, phenotype, or patient outcome.
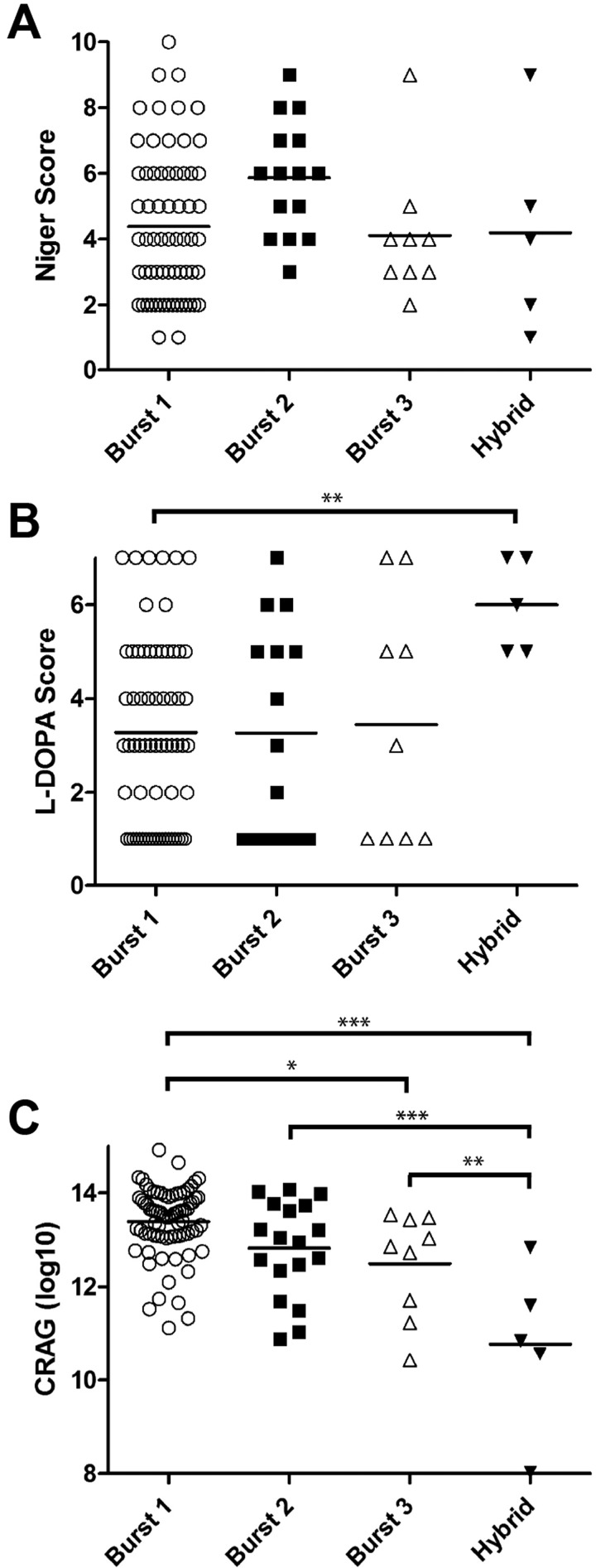
Melanin production by the clinical isolates was analyzed using media containing niger seeds or l-DOPA (l-3,4-dihydroxyphenylalanine). These media provide biphenolic precursors for melanin production along distinct pathways (27). A relative scoring method based on similarity to reference strains with weak (JEC20) and strong (KN99α) melanization was employed to analyze melanin production over 48 h (see Fig. S3A in the supplemental material). K-means clustering analysis for both types of media showed separation of the melanin production into 10 major clusters on niger seed medium and 7 major clusters on l-DOPA medium (Fig. S3B). The Burst and hybrid groups did not differ significantly in their ability to produce melanin on niger seed medium (Fig. 3A). In contrast, while no association between genotype and melanin production on l-DOPA was observed for the Burst groups, the hybrid group had significantly higher melanin production on l-DOPA medium (P = 0.001 by the Tukey-adjusted t test) (Fig. 3B).
FIG 3 .
Cryptococcal virulence factors and genotype. (A and B) Association between Burst groups and temporal melanization as indicated by K-means cluster rank on niger seed (A) or l-DOPA (B) medium. (C) Association between Burst groups and shed capsule as measured by cryptococcal antigen titer. Each symbol represents the value for the individual strain. The short vertical bars represent the means for the different groups. Mean values that are statistically significantly different are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Capsule production by the UgCl isolates was monitored in two ways for comparison to the genotype of the isolate. First, in vivo capsule release into patient CSF at the time of presentation with meningitis was measured using a qualitative cryptococcal antigen assay (CRAG) agglutination assay. No association between clinical CRAG titer and Burst genotype was observed (P = 0.32 by analysis of variance [ANOVA]). This result could be confounded by differences in the fungal burden in patients, resulting in inaccurate measures of capsule production. Consequently, an in vitro quantitative CRAG assay was performed to more precisely determine the capacity of each strain to produce and shed capsule. Burst group 1 strains had the highest capacity to produce and secrete capsule (Fig. 3C). The hybrids produced significantly less capsule than the rest of the Burst groups (P = 0.005 by Tukey-adjusted t tests). Importantly, Burst group 1 showed elevated capsule release compared to Burst group 3 (P = 0.02 by Tukey-adjusted t test) and higher (but not statistically significant) capsule release compared to Burst group 2 strains. These data implicate elevated capsule production as one potential mechanism by which Burst group 1 isolates could cause increased mortality in patients.
Patient/host immune response.
The observation that Burst group 1 isolates were associated with higher mortality and shed more capsule than the other Burst groups led us to hypothesize that the capsule from these strains could influence the type of immune response during infection. To directly test this hypothesis, we developed an ex vivo cytokine release assay to measure the responses to capsule and cell wall preparations obtained from representative Burst group 1 and 3 isolates. Blood samples from 18 healthy volunteers were incubated with Burst group 1 and 3 antigens, cytokine production was quantified by multiplex assay, and differences between each volunteer’s response to the Burst group 1 and 3 antigens was determined by a paired t test.
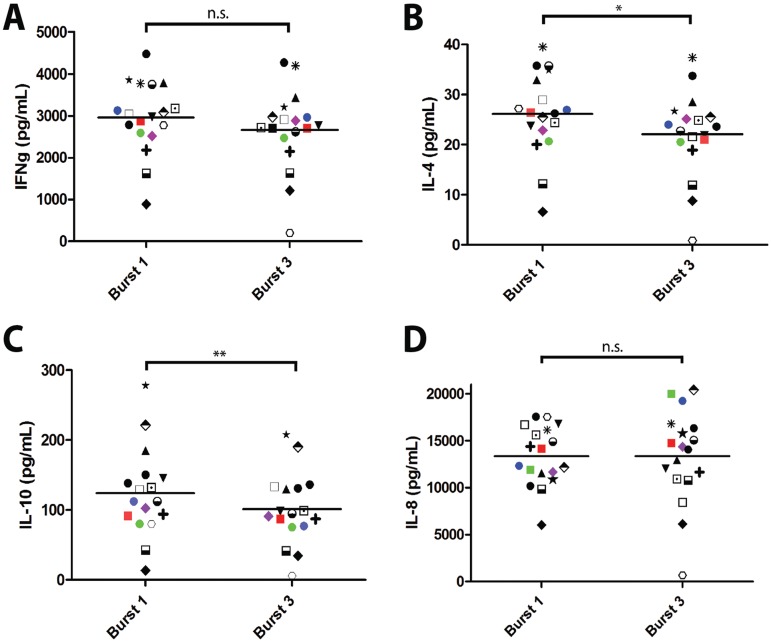
Release of the Th1 cytokine IFN-γ was similar between the Burst group 1 and 3 capsule antigens (P = 0.08 by a paired t test) (Fig. 4A). In contrast, production of Th2 cytokines IL-4 and IL-10 was significantly higher in response to capsule antigens from Burst group 1 strains compared to Burst group 3 strains (P = 0.02 and P = 0.003 by a paired t test, respectively) (Fig. 4B and C). The amount of capsule antigen (measured by CRAG assay) was standardized in these assays, and both capsule antigens elicited equivalent dose-dependent IL-8 chemokine responses (P = 0.99 by a paired t test) (Fig. 4D; see Fig. S4B in the supplemental material). Thus, the observed differences in the immune responses to capsule antigen were due to intrinsic properties of the capsule and not the quantity of capsule generated by the Burst group 1 and 3 isolates.
FIG 4 .
Immune response of healthy volunteers to ex vivo stimulation with capsule antigens from genetically distinct strains. (A to D) Production of IFN-γ (IFNg) (A), IL-4 (B), IL-10 (C), and IL-8 (D) cytokines in response to shed capsule antigen from a representative clonal cluster strain within each Burst group. Antigens were standardized to a physiologically relevant final assay CRAG titer of 1:1,024 as determined by cryptococcal antigen agglutination assay. Data are representative from assays of 6 independent strains. Mean values that are statistically different are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.005. Mean values that are not statistically different are indicated by bars labeled n.s. (for nonsignificant) with P = 0.08 (A) and P = 0.99 (D).
The cell wall antigen preparations did not elicit robust cytokine responses, but dose-dependent IL-8 chemokine responses equivalent to those observed with the capsule antigens were still detected (see Fig. S4 in the supplemental material). These data show that capsule, but not cell wall, from Burst group 1 isolates elicits a stronger Th2 cytokine response than capsule from Burst group 3 isolates.
DISCUSSION
We report the association between cryptococcal genotype and clinical outcome with further determination of the effect of virulence potential on disease severity in a cohort of Ugandans with AIDS and culture-positive CM. Our investigation revealed that different genotypes are associated with differences in patient survival. Cryptococcal genotypes associated with high patient mortality exhibited increased capsule (Burst group 1) or melanin (hybrid strains) production, which are two known virulence factors. We showed that capsule differences between genotypes also may have affected the host immune response, because capsule from genotypes associated with high mortality elicited a greater Th2 cytokine response.
Previous studies noted genotypic diversity in clinical isolates from Southern Africa (e.g., Botswana, South Africa) predominantly in the VNI and VNB clades that show evidence of recombination (24, 28, 29). In contrast, clinical isolates from other regions of the world show significant clonality and are typically members of the VNI and VNII clades (29). The UgCl strains had the same SOD1 locus observed in other African cohorts (24, 29), but the Ugandan cohort had significant clonality, and the predominant clade has diverged from the VNI, VNII, and VNB reference alleles. These data suggest that cryptococcal populations in Uganda are isolated and highly clonal and that the unique Burst group 1 genotypes may be responsible for the unusually high mortality rates observed in Ugandan patient cohorts. The study by Miglia et al. found several sequence types associated with specific geographic regions in South Africa (28). Our data showing no geographic distribution of the UgCl strains in the Ugandan patient population suggest that patient geographic location within Uganda cannot be used to predict infection with the different Ugandan genotypes.
MLST has been used extensively to analyze both global and regional population structures in both C. neoformans and C. gattii (23–25). Yet, the ability of MLST to resolve clinically relevant differences between strains remained unclear. The data presented here show that MLST can be used to distinguish differences in clinical outcome and provide insight into factors affecting disease development. The Ugandan isolates were relatively homogenous with extensive clonality by MLST genotyping and with few polymorphisms between genotypes. Because of this homogeneity, most C. neoformans var. grubii isolates in the cohort could be stratified into one of three subpopulations based on Burst analysis. Each Burst group had multiple patients with identical MLST genotypes that could be compared. This clustering of the data revealed connections between genotype, clinical outcome, and virulence factor production that may not be readily apparent in more genotypically diverse populations.
Coinfection with multiple strains of C. neoformans has been observed. Previous studies identified coinfections based on the phenotypic traits (30), mating types (25), or genotypes of serial isolates (31). Desnos-Ollivier estimated that coinfections could account for 20% of cryptococcal disease based on studies of a French cohort (25). Similarly, 17% of the Ugandan patients screened for coinfection were infected with multiple genotypes. The effect of coinfection on clinical presentation and patient outcome remains to be determined. In this study, only 4 patients had confirmed coinfections, and only two of the patients had infections with evolutionarily distinct strains in different Burst groups. Unfortunately, because the vast majority of the clinical isolates used here were colony purified before storage, coinfection could not be controlled in this study, and the effect of coinfection on clinical outcome remains unknown. If coinfection rates are almost 20%, as the available data suggest, then coinfection could lead to a 20% error rate in accurately predicting the host response to infection. In this case, our results showing associations between genotype and clinical outcome could underestimate the differences between genotypes. The differences might be more pronounced if coinfections were identified and excluded from the analysis. Future studies in which coinfections and single infections are analyzed for their clinical outcome and host response will be necessary to define the effect coinfection has and whether the host response is cumulative, associated with the predominant strain, or associated with the most antigenic strain. However, the high prevalence of coinfection suggests that single colony-purified isolates may not accurately represent C. neoformans infections. This has broad implications for many types of studies, including studies assessing antifungal susceptibility.
The genotype of the cryptococcal strain was associated with differences in clinical outcome. Patients infected with Burst group 1 or 2 isolates had higher mortality rates and shorter survival times than patients infected with Burst group 3 isolates (P = 0.02); though, admittedly, there were few subjects with Burst group 3 isolates. One possible explanation for the low prevalence of Burst group 3 isolates is that they may be poor pathogens and do not cause severe meningitis as frequently. This hypothesis is consistent with our results showing reduced capsule production and stimulation of a protective immune response by Burst group 3 strains that would limit disease progression.
To understand the connection between cryptococcal genotype and patient outcome, we analyzed virulence factor production by the different genotypes. We focused on three widely acknowledged virulence factors of Cryptococcus: mating type, melanin, and capsule. While global populations of C. neoformans var. grubii are predominantly α mating type, a high prevalence of a mating type strains were observed in strains from Botswana (32). Most of the Ugandan isolates were α mating type, which is consistent with global populations, with only 2% a mating type in the Ugandan cohort. Thus, differences in mating type are unlikely to explain the higher lethality associated with Burst groups 1 and 2 relative to Burst group 3 strains.
In assessing melanin production, we found no differences between genotypes when the strains were cultured on niger seed medium. However, the hybrid strains produced significantly more melanin when l-DOPA was provided as the biphenolic precursor. Although we found increased melanin production by hybrid strains when these isolates were grown on l-DOPA-containing media in vitro, we cannot draw conclusions about melanin production by these strains in human infections in vivo. l-DOPA, however, is prevalent in the central nervous system (21); thus, the increased melanin production of the hybrid strains in response to l-DOPA could potentially occur in vivo and promote virulence, leading to increased mortality.
Capsule production was measured both in vivo and in vitro. The in vitro data proved superior for a number of reasons. First, the high degree of variability in the in vivo CRAG titers and small cohort made the statistical comparisons underpowered. Second, many uncontrolled factors, including duration of infection and amount of fungal proliferation, affect the amount of capsule present in the in vivo samples. These factors likely resulted in the high variability of the in vivo CRAG titers. The number of cells and duration of culture were controlled more rigorously in the in vitro assay. Consequently, we were able to stratify the Burst groups and hybrids based on capsule shedding. Notably, the high-lethality Burst group 1/2 strains shed the greatest amount of capsule, whereas the less-lethal Burst group 3 strains shed significantly less capsule. The observation that capsule differences between genotypes are statistically associated with differences in patient mortality suggests that genetic diversity of the cryptococcal capsule may directly influence clinical outcome and immune response in humans.
Our findings suggested that Ugandan hybrid strains were associated with increased mortality. Hybrid strains have attenuated virulence in mouse models of cryptococcal infection (33) and often produce less severe infections in humans (21). Thus, the 100% mortality observed with the Ugandan hybrid strains was surprising. A larger cohort of patients infected with hybrid strains compared to C. neoformans var. grubii strains is needed to further explore differences between the infections. The hybrids exhibited high melanin production on l-DOPA medium but shed the least amount of capsule compared to the C. neoformans var. grubii isolates. The genetic divergence of the hybrids relative to the nonhybrids, combined with the low number of hybrids analyzed, prevents meaningful deduction in this regard. It is possible that the high lethality of the hybrids was a consequence of multiple factors, possibly unrelated to capsule or melanin production, and this will be the focus of future investigations.
The current study demonstrates that strain genotype may be a significant factor in determining the immune response in patients with AIDS and CM. Protective immunity to Cryptococcus requires several elements. Innate cells such as macrophages, monocytes, and dendritic cells produce tumor necrosis factor alpha (TNF-α), chemokine (C-C motif) ligand 2 (CCL2) (monocyte chemotactic peptide 1 [MCP-1]), CCL3 (macrophage inflammatory protein 1 [MIP-1α]), and IL-12 to recruit and license additional leukocytes to the site of infection (34–36). The innate immune system alone is not sufficient to clear cryptococcal infections, as assistance is required from adaptive immunity with helper T cells. The priming and differentiation of CD4+ T cells to the Th1 lineage is crucial for a protective immune response to cryptococcal infection (37). IFN-γ produced by Th1 cells not only acts in a positive-feedback loop to stimulate the production of more Th1 cells but also classically activates macrophages to increase reactive oxygen production and kill internalized cryptococcal cells (38–40). Conversely, an IL-4-driven Th2 adaptive response generates alternatively activated macrophages, is nonprotective, results in greater fungal burden, and allows dissemination (41). The Th1/Th2 balance paradigm used to define interactions between Mycobacterium tuberculosis and the human host may also apply to Cryptococcus (42, 43). The pathogen competes against the host to shift a protective Th1 response to a pathogenic Th2 response. How Cryptococcus affects this balance is an active area of research, and capsule production is one potential strategy that could be used by Cryptococcus.
An ex vivo antigen stimulation assay was used to determine the effect of genetic diversity within the cryptococcal capsule on the Th1/Th2 balance in humans. High-lethality Burst group 1 isolates stimulated greater IL-4 and IL-10 production compared to less-lethal Burst group 3 isolates. IL-4 is a powerful inducer of Th2 development, serving as a positive-feedback signal for more IL-4 production as well as a negative-feedback signal for Th1 responses. Similarly, IL-10 is a suppressive cytokine that has antagonistic effects on Th1 responses and development (13). These data suggest that the high-lethality Burst group 1 isolates shift the Th1/Th2 balance in favor of the pathogenic Th2 response. In contrast, the less-lethal Burst group 3 isolates are unable to elicit a robust Th2 response, allowing the Th1/Th2 balance to shift to the more protective Th1 response.
The capsule antigen preparations used in this study were a heterogeneous mix of microvesicles, proteins, immunogenic lipids, and capsule polysaccharide. Therefore, it is likely that T cell antigens, mainly proteins, present in the preparation were processed and presented by APCs to generate the Th1 protective immune response observed in all of the preparations. In contrast, capsule polysaccharide is widely implicated in host immune evasion and can influence host responses in several ways. Capsule can directly inhibit opsonization, phagocytosis, and lysosomal degradation within phagosomes (44–46). In addition, shed capsule can disrupt cytokine signaling and lymphocytes (6, 47, 48). The major component of capsule polysaccharide, glucuronoxylomannan (GXM), can also promote secretion of Th2 cytokines by monocytes (49). Burst group 1 strains produce more polysaccharide than Burst group 3 strains, yet the amount of polysaccharide used in the ex vivo assay was held constant. Thus, the differences in immune response to the capsule preparations observed in the ex vivo assay were due solely to genetic differences between the Burst group 1 and 3 strains that affect the structure, function, and/or composition of the capsule polysaccharide or other components of the preparation. The fact that Burst group 1 strains also produce more capsule polysaccharide suggests that the actual effect on the Th1/Th2 balance is even greater than observed in the ex vivo antigen stimulation assay. A shift in the Th1/Th2 balance in favor of a pathogenic Th2 response results in greater fungal burden, dissemination, and ultimately death in mouse models of cryptococcosis (50, 51). Our data show strains capable of shifting the Th1/Th2 balance toward a Th2 cytokine response in humans are also associated with high mortality rates.
The studies presented here were designed to analyze the effect of two parameters on the immune response and clinical outcome: capsule and cell wall. The capsule—both the amount of capsule and the type of capsule—differed between genotypes and affected the immune response. In contrast, while melanin production was found to differ in the hybrids, no effect of cell wall components on the immune response was observed. Thus, the combination of the melanin and cell wall cytokine induction experiments suggest that alterations in cell wall and melanin production can differ between strains, but the role, if any, cell wall components play in disease outcome remains unclear.
The high overall mortality rate in the Ugandan culture-positive cohort resulted in too few surviving patients to analyze the effect of strain genotype on subsequent development of immune reconstitution inflammatory syndrome (IRIS) following antiretroviral therapy. A larger cohort of culture-positive patients that initiate ART will be needed to examine associations between strain genotype and risk for IRIS. IRIS patients have an aberrant immune response following reconstitution of their CD4+ T cell population. Current theories suggest that this aberrant response is due to uncontrolled reconstitution of the immune system or continued prevalence of a highly reactive antigen (52). Our data show that Burst group 1 strains produce abundant, Th2-reactive, capsule antigen. Thus, Burst group 1 strains may also be more likely to elicit IRIS in patients who survive the initial infection compared to Burst group 3 strains with lower and less reactive antigen.
Taken together, the data presented show that strain genotype is associated with disease outcome. The observation that strain genotype affects the immune response to cryptococcal infections suggests that genotype can be clinically informative and leads to interesting possibilities for new treatment strategies in patients with AIDS and CM. Treatment regimens targeted at the unique combination of pathogen and host may improve clinical outcomes. For example, new treatments such as IFN-γ (53) or other approaches that increase Th1 cytokines in patients infected with those Cryptococcus strains that elicit predominant Th2 responses may restore proper Th1/Th2 balance and allow better control of the infection.
MATERIALS AND METHODS
Ethical statement and data.
Clinical samples were collected during an observational, prospective study of Ugandan AIDS patients with CM designed to study immune reconstitution inflammatory syndrome following ART initiation (3, 54, 55). Each patient had only one episode of CM. This study was reviewed and approved by the institutional review boards of the University of Minnesota, Makerere University, and the Uganda National Council of Science and Technology. Written informed consent was obtained from all subjects, and all data were deidentified.
Strains and media.
C. neoformans clinical isolates (Ugandan clinical [UgCl] isolates) from CSF samples were colony purified and stored as glycerol stocks. Cryptococcus and Candida albicans reference strains (56–62; see Table S1 in the supplemental material) were also stored as glycerol stocks. All strains were grown on yeast peptone dextrose (YPD) medium prior to subsequent analysis. Complete Dulbecco’s modified Eagle’s medium (DMEM+C), niger seed, l-DOPA, canavanine glycine bromothymol blue (CGB), and Christensen’s urea media were used as previously described (63, 64).
MLST.
Genomic DNA extraction was performed as described previously (65). Eight loci were amplified and sequenced—seven International Society for Human and Animal Mycology (ISHAM) consensus loci CAP59, GPD1, IGS1, LAC1, PLB1, SOD1, and URA5 (23) and the optional highly polymorphic TEF1 locus (24). An additional LAC1Ra primer was developed to replace the consensus primer, which was not sufficiently specific for LAC1 amplification in our samples. All primers are listed in Table S2 in the supplemental material. MLST PCR products were resolved by agarose gel electrophoresis, excised, purified, and sequenced. Sequence assembly was performed with Sequencher 4.9 software (Genecodes, Ann Arbor, MI). Exported sequences were truncated to the consensus MLST ends (23). Sequences for each locus were aligned using ClustalW software (66). All polymorphisms were verified using the sequence chromatograms, and alignments were manually edited to resolve ambiguities. Multiple alignments were processed with PAUP4.0b10 software (Sinauer Associates, Sunderland, MA) to produce maximum parsimony trees using a heuristic algorithm with 1,000 random sequence additions and gaps treated as the fifth base. A total of 4,430 characters were analyzed: 4,315 characters were constant, 71 variable characters were parsimony uninformative, and 44 characters were parsimony informative. Sequences for all loci for each UgCl sample were concatenated and subjected to maximum parsimony, maximum likelihood, and bootstrap analyses using PAUP4.0b10 software. Nonredundant allelic sets were compiled using the Non-redundant databases (NRDB) web tool (http://pubmlst.org). Novel allele sequences were deposited in MLST.net (http://www.MLST.net), BioloMICS.net, and BLAST public databases. Sequence types were identified as combinations of allelic types for individual loci based on NRDB data. Sequence types were subjected to Burst analysis (26) to produce a putative evolutionary relationship between clusters of closely related sequence types. A minimum spanning tree was produced from the sequence types with Prim’s algorithm combined with Burst clustering.
Phenotypic analyses.
Cryptococcal yeasts were plated onto either Christensen’s urea or CGB plates and incubated concurrently with control strains at room temperature for 24 to 48 h. Plates were visually inspected for color changes. C. gattii strains were used as positive controls for the CGB test, and C. neoformans KN99α was used as a positive control for Christensen’s urea and a negative control for CGB test. Bright-field microscopy with a Zeiss Axioplan microscope with an attached AxioCam camera (Carl Zeiss, Germany) was used for analysis of cellular morphology.
Melanin production was measured by comparing pigmentation of UgCl strains grown on niger seed and l-DOPA media (67) to C. neoformans reference strains with strong (KN99α) and weak (JEC20) melanization. Cell suspensions were adjusted to 106 and 107 yeasts/ml using a hemocytometer, plated onto YPD medium, grown at 25°C in the dark, and examined at 12, 18, 36, and 48 h. Melanin assays were performed in triplicate. The absolute production of melanin varied in the replicates, but the relative amounts of melanin were highly reproducible. Relative melanization scores of zero (no pigmentation), one (equal to JEC20), two (between JEC20 and KN99α), three (equal to KN99α), or four (more than KN99α) were assigned to UgCl strains based on comparison with the reference strains grown on the same plate. C. albicans strain SC5314 was used as a negative melanization control.
Cryptococcal antigen assay (CRAG) was performed at the time of diagnosis using the Murex Cryptococcus latex agglutination assay (Murex Diagnostics, Norcross, GA) according to the manufacturer’s specifications or following in vitro culture with the Premier cryptococcal antigen kit (Meridian Bioscience, Cincinnati, OH) according to the manufacturer’s specifications with a modified procedure for sample preparation. Briefly, DMEM+C was inoculated and incubated at 30°C. Cultures with an optical density at 600 nm (OD600) of 0.90 to 1.10 were centrifuged at 10,000 rpm, and the supernatant was serially diluted from 1:10 to 1:6,250 with the provided “DIL” solution to obtain an absorbance at 450 nm of 0.14 to 1.50. The enzyme immunoassay value for each sample was calculated from the A450 value according to the manufacturer’s instructions and normalized to an OD600 of 1.0. PCR with differential PAK1, GPA1, and Ste20 primers was used for determination of serotype and mating type as previously described (68).
Ex vivo antigen stimulation.
Antigens were prepared from the cell wall and culture supernatants of six strains, including the control H99 strain and five strains representative of the mean CRAG data points within each of the MLST clonal cluster genotypic groups. A capsule antigen preparation was prepared from culture supernatant from each strain grown in DMEM+C. The supernatant was decanted, frozen at −80°C, and lyophilized. The lyophilized sample was reconstituted in phosphate-buffered saline (PBS), and the CRAG titer was measured. A cell wall antigen preparation was prepared from the yeasts. The cells were flash frozen in liquid nitrogen, combined with glass beads, and vortexed vigorously for 2 h at 4°C to disrupt the cells. The insoluble fraction (i.e., cell wall) was analyzed for CRAG and for protein (BCA protein assay [BCA stands for bicinchoninic acid]; Thermo Fisher Scientific, Rockford, IL) concentrations. Endotoxin levels in all antigen preparations were undetectable (<0.06 U/ml) by Limulus amebocyte lysate assay (Associates of Cape Cod, East Falmouth, MA).
Whole-blood samples for the cryptococcal cytokine release assay were obtained from healthy individuals in North America. Peripheral blood samples from each subject were drawn into lithium heparin tubes, diluted 2-fold with PBS, and dispensed into a tissue culture plate. Cell wall and capsule antigens were added to each well to yield a physiologically comparable glucuronoxylomannan (GXM) CRAG titer of 1:256. PBS was used as the negative control. The plates were incubated at 37°C in 5% CO2 for 20 h. After incubation, the plasma was separated from the cells and stored at 4°C until cytokine analysis. Cytokines were quantified in duplicate for interleukin 2 (IL-2), IL-4, IL-6, IL-8, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) using a Luminex 100 instrument (Bio-Rad, Hercules, CA). Negative controls incubated in PBS were subtracted from the raw data to determine the net response.
Statistics.
The means for groups of isolates were compared by analysis of variance (ANOVA) and t tests, and medians were compared using Kruskal-Wallis rank sum tests and Mann-Whitney U tests. The chi-square and Fisher’s exact tests were used to evaluate the frequencies of categorical variables. Tukey adjustments were made to correct for multiple comparisons. The Mantel-Cox log rank test was used to compare survival data. All analyses were performed with SAS (SAS Institute Inc., Cary, NC).
SUPPLEMENTAL MATERIAL
Maximum parsimony trees of individual MLST loci. Download Figure S1, PDF file, 1.5 MB.
Geographical distribution of culture-positive patients presenting at Mulago Hospital in Kampala, Uganda. No association between geographic location and strain genotype was observed. (A) Districts within Uganda with culture-positive patients. (B) Parishes within the Kampala district with culture-positive patients. Download Figure S2, PDF file, 1.3 MB.
Melanin production. (A) Melanization of representative UgCl strains relative to controls on niger seed and l-DOPA media. (B) Temporal representation of strain melanization on niger seed and l-DOPA media using K-means clustering of melanin production from 12 to 48 h. Download Figure S3, PDF file, 1 MB.
Immune response of healthy volunteers to ex vivo stimulation with cell wall antigens. (A) Production of IL-4, IL-10, IFN-γ, and IL-8 cytokines in response to cell wall antigen from a representative clonal cluster strain within each Burst group. Antigens were standardized to a final assay concentration of 5 µg of protein per well as determined by the Bradford protein assay. (B) IL-8 dose-response curve to diluted (0.5), undiluted (1.0), and concentrated (2.0) capsule antigen. The best-fit linear regression line is red (P < 0.0001, R2 = 0.79, and β = 3401). Units are fluorescence intensity (FI) as reported by the Luminex multiplex instrument. Download Figure S4, PDF file, 0.8 MB.
Reference strains
Primers, PCR conditions, and sequence ends used for MLST analysis
ACKNOWLEDGMENTS
This work was supported by NIH grants AI070152, AI089244, AI073192, AI078750, and AI096925; the University of Minnesota (UMN) Office of International Programs; UMN Center for Infectious Diseases and Microbiology Translational Research; UMN Academic Health Center Collaborative and Tibotec REACH initiative grants; and the Minnesota Medical Foundation Robert and Mabel Bohjanen Immune Reconstitution Research Fund.
Footnotes
Citation Wiesner DL, et al. 2012. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3(5):e00196-12. doi:10.1128/mBio.00196-12.
REFERENCES
- 1. Joint United Nations Programme on HIV/AIDS. UNAIDS 2011. World AIDS Day Report. Joint United Nations Programme on HIV/AIDS (U.N. AIDS), Geneva, Switzerland [Google Scholar]
- 2. Park BJ, et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 3. Boulware DR, et al. 2010. Clinical features and serum biomarkers in HIV immune reconstitution inflammatory syndrome after cryptococcal meningitis: a prospective cohort study. PLoS Med. 7:e1000384 http://dx.doi.org/10.1371/journal.pmed.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bicanic T, et al. 2009. Immune reconstitution inflammatory syndrome in HIV-associated cryptococcal meningitis: a prospective study. J. Acquir. Immune Defic. Syndr. 51:130–134 [DOI] [PubMed] [Google Scholar]
- 5. Perfect JR. 2005. Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol. Med. Microbiol. 45:395–404 [DOI] [PubMed] [Google Scholar]
- 6. Vecchiarelli A. 2000. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 38:407–417 [DOI] [PubMed] [Google Scholar]
- 7. Kozel TR, Wilson MA, Pfrommer GS, Schlageter AM. 1989. Activation and binding of opsonic fragments of C3 on encapsulated Cryptococcus neoformans by using an alternative complement pathway reconstituted from six isolated proteins. Infect. Immun. 57:1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozel TR, Gotschlich EC. 1982. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 129:1675–1680 [PubMed] [Google Scholar]
- 9. Beenhouwer DO, Shapiro S, Feldmesser M, Casadevall A, Scharff MD. 2001. Both Th1 and Th2 cytokines affect the ability of monoclonal antibodies to protect mice against Cryptococcus neoformans. Infect. Immun. 69:6445–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddiqui AA, et al. 2005. IFN-γ at the site of infection determines rate of clearance of infection in cryptococcal meningitis. J. Immunol. 174:1746–1750 [DOI] [PubMed] [Google Scholar]
- 11. Hernandez Y, et al. 2005. Distinct roles for IL-4 and IL-10 in regulating T2 immunity during allergic bronchopulmonary mycosis. J. Immunol. 174:1027–1036 [DOI] [PubMed] [Google Scholar]
- 12. Stenzel W, et al. 2009. IL-4/IL-13-dependent alternative activation of macrophages but not microglial cells is associated with uncontrolled cerebral cryptococcosis. Am. J. Pathol. 174:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murphy K. 2011. Janeway’s immunobiology, 8th ed. Garland Science, New York, NY. [Google Scholar]
- 14. Retini C, et al. 2001. Interdependency of interleukin-10 and interleukin-12 in regulation of T-cell differentiation and effector function of monocytes in response to stimulation with Cryptococcus neoformans. Infect. Immun. 69:6064–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. 2005. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 187:5709–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruiz-Garbajosa P, et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dagerhamn J, et al. 2008. Determination of accessory gene patterns predicts the same relatedness among strains of Streptococcus pneumoniae as sequencing of housekeeping genes does and represents a novel approach in molecular epidemiology. J. Clin. Microbiol. 46:863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang GH, et al. 2010. Sequence analysis of six enterovirus 71 strains with different virulences in humans. Virus Res. 151:66–73 [DOI] [PubMed] [Google Scholar]
- 19. Ngamskulrungroj P, Serena C, Gilgado F, Malik R, Meyer W. 2011. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clin. Microbiol. Infect. 17:251–258 [DOI] [PubMed] [Google Scholar]
- 20. Harris JR, et al. 2011. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin. Infect. Dis. 53:1188–1195 [DOI] [PubMed] [Google Scholar]
- 21. Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, DC: . [Google Scholar]
- 22. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. 2011. Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 23. Meyer W, et al. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desnos-Ollivier M, et al. 2010. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. mBio. 1:e00091-10 http://dx.doi.org/10.1128/journal.pmed.00091-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Staib F, Bethäuser G. 1968. Demonstration of Cryptococcus neoformans in the dust of a dovecote. Mykosen 11:619–624 [PubMed] [Google Scholar]
- 28. Miglia KJ, et al. 2011. Analyses of pediatric isolates of Cryptococcus neoformans from South Africa. J. Clin. Microbiol. 49:307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Litvintseva AP, et al. 2011. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS One 6:e19688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mandal P, Banerjee U, Casadevall A, Nosanchuk JD. 2005. Dual infections with pigmented and albino strains of Cryptococcus neoformans in patients with or without human immunodeficiency virus infection in India. J. Clin. Microbiol. 43:4766–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haynes KA, et al. 1995. Involvement of multiple Cryptococcus neoformans strains in a single episode of cryptococcosis and reinfection with novel strains in recurrent infection demonstrated by random amplification of polymorphic DNA and DNA fingerprinting. J. Clin. Microbiol. 33:99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Litvintseva AP, et al. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2:1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin X, Nielsen K, Patel S, Heitman J. 2008. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect. Immun. 76:2923–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huffnagle GB, et al. 1996. Afferent phase production of TNF-α is required for the development of protective T cell immunity to Cryptococcus neoformans. J. Immunol. 157:4529–4536 [PubMed] [Google Scholar]
- 35. Olszewski MA, et al. 2000. The role of macrophage inflammatory protein-1 alpha/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J. Immunol. 165:6429–6436 [DOI] [PubMed] [Google Scholar]
- 36. Kawakami K, Tohyama M, Xie Q, Saito A. 1996. IL-12 protects mice against pulmonary and disseminated infection caused by Cryptococcus neoformans. Clin. Exp. Immunol. 104:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huffnagle GB, Yates JL, Lipscomb MF. 1991. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J. Exp. Med. 173:793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flesch IE, Schwamberger G, Kaufmann SH. 1989. Fungicidal activity of IFN-γ-activated macrophages: extracellular killing of Cryptococcus neoformans. J. Immunol. 142:3219–3224 [PubMed] [Google Scholar]
- 39. Hoag KA, Lipscomb MF, Izzo AA, Street NE. 1997. IL-12 and IFN-γ are required for initiating the protective Th1 response to pulmonary cryptococcosis in resistant C.B-17 mice. Am. J. Respir. Cell Mol. Biol. 17:733–739 [DOI] [PubMed] [Google Scholar]
- 40. Hardison SE, et al. 2010. Pulmonary infection with an interferon-γ-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am. J. Pathol. 176:774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jain AV, et al. 2009. Th2 but not Th1 immune bias results in altered lung functions in a murine model of pulmonary Cryptococcus neoformans infection. Infect. Immun. 77:5389–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koguchi Y, Kawakami K. 2002. Cryptococcal infection and Th1-Th2 cytokine balance. Int. Rev. Immunol. 21:423–438 [DOI] [PubMed] [Google Scholar]
- 43. Chayakulkeeree M, Perfect JR. 2006. Cryptococcosis. Infect. Dis. Clin. North Am. 20:507–544 [DOI] [PubMed] [Google Scholar]
- 44. Kozel TR, Mastroianni RP. 1976. Inhibition of phagocytosis by cryptococcal polysaccharide: dissociation of the attachment and ingestion phases of phagocytosis. Infect. Immun. 14:62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitchell TG, Friedman L. 1972. In vitro phagocytosis and intracellular fate of variously encapsulated strains of Cryptococcus neoformans. Infect. Immun. 5:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bulmer GS, Sans MD. 1968. Cryptococcus neoformans: inhibition of phagocytosis. J. Bacteriol. 95:5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yauch LE, Lam JS, Levitz SM. 2006. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog. 2:e120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Retini C, Vecchiarelli A, Monari C, Bistoni F, Kozel TR. 1998. Encapsulation of Cryptococcus neoformans with glucuronoxylomannan inhibits the antigen-presenting capacity of monocytes. Infect. Immun. 66:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mariano Andrade R, Monteiro Almeida G, Alexandre DosReis G, Alves Melo Bento C. 2003. Glucuronoxylomannan of Cryptococcus neoformans exacerbates in vitro yeast cell growth by interleukin 10-dependent inhibition of CD4+ T lymphocyte responses. Cell. Immunol. 222:116–125 [DOI] [PubMed] [Google Scholar]
- 50. Kawakami K, et al. 1999. Interleukin-4 weakens host resistance to pulmonary and disseminated cryptococcal infection caused by combined treatment with IFNγ-inducing cytokines. Cell. Immunol. 197:55–61 [DOI] [PubMed] [Google Scholar]
- 51. Müller U, et al. 2012. Lack of IL-4 receptor expression on T helper cells reduces T helper 2 cell polyfunctionality and confers resistance in allergic bronchopulmonary mycosis. Mucosal Immunol. 5:299–310 [DOI] [PubMed] [Google Scholar]
- 52. Wiesner DL, Boulware DR. 2011. Cryptococcus-related immune reconstitution inflammatory syndrome (IRIS): pathogenesis and its clinical implications. Curr. Fungal Infect. Rep. 5:252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jarvis JN, et al. 2012. Adjunctive IFNγ immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS 26:1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kambugu A, et al. 2008. Outcomes of cryptococcal meningitis in Uganda before and after the availability of highly active antiretroviral therapy. Clin. Infect. Dis. 46:1694–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boulware DR, et al. 2010. Paucity of initial cerebrospinal fluid inflammation in cryptococcal meningitis is associated with subsequent immune reconstitution inflammatory syndrome. J. Infect. Dis. 202:962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nielsen K, et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Toffaletti DL, Rude TH, Johnson SA, Durack DT, Perfect JR. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Meyer W, et al. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Varma A, Kwon-Chung KJ. 1998. Construction of stable episomes in Cryptococcus neoformans. Curr. Genet. 34:60–66 [DOI] [PubMed] [Google Scholar]
- 61. Fraser JA, et al. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364 [DOI] [PubMed] [Google Scholar]
- 62. Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 63. Christensen WB. 1946. Urea decomposition as a means of differentiating Proteus and Paracolon cultures from each other and from Salmonella and Shigella types. J. Bacteriol. 52:461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klein KR, et al. 2009. Identification of Cryptococcus gattii by use of l-canavanine glycine bromothymol blue medium and DNA sequencing. J. Clin. Microbiol. 47:3669–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu D, Coloe S, Baird R, Pederson J. 2000. Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Y, Aisen P, Casadevall A. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lengeler KB, Cox GM, Heitman J. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum parsimony trees of individual MLST loci. Download Figure S1, PDF file, 1.5 MB.
Geographical distribution of culture-positive patients presenting at Mulago Hospital in Kampala, Uganda. No association between geographic location and strain genotype was observed. (A) Districts within Uganda with culture-positive patients. (B) Parishes within the Kampala district with culture-positive patients. Download Figure S2, PDF file, 1.3 MB.
Melanin production. (A) Melanization of representative UgCl strains relative to controls on niger seed and l-DOPA media. (B) Temporal representation of strain melanization on niger seed and l-DOPA media using K-means clustering of melanin production from 12 to 48 h. Download Figure S3, PDF file, 1 MB.
Immune response of healthy volunteers to ex vivo stimulation with cell wall antigens. (A) Production of IL-4, IL-10, IFN-γ, and IL-8 cytokines in response to cell wall antigen from a representative clonal cluster strain within each Burst group. Antigens were standardized to a final assay concentration of 5 µg of protein per well as determined by the Bradford protein assay. (B) IL-8 dose-response curve to diluted (0.5), undiluted (1.0), and concentrated (2.0) capsule antigen. The best-fit linear regression line is red (P < 0.0001, R2 = 0.79, and β = 3401). Units are fluorescence intensity (FI) as reported by the Luminex multiplex instrument. Download Figure S4, PDF file, 0.8 MB.
Reference strains
Primers, PCR conditions, and sequence ends used for MLST analysis