ABSTRACT
SAR11 is an ancient and diverse clade of heterotrophic bacteria that are abundant throughout the world’s oceans, where they play a major role in the ocean carbon cycle. Correlations between the phylogenetic branching order and spatiotemporal patterns in cell distributions from planktonic ocean environments indicate that SAR11 has evolved into perhaps a dozen or more specialized ecotypes that span evolutionary distances equivalent to a bacterial order. We isolated and sequenced genomes from diverse SAR11 cultures that represent three major lineages and encompass the full breadth of the clade. The new data expand observations about genome evolution and gene content that previously had been restricted to the SAR11 Ia subclade, providing a much broader perspective on the clade’s origins, evolution, and ecology. We found small genomes throughout the clade and a very high proportion of core genome genes (48 to 56%), indicating that small genome size is probably an ancestral characteristic. In their level of core genome conservation, the members of SAR11 are outliers, the most conserved free-living bacteria known. Shared features of the clade include low GC content, high gene synteny, a large hypervariable region bounded by rRNA genes, and low numbers of paralogs. Variation among the genomes included genes for phosphorus metabolism, glycolysis, and C1 metabolism, suggesting that adaptive specialization in nutrient resource utilization is important to niche partitioning and ecotype divergence within the clade. These data provide support for the conclusion that streamlining selection for efficient cell replication in the planktonic habitat has occurred throughout the evolution and diversification of this clade.
IMPORTANCE
The SAR11 clade is the most abundant group of marine microorganisms worldwide, making them key players in the global carbon cycle. Growing knowledge about their biochemistry and metabolism is leading to a more mechanistic understanding of organic carbon oxidation and sequestration in the oceans. The discovery of small genomes in SAR11 provided crucial support for the theory that streamlining selection can drive genome reduction in low-nutrient environments. Study of isolates in culture revealed atypical organic nutrient requirements that can be attributed to genome reduction, such as conditional auxotrophy for glycine and its precursors, a requirement for reduced sulfur compounds, and evidence for widespread cycling of C1 compounds in marine environments. However, understanding the genetic variation and distribution of such pathways and characteristics like streamlining throughout the group has required the isolation and genome sequencing of diverse SAR11 representatives, an analysis of which we provide here.
Introduction
Alphaproteobacteria of the SAR11 clade are the most abundant group of planktonic cells in marine systems, typically accounting for ~25% of prokaryotic cells in seawater worldwide (1, 2). In addition to their importance to marine biogeochemical cycles, these highly successful organisms, along with genomes from Prochlorococcus (3), provided the first compelling evidence for the theory that streamlining selection has shaped the evolution of some major lineages of marine bacterioplankton. Cultivation of the temperate coastal SAR11 isolate “Candidatus Pelagibacter ubique” strain HTCC1062 and the subsequent sequencing of its genome revealed it possesses many unusual features for a free-living organism, including an extremely small, streamlined genome with few paralogs, no pseudogenes, and many missing genes and pathways that are otherwise common in bacteria (4, 5). However, the SAR11 clade is phylogenetically diverse, spanning 18% 16S rRNA gene divergence (6) and encompassing at least a dozen ecotypes that are identified by their unique distributions in the environment (7–11; K. L. Vergin et al., submitted for publication). Wilhelm et al. (12) drew the conclusion that SAR11 genomes are highly conserved in gene content and synteny by comparing SAR11 genome sequences with fragmentary SAR11 sequence data extracted from Global Ocean Survey (GOS) metagenomes and measuring conservation of synteny and variation in gene-gene boundaries. They found that 96% of homologous fragments were conserved in gene order relative to the HTCC1062 genome. They also reported greater genomic rearrangement at operon boundaries than within operons, as well as hypervariable regions (HVRs), also termed genomic islands in Prochlorococcus (13), that appeared to have conserved locations within the genome, possibly allowing these cells to acquire novel genetic material with adaptive significance (12, 14).
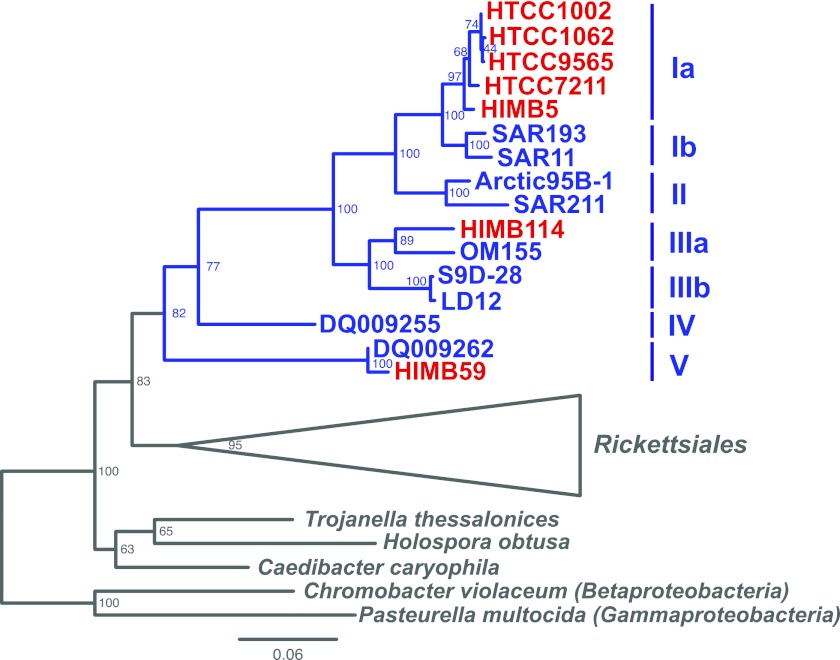
Comparative genomics with more strains offers a means to understand the evolutionary history of SAR11, to confirm predictions, such as those of Wilhelm et al. (12), and to understand the functional significance of SAR11 ecotype diversity in the oceans today. For example, it has been uncertain whether proteorhodopsin (PR) (15, 16), C1 and methyl group oxidation (17), or the requirements for reduced sulfur (18) and glycine/serine (19) are found throughout the clade. Advancements in high-throughput culturing techniques (20, 21) and knowledge obtained from our previous work has recently resulted in the successful culturing of representatives of SAR11 that span three divergent phylogenetic lineages of the proposed family “Pelagibacteraceae” (6) (Fig. 1). Five SAR11 strains (HTCC1062, HTCC1002, HTCC9565, HTCC7211, and HIMB5) form a group of closely related lineages (16S identity ≥ 98%; ANI [average nucleotide identity] ≥ 75%) within SAR11 subclade Ia, which is ubiquitous in geographic distribution (1, 2). Strain HIMB114 is more distantly related (88% 16S identity with HTCC1062) and is part of the subclade IIIa, which is a sister group to the freshwater SAR11 subclade IIIb/LD12 lineage (Vergin et al., submitted). The subclade Va strain HIMB59 is very distantly related (82% 16S identity with HTCC1062) but has been classified as a SAR11 strain based on monophyletic grouping with the other SAR11 strains using both 16S (Vergin et al., submitted) (Fig. 1) and concatenated protein phylogenies (6). Here we present a detailed comparative analysis of these seven SAR11 genomes that provides new insight into the genome features and genetic content of this diverse group of globally abundant organisms.
FIG 1 .
16S phylogenetic tree of the SAR11 clade (blue), showing a subset of major subclades defined here and elsewhere (6, 7) and the genomes included in this study (red). Bootstrap support is displayed at the nodes. Scale bar indicates 0.06 changes per position.
RESULTS
General genome features.
The strains in this study were isolated from surface seawater of disparate origin: HTCC1062 and HTCC1002 from the temperate coastal Northeast Pacific (4), HTCC9565 from the temperate open ocean of the Northeast Pacific, HTCC7211 from the Sargasso Sea in the subtropical Atlantic (21), and HIMB5, HIMB114, and HIMB59 from the coastal tropical North Pacific (Table 1; see also Table S8 at http://giovannonilab.science.oregonstate.edu/publications). The genomes of HTCC1062, HTCC1002, HTCC7211, HIMB5 and HIMB59 are closed, while the genomes of HIMB114 and HTCC9565 consist of scaffolds with one and three contigs, respectively. Based on synteny with the other genomes of subclade Ia, the amount of missing information for the HTCC9565 genome is estimated to be from <1 to ca. 5.5 kbp. While the degree of completion of the HIMB114 genome is more difficult to estimate, a second recently sequenced subclade IIIa genome is complete at 1.285 Mbp (22), which is less than 50 kbp larger than the current HIMB114 sequence. The presence of a compact (mean genome size of 1.337 ± 0.08 Mbp), low G+C (28.6 to 32.3%) genome is a unifying characteristic of the SAR11 clade (Table 1). The genomes code for between 1,357 and 1,576 genes, one copy of the 5S, 16S, and 23S ribosomal RNA genes, and 30 to 35 tRNAs (see Table S1 at the above URL). No pseudogenes were identified in any of the strains.
TABLE 1 .
Characteristics of SAR11 genomes used in this study
| Characteristic | Value or description for strain |
||||||
|---|---|---|---|---|---|---|---|
| HTCC1062 | HTCC1002 | HTCC9565 | HTCC7211 | HIMB5 | HIMB114 | HIMB59 | |
| Subclade | Ia | Ia | Ia | Ia | Ia | IIIa | Va |
| Environment | Coastal, temperate | Coastal, temperate | Open ocean, temperate | Open ocean, subtropic | Coastal, tropic | Coastal, tropic | Coastal, tropic |
| Origin | NE Pacific | NE Pacific | NE Pacific | Sargasso Sea, Atlantic | N. Pacific | N. Pacific | N. Pacific |
| Size (Mbp) | 1.309 | 1.323 | 1.280 | 1.457 | 1.343 | 1.237 | 1.410 |
| Status | Closed | Closed | 3 contigs | Closed | Closed | 1 contig | Closed |
| GC content (%) |
29.7 | 29.8 | 28.9 | 29.0 | 28.6 | 29.6 | 32.3 |
| Total no. of genes |
1,394 | 1,423 | 1,386 | 1,576 | 1,467 | 1,357 | 1,532 |
| No. of protein coding genes |
1,354 | 1,387 | 1,352 | 1,541 | 1,431 | 1,321 | 1,493 |
| % SAR11 corea |
54.0 | 52.6 | 54.0 | 48.5 | 51.0 | 56.4 | 51.8 |
| % SAR11 uniqueb |
1.4 | 3.6 | 7.1 | 10.7 | 7.8 | 13.1 | 26.2 |
| % subclade Ia corea |
80.8 | 78.4 | 80.7 | 72.9 | 75.9 | ||
| % subclade Ia uniquea |
1.4 | 3.9 | 7.6 | 12.8 | 10.9 | ||
Percentage of total genes within the SAR11 or SAR11 subclade Ia core.
Percentage of total genes unique to individual strains compared with all seven SAR11 genomes (SAR11 unique) or five SAR11 subclade Ia genomes (subclade Ia unique).
The core and pan-genome of the Pelagibacteraceae.
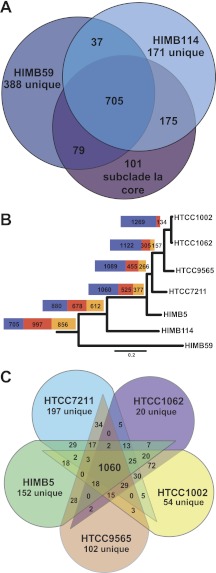
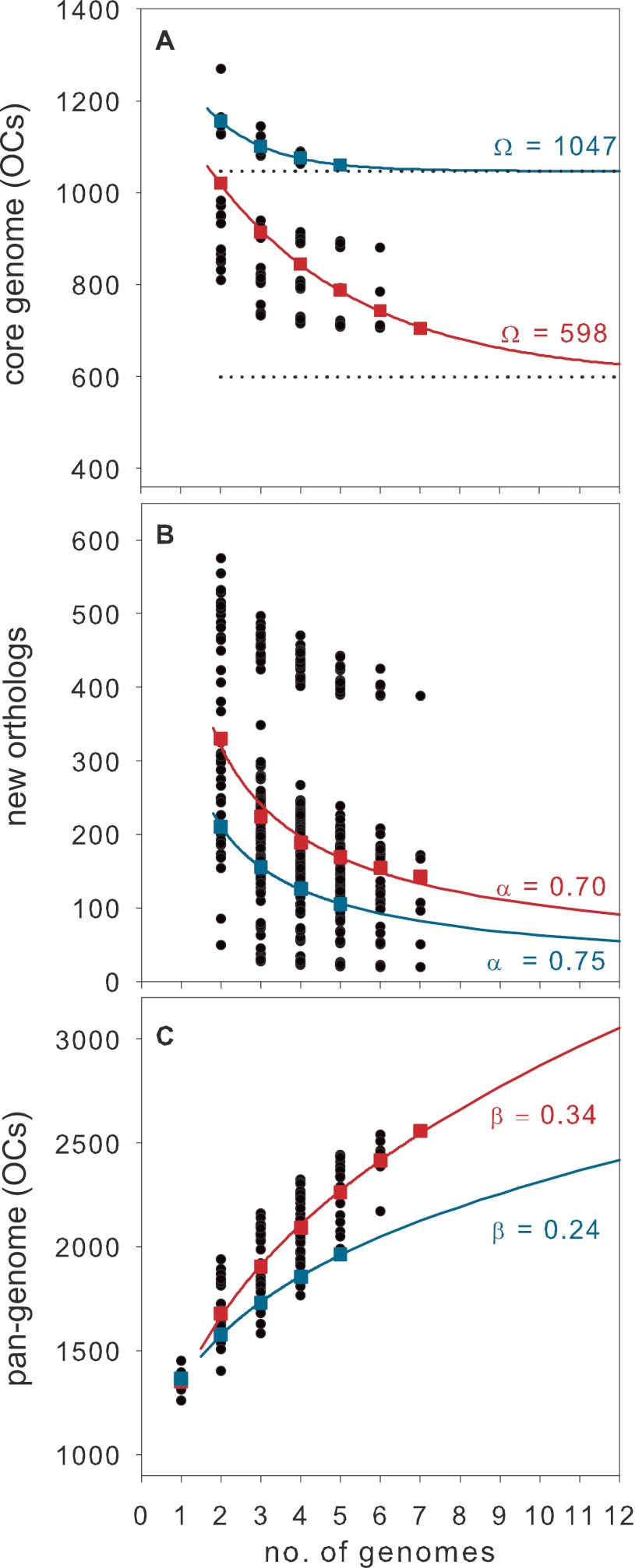
We investigated the SAR11 pan-genome, the total set of genes found in all seven genomes, by examining orthologous clusters (OCs) and excluding paralogs and non-protein-coding genes. The Pelagibacteraceae pan-genome contains a total of 2,558 predicted OCs, with a conserved core genome of 705 OCs present in all SAR11 strains (Fig. 2A). The “flexible” genome (genes found in one or more but not all genomes) contains 1,853 OCs, 997 unique and 856 shared non-core (Fig. 2B). The contribution of core, unique, and shared non-core OCs to the SAR11 pan-genome changes considerably at different levels of phylogenetic similarity, with the number of orthologs in the core genome (blue boxes) negatively correlated with evolutionary distance (Fig. 2B). When considering only the five SAR11 subclade Ia genomes, the pan-genome of 1,962 OCs consists of an even more conserved core genome of 1,060 OCs (Fig. 2B and C). The actual numbers of genes in the core and flexible genomes differ by strain due to paralogs, detailed below. The predicted size of the core SAR11 and subclade Ia genomes was extrapolated by fitting an exponential decay function to the average number of core OCs calculated for the sequential addition of the seven genome sequences (Fig. 3A), resulting in a predicted SAR11 core genome of 598 OCs. When the five SAR11 subclade Ia genomes are considered separately, the predicted core genome of 1,047 OCs closely matches the observed core genome size of 1,060, suggesting that the current subclade Ia core genome is well defined by the available genomes.
FIG 2 .
(A) Venn diagram showing the number of OCs shared between the SAR11 subclade Ia core genome, HIMB114, and HIMB59. (B) The relative contribution of core (blue), shared non-core (orange), and unique (red) orthologs to the pan-genome at each level of divergence. The total size of each bar is proportional to the total number of orthologs in the pan-genome. The scale bar indicates 0.2 changes per position. The tree was redrawn based on the work of Thrash et al. (6). (C) Venn diagram showing the number of shared OCs among the five strains of SAR11 subclade Ia.
FIG 3 .
SAR11 pan-genome analysis. The number of core genes (A), new orthologs (B), or total genes (pan-genome) (C) is plotted versus the sequential addition of genomes 7!(N!(7 − N)!). Squares show average values for all members of SAR11 (red) and SAR11 subclade Ia (blue). In panel A, the curve represents the least-squares fit of the average values to an exponential decay function, and the dotted line indicates the asymptotic values predicted for the SAR11 and SAR11 subclade Ia core genome size. Curves in panels B and C are from power law regression analyses.
To model the global SAR11 pan-genome, we applied the method described previously by Tettelin et al. (23), which predicts the number of new orthologs expected to be discovered with each additional sequenced genome, as well as to what degree the SAR11 pan-genome is open, meaning how many unique genes will be identified with each new sequenced strain. The number of new orthologs decreased as more strains were compared, resulting in an average value of 142 new orthologs per genome when all seven sequenced genomes are considered (Fig. 3B). When the five SAR11 subclade Ia genomes were analyzed separately, the number of new orthologs added by the 5th strain was 105 on average. Power law regression analyses of the average number of new SAR11 orthologs and average total pan-genome size resulted in values of α = 0.70 and β = 0.34 for the exponents (Fig. 3B and C). These values agree reasonably with the relation α = 1 − β as required by Heaps’ law applied to the pan-genome model (24), and α ≤ 1 indicates an open pan-genome for SAR11. While the SAR11 subclade Ia pan-genome is also open (α = 0.75 and β = 0.24), its smaller size and lower rate of growth reflect that this group is better defined by the current genomes than the entire clade.
Comparison of total conserved gene content to that of other bacterial groups.
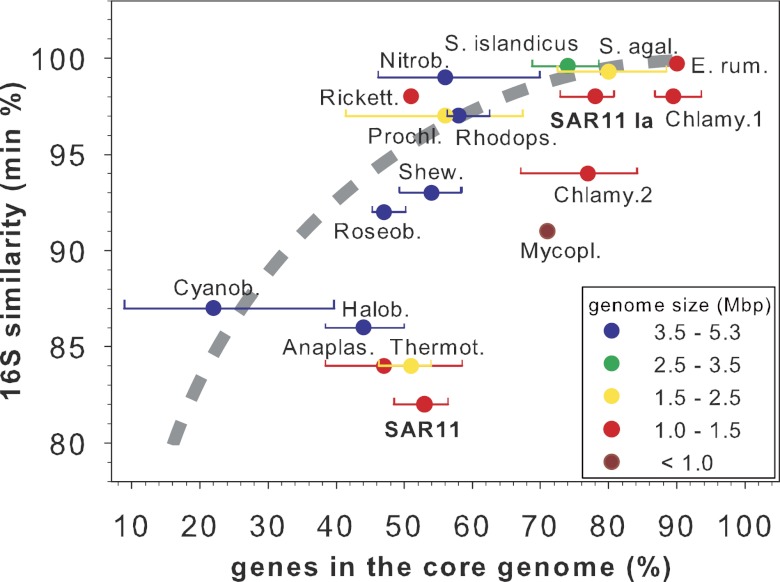
To put the relative conservation of the SAR11 core genome in perspective, we compared our results to those from other comparative genome studies, including studies of environmentally relevant prokaryotes and those with similar genome sizes (Fig. 4; see also Table S2 at http://giovannonilab.science.oregonstate.edu/publications). Here we considered all genes, including paralogs, since ignoring duplications would artificially inflate the amount of conservation, and calculated the number of core genes as a percentage of total genes for each SAR11 strain (Table 1). Pairwise average amino acid identity (AAI) for the SAR11 clade follows the general trend for bacteria (Fig. 5A; see also Table S3 at the above URL) (25, 26) and, based on 16S rRNA gene comparisons (18%) and AAI comparisons, spans order-level divergence. In spite of this, the SAR11 core genome represents 48 to 56% of the total gene repertoire per strain and is similar in proportion to that of bacterial genera like Shewanella (7% 16S rRNA gene divergence) (27) (Fig. 4). The core genomes for groups with similar divergence at the 16S rRNA gene (Cyanobacteria [28], Halobacteriaceae [29], Thermotogales [30], and Anaplasmataceae [31]) have smaller average conservation than the SAR11 core genome. The most comparable values are those for the Anaplasmataceae, composed of obligate intracellular symbionts with an even smaller average genome size than SAR11, and the thermophilic/hyperthermophilic Thermotogales group. However, these two groups are less divergent than SAR11 in the 16S rRNA gene (16%).
FIG 4 .
Comparison of the minimal 16S rRNA gene similarity, core genome conservation, and average genome size for relevant groups of the Bacteria and Archaea. Averages (circles) within the range (lines) of genes in the core genome as percentages of total genes or total protein coding genes as specified in the original publication are shown. Circles without lines had insufficient information to calculate a range. 16S rRNA gene similarities were calculated with the megablast using default settings. The color code indicates average genome sizes. The dotted curve represents approximate average values taken from Fig. 1a in reference 25. The number of genomes compared per study and the average number of core genes can be found at http://giovannonilab.science.oregonstate.edu/publications. Anaplas., Anaplasmataceae (31); Chlamy., Chlamydiaceae; Chlamy.1, Chlamydophila psittaci, Chlamydia abortus, Chlamydia caviae, and Chlamydophila felis; Chlamy.2, C. psittaci, C. abortus, Chlamydophila pneumoniae, and Chlamydia trachomatis (35); Cyanob., cyanobacteria (28); E. rum., Ehrlichia ruminantium (36); Halob., Halobacteriaceae (29); Mycopl., Mycoplasma (89); Nitrob., Nitrobacter (90); Prochl., Prochlorococcus (32); Rhodops., Rhodopseudomonas (33); Rickett., Rickettsia (34); Roseob., Roseobacter clade (47); Shew., Shewanella (27); S. agal., Streptococcus agalactiae (23); S. islandicus, Sulfolobus islandicus (37); Thermot., Thermotogales (30).
FIG 5 .
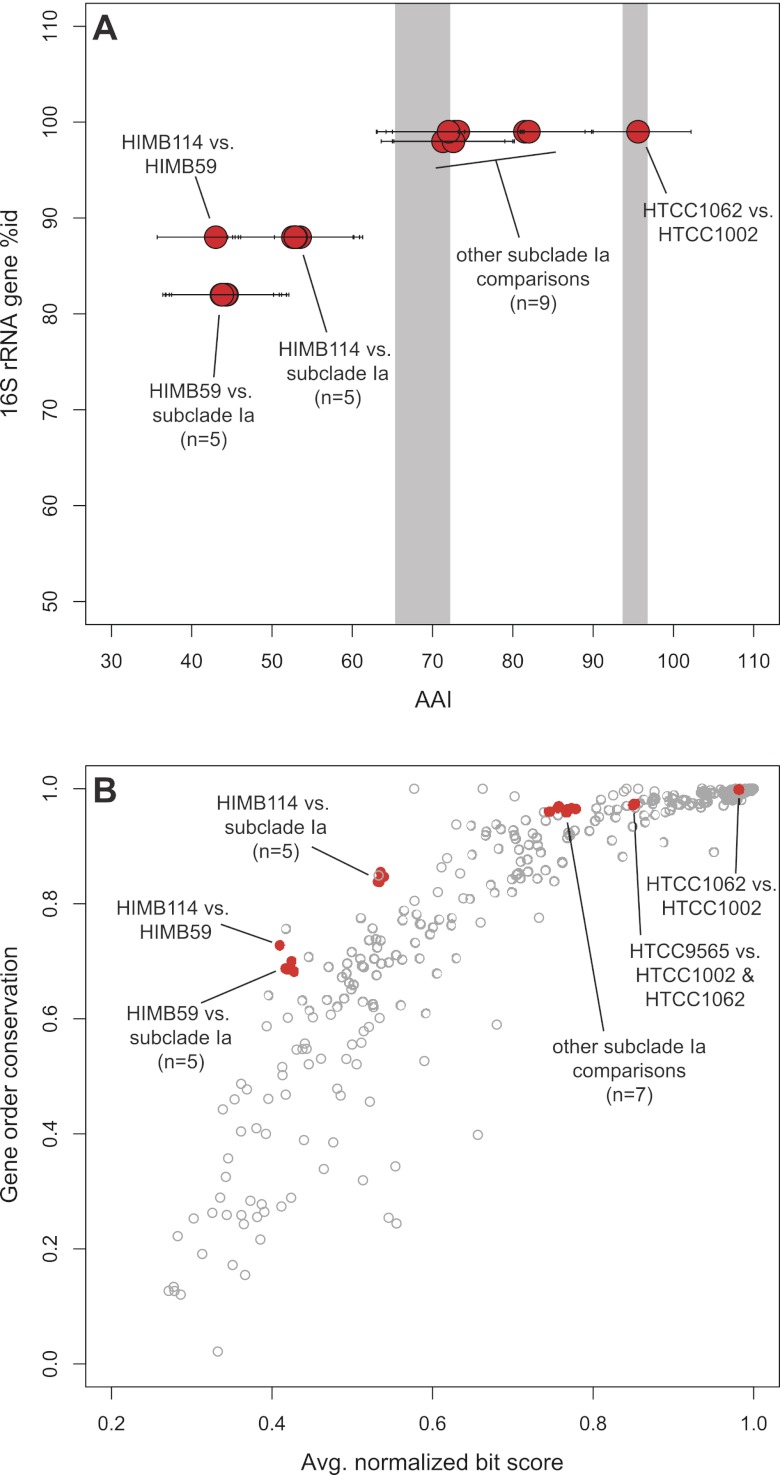
(A) 16S rRNA gene identity versus average amino acid identity (AAI). AAI for each pairwise comparison is plotted for all shared genes. Error bars are standard errors; “n” is the number of pairwise comparisons in a group of points. Shaded regions are an approximation of data from the work of Konstantinidis and Tiedje, 2007 (25), delineating proposed (left to right) genus and species boundaries based on AAI versus 16S rRNA identity. (B) Gene order conservation versus average normalized bit score of protein-coding genes. The data are from Fig. 2 of the work of Yelton et al. (38), with our new analyses of the SAR11 genomes overlaid in red. Gene order conservation is defined as the fraction of genes shared by any two organisms that are syntenic (39); “n” is the same as in panel A.
The core genomes of free-living microorganisms with a degree of 16S rRNA gene divergence similar to that of SAR11 subclade Ia (2%), such as Prochlorococcus (3%) (32) or Rhodopseudomonas (3%) (33), are considerably less conserved (Fig. 4). The core genome of 10 obligately intracellular Rickettsia strains, which are phylogenetically closely related to the SAR11 lineage and possess similarly small genome sizes (~1.2 Mbp), is also much less conserved than the SAR11 subclade Ia core genome (34). The only groups with more average core genome conservation were either less divergent at the 16S rRNA gene, obligate intracellular organisms, or both (23, 35, 36). Although the AAI values for SAR11 subclade Ia are appropriate for a genus (25) (see Table S3 at http://giovannonilab.science.oregonstate.edu/publications), core genome conservation within subclade Ia is more similar to that of single bacterial species (e.g., Sulfolobus islandicus and Streptococcus agalactiae [23, 37]).
Synteny.
The conservation of gene order (synteny) within genomes can be a strong indicator of conserved gene function and relatedness. Previous studies have demonstrated that synteny decreases with phylogenetic distance, although this relationship varies depending on the group examined (38–40). A comparison of gene order conservation versus genome sequence similarity (average bit score of protein-coding orthologs) demonstrated that the SAR11 strains are on the extreme maximum edge of the range described by Yelton et al. (38), indicating much higher gene order conservation than most other organisms (Fig. 5B), consistent with predictions (12).
Genome organization.
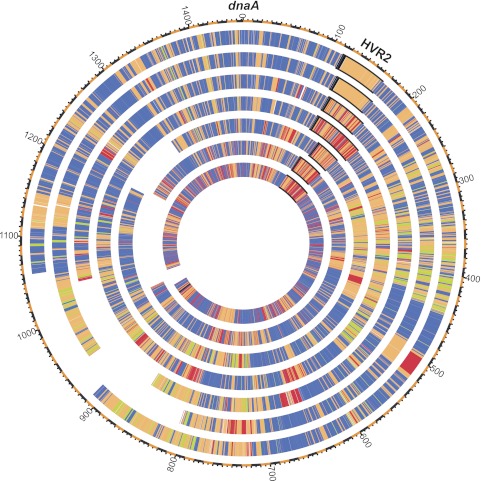
To visualize global genome organization of core, additional subclade Ia core, shared non-core, and unique genes, we ordered the seven SAR11 genomes by colocalizing them at dnaA, adjacent to the origin in HTCC1062 (5), moving clockwise toward dnaN (Fig. 6). Consistent with our calculations showing high conservation of synteny, core (and additional subclade Ia core) genes are grouped in blocks throughout each genome (blue and green areas of Fig. 6). Shared non-core and unique genes are scattered throughout the genomes, though some areas of dense groupings are evident (Fig. 6; see also Fig. S1 in the supplemental material).
FIG 6 .
Circular representation of SAR11 genomes. The genomes are arranged in order from the outermost to the innermost as follows: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, and HIMB59. Organisms are aligned with 0 at dnaA, sequences going clockwise to dnaN and continuing in the order in which they are presented at IMG. Blue, core SAR11 genes; bright green, additional SAR11 subclade Ia core genes; orange, shared non-core genes, red, unique genes; black, rRNA genes. The outer scale is measured in units of 10-kbp increments. HVR2 is highlighted in black. Gaps in complete genomes were necessary to display the genomes in this manner due to the disparity of genome sizes.
Previous work revealed HVRs—islands of low genomic recruitment of metagenomic data sets—in SAR11 (12, 14, 41). HVR2 from the work of Wilhelm et al. (12) is conserved in all seven SAR11 genomes, bounded by the 16S rRNA, tRNAIle-GAT, tRNAAla-TGC, 23S rRNA cassette on one side and 5S rRNA on the other in all genomes except HIMB59, which has HVR2 bounded by tRNASer-GGA and tRNAAla-GGC genes. In HIMB59, the rRNA genes are in the same order but include the 5S rRNA as part of the operon (16S rRNA, tRNAIle-GAT, tRNAAla-TGC, 23S rRNA, and 5S rRNA) and are on the other side of the genome from HVR2. Nevertheless, in all strains, the HVR2 region remains similar in both size and location to the dnaAN locus (Fig. 6; see also Table S4 at http://giovannonilab.science.oregonstate.edu/publications) and comprises ~50 protein coding genes except in HTCC7211 and HIMB59, where it comprises 83 and 74 genes, respectively. Consistent with initial observations (12), genes commonly found in HVR2 include glycosyltransferases, unknown membrane proteins, hypothetical proteins, and methyltransferases (see Table S1 at the above URL). Probably because HTCC1062 and HTCC1002 are the most closely related strains (AAI, ~96%; ANI, 98%) (Fig. 5; see also Tables S3 and S5 at the above URL), they share all genes in HVR2. However, the remaining isolates contain large numbers of unique genes in this region (Fig. 6; see also Fig. S1 in the supplemental material), including some that appear to confer strain-specific metabolic abilities, such as sulfur metabolism genes unique to HTCC9565 and sugar transporters and phosphofructokinase in HIMB59 that may be indicative of a unique niche for this strain (discussed further below). HVR2 also contains a concentration of paralogs for most strains (see Fig. S2). In contrast to the conserved location of HVR2, HVR1, -3, and -4, identified by Wilhelm et al. (12), are not conserved in other SAR11 strains outside of HTCC1062 and HTCC1002, although each of the other subclade Ia genomes (HTCC9565, HTCC7211, and HIMB5) possesses distinct HVR-like regions where unique genes are clustered (Fig. 6; see also Fig. S1).
Paralogs.
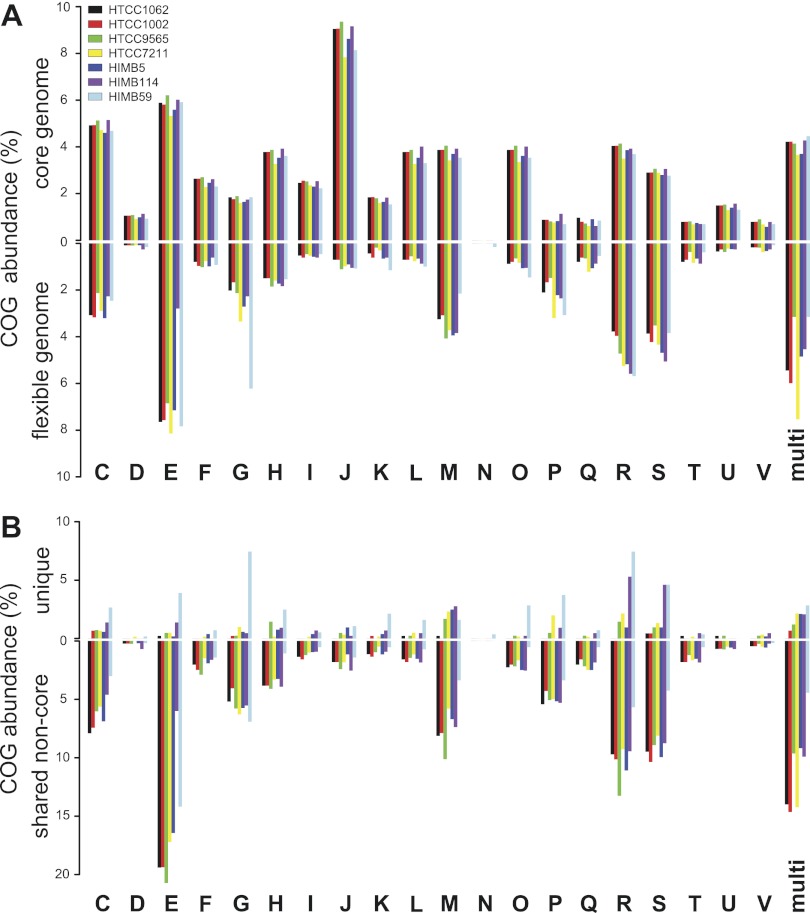
One conspicuous feature previously identified within the streamlined genome of HTCC1062 was a low incidence of paralogs (5). Consistent with this finding and the general trend of decreasing numbers of paralogs with smaller genome size in Bacteria (42), the SAR11 genomes range from 4.3 to 15.2% of protein-coding genes as paralogs, averaging 7.8% (see Table S6 at http://giovannonilab.science.oregonstate.edu/publications). The proportion of strain-specific paralogs ranges from 18 to 61% of the total paralogous genes per genome, with similar distributions across genes in different categories of the pan-genome (Fig. 7A). Inparalogs and outparalogs are defined as duplications after or before a given speciation event, respectively (43). In this study, we characterized in- versus outparalogs with phylogenetic trees to determine the number of gene duplications that occurred relative to the divergence of the SAR11 clade. Of >80% of paralogs with a reliable phylogenetic assignment, <19% are classified as outparalogs (see Table S1 at the above URL). Thus, the majority of gene duplications are predicted to have occurred since the divergence of the SAR11 lineages from a last common ancestor. Paralogs are concentrated in a few COG (Clusters of Orthologous Groups) functional categories: energy production and conversion (C), amino acid transport and metabolism (E), carbohydrate transport and metabolism (G), cell wall/membrane/envelope biogenesis (M), and those with mixed designations (Fig. 7B; see also Fig. S3 in the supplemental material). Amino acid transport and metabolism (E) accounts for the largest number of paralogs in the seven SAR11 genomes, of which 38% are found only in HIMB59 (see Table S1 at the above URL).
FIG 7 .
Paralogs in SAR11. (A) The distribution of paralogs as a function of total protein-coding genes. Blue, core genes; green, additional SAR11 subclade 1a core genes; orange, shared non-core genes; red, unique genes; grey, single-copy genes. (B) Distribution of paralogs by strain according to COG category.
Conserved gene content of the Pelagibacteraceae.
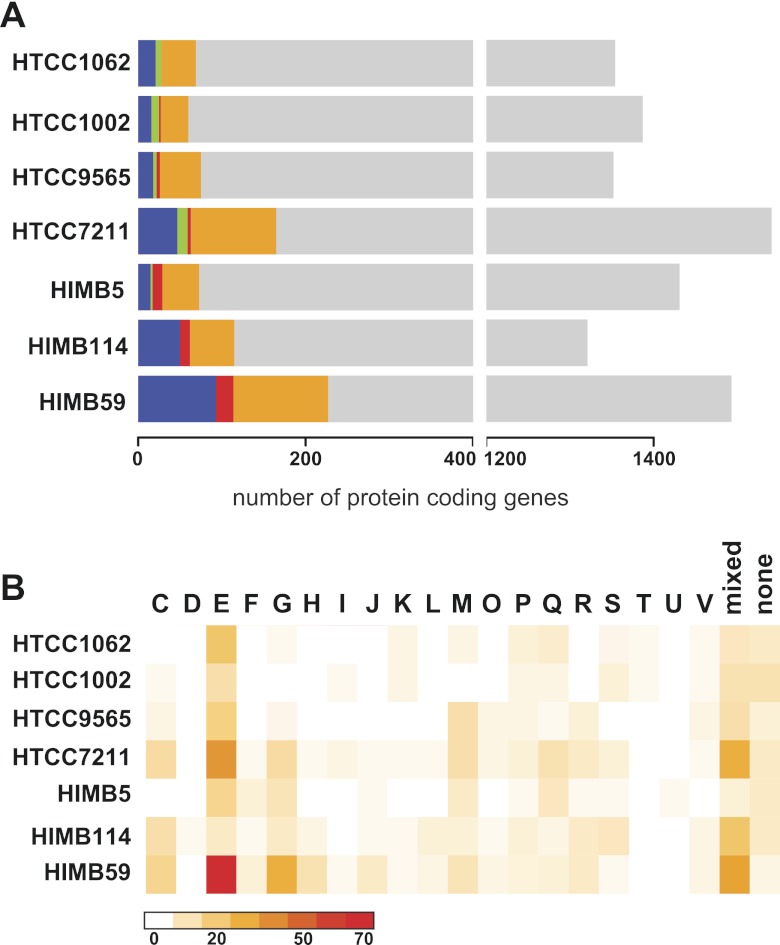
Similar to the core genomes of Prochlorococcus, the Roseobacter clade, and Shewanella, the SAR11 core genome possesses a high proportion of genes coding for proteins involved in housekeeping functions and central metabolism, with a small fraction (2.1 to 3.7%) of core SAR11 genes not assigned to COG functional categories (Fig. 8). The uncategorized fraction in the flexible genome is much higher (23.5 to 34.2%) due to a larger proportion of putative and hypothetical genes. Compared to the core genome, the SAR11 flexible genome includes an overrepresentation of genes assigned to the COG categories amino acid transport and metabolism (E), carbohydrate transport and metabolism (G), inorganic ion transport and metabolism (P), and general (R) and unknown (S) functions (Fig. 8A).
FIG 8 .
Relative abundance and distribution of selected COG categories within SAR11 core and flexible genomes (A) or SAR11 shared non-core and unique genes (B).
Generally, SAR11 cells are predicted to share a typical electron transport chain and a complete tricarboxylic acid cycle. In addition, all of the SAR11 genomes encode putative genes for the biosynthesis of most of the 20 standard amino acids and some but not all vitamins and cofactors that are predicted to be required (see supplemental text at http://giovannonilab.science.oregonstate.edu/publications). All strains lack a phosphoenolpyruvate:sugar phosphotransferase transport system (PTS) but have a complete non-oxidative portion and an incomplete oxidative portion of the pentose phosphate shunt.
All SAR11 genomes encode proteorhodopsin (PR), which is found in two groups that are specific for different light wavelengths: green and blue absorbing (GPR and BPR, respectively) (44, 45). Whereas GPRs have been found to be highly abundant in the North Atlantic and surface waters of the Mediterranean Sea, BPRs dominate the open ocean, such as in the Sargasso Sea (14, 46). The two SAR11 strains isolated from open ocean sites, HTCC7211 (Sargasso Sea) and HTCC9565 (northeastern Pacific), contain BPR, while the remaining coastal strains encode GPRs. Strain HIMB114 encodes an additional divergent PR gene with a currently unknown function and absorption spectrum. Putative genes for the biosynthesis of retinal from β-carotene, crtIBY and blh (15), are present in all seven SAR11 genomes. Thus, the conservation of PR and associated genes in all of the genomes examined in this study demonstrates an important adaptive role for this gene across the SAR11 clade, where it potentially facilitates survival by providing ATP during periods of carbon limitation, as demonstrated for HTCC1062 (16).
In SAR11, a high proportion (13 to 16%) of all protein-coding genes encode transport proteins, in comparison to all open reading frames (ORFs) in the Bacteria (~9%) and the Roseobacter clade (6%) (47, 48). Within this fraction, the most abundant form of transporters are primary active transporters, including ABC (ATP-binding cassette) and electrochemical potential-driven transporters (see Table S7 at http://giovannonilab.science.oregonstate.edu/publications). The core set of transporters found in all SAR11 genomes includes ABC transport systems for general l-amino acids (encoded by yhdWXYZ), iron(III) (sfuABC), lipoprotein release (ycfUV), multidrug/antibiotic (yadGH), and heme export (ccmD). Electrochemical potential-driven transporters for potassium (trkA) and mannitol/chloroaromatic compounds (tripartite ATP-independent periplasmic [TRAP] type), a channel transport system for ammonium (amtB), and several incompletely characterized transport systems are also present in all seven SAR11 genomes.
Strain variation within the pan-genome of the Pelagibacteraceae.
In spite of the highly conserved gene content in SAR11, we observed variability in some notable genes and pathways that have been considered enigmatic for the type strain, HTCC1062, and which may serve as lineage-specific adaptations. Glucose oxidation by a proposed variant of the Entner-Doudoroff (ED) pathway (49) is predicted only for HIMB5, HTCC1002, and HTCC1062. HIMB59 is the only genome predicted to have a complete Embden-Meyerhof-Parnas (EMP) glycolysis pathway and genes for metabolism of other sugars (see supplemental text at http://giovannonilab.science.oregonstate.edu/publications). Furthermore, an expansion of transporter paralogs in COG category G indicates that this microorganism may be adapted to use a variety of sugar compounds. Recent work has demonstrated that subclade Va organisms bloom at the surface in the Sargasso Sea during the same time periods as subclade Ia organisms there (Vergin et al., submitted), and thus carbohydrate utilization may allow HIMB59-type strains to co-occur with the numerically dominant SAR11 subclade Ia. However, HIMB59 does not have genes for the glyoxylate bypass, which is a conserved feature of subclade Ia genomes. SAR11 genes for metabolism of one-carbon and methylated compounds (17) are conserved in subclade Ia organisms, although they have variable distribution in the other two strains (see Fig. S4 in the supplemental material; see also supplemental text at http://giovannonilab.science.oregonstate.edu/publications). HIMB5 and HIMB114 contain putative copies of aerobic carbon monoxide dehydrogenase (CODH) genes (coxSLM) but, as with other SAR11 strains, no genes for carbon fixation. HIMB114 contains a complete serACB operon, which implies that this strain may be able to synthesize glycine de novo, in contrast with HTCC1062 (19) and other SAR11 subclade Ia organisms (see supplemental text at the above URL). Sulfur and phosphate metabolism are also not conserved among SAR11 strains. Genes for dimethylsulfoniopropionate (DMSP) transport and demethylation are missing from HIMB114, whereas HTCC9565 is the only strain that contains a predicted copy of sulfate adenylyltransferase (encoded by sat), which catalyzes the first step of sulfate reduction. Five of the seven strains contain the predicted high-affinity phosphate operon (pstSCAB-phoBU), but only the HTCC7211 genome encodes genes for production and use of polyphosphate (ppx and ppk) as well as phosphonate transport (phnCDEE2) and degradation (phnGHIJKLMN and phnZX). The Pacific isolates HIMB114 and HIMB59 also possess genes for phosphonate transport, but phosphonate mineralization is likely to be restricted to specific compounds (see supplemental text at the above URL).
DISCUSSION
The observation of small genomes in SAR11 strains of the Ia subclade provided strong support for streamlining theory and showed that streamlined heterotrophs could be highly successful interacting with complex organic carbon originating from phytoplankton communities (5, 12). New results reported here extend this observation by showing that small genomes, high synteny, and conservation of the core genome are consistent qualities of isolates spanning the SAR11 clade. It was previously hypothesized that paralogs of HTCC1062 were of ancient origin (5). However, our current phylogenetic designation of the majority of duplications as inparalogs indicates that most gene duplication has happened since the divergence of the SAR11 lineage. The overall paucity of paralogs in SAR11 genomes compared to those of other bacteria, especially those with free-living lifestyles (50), provides further support for the hypothesis that a streamlined genome was a feature of the last common ancestor of SAR11.
At ~600 genes, the core genome for SAR11 provides an estimate of the lower limit of genes essential for maintenance of the free-living state in marine environments. Genome reduction in Prochlorococcus often includes the loss of gene families that are environmentally important but not essential in all water column environments, for example, genes for the uptake of macronutrients, such as compounds of P and N (32, 51). Coleman et al. (13) noted that the pattern of gene gain and loss in Prochlorococcus for genes involved in macronutrient (P and N) acquisition is often not congruent with phylogeny, suggesting that these genes play a role in the evolution of ecotypes (52).
Similarly, we observed differential conservation of genes for iron and phosphorus metabolism in SAR11. As famously described in the “Iron Hypothesis,” in some ocean surface waters, Fe is so low that it limits primary production over broad ocean regions (53). Smith et al. (54) described complex regulatory adaptions to iron limitation in strain HTCC1062. Transcripts for most genes involved in iron metabolism increased in iron-limited cells, but only the iron-binding protein encoded by sfuC, a component of the predicted sfuABC transporter, increased in both mRNA and protein abundance during iron limitation. Two RNA-binding proteins (CspE and CspL), members of the cold-shock protein family, were postulated to play a role in a broad regulatory response that suppressed translation of nonessential transcripts. cspL, the ABC transporter genes (sfuABC), and the Fe–S synthesis operon (sufBCD) were all found in the SAR11 core genome, but the transcription factors encoded by fur and irr, which are reported to be involved in iron regulation in bacteria (55, 56), are conserved only in the SAR11 subclade Ia core genome. Iron is essential for respiration, and therefore the conservation of sfuABC and sufBCD is not surprising. The absence of iron-related regulatory genes from the SAR11 core genome suggests that iron metabolism is constitutive in some strains and that the iron regulatory system does not consistently yield benefits in fitness across the clade, which ensures that these genes will be maintained by selection.
Phosphorous is probably the most common cause of nutrient limitation in the oceans (57). It is biologically accessible in a variety of forms, most importantly as the phosphate ion but also as phosphonates, in which P and C atoms are linked directly by a bond. However, P availability relative to that of other nutrients differs across oceans. For example, the subtropical Atlantic Ocean is typically regarded as phosphate limited, whereas phosphate is thought to be more available in the central Pacific (e.g., see reference 58). Thus, in contrast to iron, phosphate limitation is probably a much less universal selective pressure, since no phosphate metabolism genes are conserved in all strains: HTCC1002 and HTCC9565 lack the high-affinity phosphate operon, and only HTCC7211contains genes for both polyphosphate and phosphonate transport and degradation (see supplemental text at http://giovannonilab.science.oregonstate.edu/publications). Recently, Coleman and Chisholm (58) found that the abundance of phosphate-related SAR11 gene content in metagenomic data sets was higher for the phosphate-depleted waters of the Atlantic than for the Pacific. Similarly, genes for the metabolism of polyphosphate have also been found to be more abundant in data sets collected from environments depleted in phosphate (59). Our data support these findings, since HTCC7211 is also the only isolate from the Atlantic Ocean, and demonstrate how the open SAR11 pan-genome provides for ecosystem-specific adaptations.
The values we report for core genome conservation and gene conservation as a function of 16S rRNA gene sequence similarity place the members of SAR11 as outliers among bacteria—the seven strains investigated shared ~50% of total gene content across 18% divergence in 16S rRNA gene sequence (~44% AAI), while the average for bacteria is ~20% shared genes at this level of divergence (25, 40). Early investigations of shared gene content by Konstantinidis and Tiedje (26) and Tamames (40) were extended later by Zaneveld et al. (60), who showed that conserved gene content tended to be higher among organisms from the same habitat. With the exception of the work of Zaneveld et al. (60), the studies referenced above did not examine the influence of common ancestry on core genome conservation. To address this issue, in Fig. 4 we present an analysis of core genome content for a selection of microbial clades. This analysis shows that for many monophyletic groups, shared gene content is much higher than the averages reported by Konstantinidis and Tiedje (25) and that some, notably the Anaplasmataceae and the Thermotogales, are close to SAR11.
Although unusual for comparative genomics studies, our findings are consistent with previous conclusions by analysis of gene-to-gene boundaries and the conservation of synteny in metagenomic data (12). By comparing the genomes of two closely related SAR11 strains with metagenome data, Wilhelm et al. (12) concluded that selection was variable across the SAR11 genome, leading to high apparent diversity in SAR11 populations by common metrics, while simultaneously maintaining a conservation of gene content and function. Comparatively high synteny across our genomes in spite of “typical” amino acid divergence with decreasing 16S rRNA identity agrees with these conclusions, since gene order conservation implies likely gene function conservation (38).
We also report the conservation of a hypervariable region (HVR2) across the clade. The presence of this variable genome region, bounded by structural RNA genes, is evidence that mechanistic restrictions to horizontal gene flow cannot be invoked to explain small genome size in SAR11 strains, providing further support for the conclusion that streamlining is a consequence of selective processes favoring genome minimization. Considering the enormous predicted population size of the SAR11 lineage globally (1), the open SAR11 pan-genome is apparently very large and is a significant genetic reservoir that can be exploited for adaptive purposes. High rates of recombination between similar SAR11 cells, calculated to be as much as 60× the mutation rate (61, 62), may provide one means of accessing such a reservoir. Genes found in HVR2 appear to augment function, analogously to genomic islands in genome-streamlined Prochlorococcus strains (13), in some cases restoring basic metabolic pathways that are not conserved across the clade. For example, the sat gene in HTCC9565, in combination with aprBA and/or APS kinase, may allow it to utilize sulfate as a sulfur source. The phosphofructokinase in HIMB59, conferring this organism with a complete glycolysis pathway, is also located in HVR2 (see Table S1 at http://giovannonilab.science.oregonstate.edu/publications).
The monophyly of the SAR11 clade has recently been called into question (63). However, in addition to the phylogenetic support for inclusion of HIMB59 as a member of the SAR11 clade both with concatenated protein and 16S rRNA gene trees (Fig. 1) (6, 64; Vergin et al., submitted), the conservation of gene content, synteny, and the HVR2 region across these strains provides additional evidence for the shared common ancestry of HIMB59 and other SAR11 strains. In fact, such unusually high conservation in these metrics across the clade raises the question of whether or not SAR11 genomes may be evolving at an unusual rate compared to those of other organisms. The depth of branching for SAR11 in the 16S rRNA gene tree is comparable to that of the nearby Rickettsiales clade (Fig. 1). AAI versus 16S rRNA gene identity follows predictions from previous observations (Fig. 5A) (25), indicating that the 16S rRNA gene is not evolving independently from the rest of the genome. Furthermore, while synteny in SAR11 genomes is higher than that in most other organisms, it is not unprecedented (Fig. 5B), falling near that in other organisms with small genomes. Thus, while unusual, all of these features are consistent with genome streamlining, which is expected to minimize genomes to a highly constrained set of genes that offer maximum fitness. Since these organisms form a monophyletic group with a depth of branching comparable to that of the Rickettsiales and have minimum AAI and 16S rRNA gene identities of ~44% and 82%, respectively, we therefore propose that the Pelagibacteraceae be expanded to a novel order, the “Pelagibacterales.” Based on the same metrics, subclade Ia organisms should be considered part of the genus “Candidatus Pelagibacter.”
Small genome size with a bias towards low GC content has also been observed in host-associated bacteria (65–67), as well as other free-living marine bacterioplankton, such as Prochlorococcus (3, 68) and OM43 (69, 70). Whereas genetic drift coupled with relaxed selection has been proposed as the driving force behind genome reduction in host-dependent bacteria, selection for a more economical lifestyle is the purported pressure for genome reduction in large populations of cells, such as those seen with Prochlorococcus and SAR11 (5, 71, 72). In principle, small cells with small genomes require fewer resources, such as carbon, nitrogen, and phosphorus, to divide and also compete more efficiently for nutrients because of their higher surface area-to-volume ratio (68, 72,73).
The paradox of genome streamlining is that small genomes are found in some planktonic marine bacteria but not others. Many common marine microbial lineages, such as the Roseobacter clade, Vibrio species, Photobacterium species, Pseudalteromas species, and Alteromonas species, have genomes of average size (47, 74–76). Plausible explanations for this paradox include differences in life cycle strategy and differences in Neµ, the product of effective population size and mutation rate (77). Commonly, concepts such as generalist versus specialist (78), r strategist versus k strategist (79), and oligotroph versus copiotroph (76) are used to explain variation in life cycle strategy, with large genomes often attributed to “generalists” (78). However, individual bacterial life cycle strategies may be complicated and elude accurate description with these concepts; for example, many Vibrio spp. alternate between specific host associations and living freely suspended in the water column (80). Moreover, small genomes suggest specialization, but the members of SAR11, which have small genomes, cannot plausibly be characterized as specialists, being one of the most successful and widely distributed chemoheterotrophic groups in the ocean (1).
Novel cultivation approaches that favor oligotrophs, such as those we pioneered, are responsible for some of the most dramatic examples of genome reduction in free-living cells (5, 20, 69). Following up on these observations, we reported unusual nutrient requirements in these strains and linked these requirements to genome reduction (16–19). We hypothesize that genome streamlining may explain why many microorganisms that are abundant in nature are difficult to cultivate. This is a testable hypothesis. It predicts that those data emerging from single-cell genomics will show that small genomes are prevalent among uncultured taxa and that unusual nutrient requirements stemming from genome reduction explain the difficulty of their cultivation.
MATERIALS AND METHODS
Isolation of SAR11 strains.
Strains HTCC1062 and HTCC1002 were isolated from the coastal Pacific Ocean, Newport, OR (4), strain 9565 was isolated from water collected above the Juan De Fuca ridge, strain HTCC7211 was isolated from the Bermuda Atlantic Time Series study site located in the Sargasso Sea (21), and strains HIMB114, HIMB5, and HIMB59 were isolated from tropical Kaneohe Bay, located on the northeastern shore of the island of O’ahu, HI (see Table S8 at http://giovannonilab.science.oregonstate.edu/publications). All strains were isolated using dilution-to-extinction methods (4, 20, 81). Following isolation, strains were grown in 100 liters of pristine seawater medium amended with low concentrations of inorganic nitrogen and phosphorus (1.0 µM NH4Cl, 1.0 µM NaNO3, and 0.1 µM KH2PO4) or nitrogen, phosphorus, organic carbon, and iron (18). Cells were collected on 0.1-µm-pore-size polyethersulfone membrane filters, and genomic DNA was isolated using a standard phenol-chloroform-isoamyl alcohol extraction protocol.
Sequencing and annotation.
Sequencing of the complete genomes of strains HTCC1062, HTCC1002, and HTCC7211 has been described previously (5, 49). The genomes of strains HIMB5, HIMB59, and HIMB114 were sequenced by the J. Craig Venter Institute as part of the Moore Foundation Microbial Genome Sequencing project (http://camera.calit2.net/microgenome/). Strain HTCC9565 was sequenced by the JGI as part of the Community Sequencing Program. MIGS environmental metadata and sequencing details can be found elsewhere (see Table S8 at http://giovannonilab.science.oregonstate.edu/publications). Functional annotation was performed with the Integrated Microbial Genomes Expert Review (IMG-ER) pipeline (82), except for the previously annotated strains HTCC1062 and HTCC1002 (see references 5 and 12, respectively), for which annotations were maintained (for details, see supplemental Methods at http://giovannonilab.science.oregonstate.edu/publications).
Genome comparisons.
We assessed homologous genes through the use of Hal, an automated pipeline for phylogenomic analysis (83). Hal initially performs an all-versus-all BLASTp analysis with all genome protein fasta files, followed by Markov clustering (MCL) at 13 inflation parameters (I). We chose to use clusters generated at I 1.5 because this is the default setting for OrthoMCL and was shown to have the highest accuracy for detecting orthologs with that software program (84). From there, we curated the clusters for accuracy with several filtering steps that flagged potential erroneous assignments of orthologs. Details of the filtering steps are provided elsewhere (see supplemental Methods at http://giovannonilab.science.oregonstate.edu/publications), along with determination of paralogs. In- and outparalogs were assessed according to the method of Sonnhammer and Koonin (43) with phylogenetic trees (see supplemental Methods). Synteny was determined using scripts and methods from Yelton et al. (38).
Core genome and pan-genome analyses.
To calculate the core and pan-genomes, as well as the unique genes per strain, we made use of the heat map table created by Hal (see Table S1 at http://giovannonilab.science.oregonstate.edu/publications). By curating this table with the alterations from the filters above, we were then able to utilize a custom program, query, written for parsing data output from Hal (83), to calculate the shared/unique gene content for any combination of strains. Gene-gene boundary calculations were completed with the data in Table S1 (see the above URL) using a custom Perl script, available on request.
The sequential inclusion of seven genomes allows 7!(N!(7 − N)!) possible combinations to calculate the core genome, new orthologs per genome, and the pan-genome. The regression analysis of the SAR11 core genome, new orthologs, and pan-genome was performed as described by Tettelin et al. (23, 24). For details, see supplemental Methods at http://giovannonilab.science.oregonstate.edu/publications.
16S phylogeny.
16S rRNA gene sequences from SAR11 organisms with sequenced genomes and clone libraries were aligned with near neighbors identified by previous phylogenetic and phylogenomic tests of the Alphaproteobacteria (6, 85, 86). Sequences were aligned with the software program NAST (87) and lane masked at greengenes (http://greengenes.lbl.gov/), and the phylogeny was determined using the RAxML software program (88) (-f a –m GTRGAMMA –N 500). Accession numbers are provided in the supplemental Methods at http://giovannonilab.science.oregonstate.edu/publications.
SUPPLEMENTAL MATERIAL
Gene content in SAR11 genomes as a function of distance from dnaA. Gene categories were plotted for 10-kbp blocks for each genome. y axes, numbers of genes; x axes, distances from dnaA in bp. Blue, core; green, additional subclade 1a core; orange, shared non-core; red, unique; black, rRNA. *, since HTCC9565 was not a single contig, the histogram assumes a length of 1.46 Mbp to match data in Fig. 5. Download Figure S1, PDF file, 0.3 MB.
Circos plot showing paralogs according to genomes. The genomes are arranged in order from the outermost to the innermost as follows: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, and HIMB59. Organisms are aligned with 0 at dnaA, sequences going clockwise to dnaN and continuing in the order in which they are presented at IMG. Blue, core SAR11 paralogs; bright green, additional subclade Ia core paralogs; orange, shared non-core paralogs; red, unique paralogs; black, rRNA genes; grey, single-copy genes. The outer scale is measured in units of 10-kbp increments. HVR2 is highlighted in black. Download Figure S2, PDF file, 1.3 MB.
Box plot of paralogs by COG category. Boxes show 25th to 75th interquartile range, with the median indicated by a dark line; whiskers indicate adjacent values to quartile ranges, and circles indicate outside values. Download Figure S3, PDF file, 0.1 MB.
Distribution of genes for C1 and methylated compound oxidation by strain. Figure modified with data from Fig. 2 in the work of Sun et al. (17). Download Figure S4, PDF file, 0.2 MB.
ACKNOWLEDGMENTS
This work was supported by a grant from the Marine Microbiology Initiative of the Gordon and Betty Moore Foundation (to S.J.G.), NSF Microbial Observatory grant no. MCB-0237713 (to S.J.G.), funding from the Center for Microbial Oceanography: Research and Education (NSF Science and Technology Center award EF-0424599) (to M.S.R.), and a Community Sequencing Project grant to S.J.G. and M.S.R. (CSP2009.797268) and is based on work supported by the NSF under award no. DBI-1003269 (to J.C.T.).
We thank Alex Boyd for his help in parsing Hal data, Christopher M. Sullivan at the Oregon State University Center for Genome Research and Biocomputing for help with the computational infrastructure, Alexis P. Yelton and Brian C. Thomas for assistance with synteny scripts, and Amy Chen for technical assistance associated with IMG-ER (Integrated Microbial Genomes Expert Review). We would also like to thank Jonathan A. Eisen and Matthew B. Sullivan for critical and helpful review of the manuscript.
J.G., J.C.T., S.J.G., and M.S.R. designed research; J.G., J.C.T., M.J.H., Z.C.L., and P.C. performed research; J.G., J.C.T., Z.C.L., S.J.G., and M.S.R. analyzed data; and J.G., J.C.T., M.J.H., S.J.G., and M.S.R. wrote the article.
Footnotes
Citation Grote J, et al. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3(5):e00252-12. doi:10.1128/mBio.00252-12.
REFERENCES
- 1. Morris RM, et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810 [DOI] [PubMed] [Google Scholar]
- 2. Schattenhofer M, et al. 2009. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ. Microbiol. 11:2078–2093 [DOI] [PubMed] [Google Scholar]
- 3. Rocap G, et al. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047 [DOI] [PubMed] [Google Scholar]
- 4. Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 5. Giovannoni SJ, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
- 6. Thrash JC, et al. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1:13 http://dx.doi.org/doi:10.1038/srep00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morris RM, et al. 2005. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol. Oceanogr. 50:1687–1696 [Google Scholar]
- 8. Carlson CA, et al. 2009. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 3:283–295 [DOI] [PubMed] [Google Scholar]
- 9. Treusch AH, et al. 2009. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 3:1148–1163 [DOI] [PubMed] [Google Scholar]
- 10. Fuhrman JA, et al. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. U. S. A. 103:13104–13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steele JA, et al. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5:1414–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ. 2007. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol. Direct 2:27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coleman ML, et al. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311:1768–1770 [DOI] [PubMed] [Google Scholar]
- 14. Rusch DB, et al. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 http://dx.doi.org/10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giovannoni SJ, et al. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82–85 [DOI] [PubMed] [Google Scholar]
- 16. Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. 2011. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS ONE 6:e19725 http://dx.doi.org/10.1371/journal.pone.0019725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun J, et al. 2011. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE 6:e23973 http://dx.doi.org/10.1371/journal.pone.0023973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tripp HJ, et al. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452:741–744 [DOI] [PubMed] [Google Scholar]
- 19. Tripp HJ, et al. 2009. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ. Microbiol. 11:230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Connon SA, Giovannoni SJ. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stingl U, Tripp HJ, Giovannoni SJ. 2007. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 1:361–371 [DOI] [PubMed] [Google Scholar]
- 22. Oh H-M, et al. 2011. Complete genome sequence of strain IMCC9063, belonging to SAR11 subgroup 3, isolated from the Arctic Ocean. J. Bacteriol. 193:3379–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11:472–477 [DOI] [PubMed] [Google Scholar]
- 25. Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10:504–509 [DOI] [PubMed] [Google Scholar]
- 26. Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187:6258–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konstantinidis KT, et al. 2009. Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc. Natl. Acad. Sci. U. S. A. 106:15909–15914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi T, Falkowski PG. 2008. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc. Natl. Acad. Sci. U. S. A. 105:2510–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson I, et al. 2011. Novel insights into the diversity of catabolic metabolism from ten Haloarchaeal genomes. PLoS ONE 6:e20237 http://dx.doi.org/10.1371/journal.pone.0020237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhaxybayeva O, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U. S. A. 106:5865–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin M, Zhang C, Gibson K, Rikihisa Y. 2009. Analysis of complete genome sequence of Neorickettsia risticii: causative agent of Potomac horse fever. Nucleic Acids Res. 37:6076–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kettler GC, et al. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3:e231 http://dx.doi.org/10.1371/journal.pgen.0030231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oda Y, et al. 2008. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc. Natl. Acad. Sci. U. S. A. 105:18543–18548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillespie JJ, et al. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE 3:e2018 http://dx.doi.org/10.1371/journal.pone.0002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voigt A, Schöfl G, Saluz HP. 2012. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS ONE 7:e35097 http://dx.doi.org/10.1371/journal.pone.0035097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frutos R, et al. 2006. Comparative genomic analysis of three strains of Ehrlichia ruminantium reveals an active process of genome size plasticity. J. Bacteriol. 188:2533–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106:8605–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yelton AP, et al. 2011. A semi-quantitative, synteny-based method to improve functional predictions for hypothetical and poorly annotated bacterial and archaeal genes. PLoS Comput. Biol. 7:e1002230 http://dx.doi.org/10.1371/journal.pcbi.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rocha EP. 2006. Inference and analysis of the relative stability of bacterial chromosomes. Mol. Biol. Evol. 23:513–522 [DOI] [PubMed] [Google Scholar]
- 40. Tamames J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2:research0020–research0020 11 http://dx.doi.org/doi:10.1186/gb-2001-2-6-research0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodriguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828–836 [DOI] [PubMed] [Google Scholar]
- 42. Pushker R, Mira A, Rodríguez-Valera F. 2004. Comparative genomics of gene-family size in closely related bacteria. Genome Biol. 5:R27 http://dx.doi.org/10.1186/gb-2004-5-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonnhammer EL, Koonin EV. 2002. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18:619–620 [DOI] [PubMed] [Google Scholar]
- 44. Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786–789 [DOI] [PubMed] [Google Scholar]
- 45. Man D, et al. 2003. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22:1725–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sabehi G, et al. 2007. Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 1:48–55 [DOI] [PubMed] [Google Scholar]
- 47. Moran MA, et al. 2007. Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ren Q, Paulsen IT. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1:e27 http://dx.doi.org/10.1371/journal.pcbi.0010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. 2010. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ. Microbiol. 12:490–500 [DOI] [PubMed] [Google Scholar]
- 50. Gevers D, Vandepoele K, Simillon C, Van de Peer Y. 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12:148–154 [DOI] [PubMed] [Google Scholar]
- 51. Luo H, Friedman R, Tang J, Hughes AL. 2011. Genome reduction by deletion of paralogs in the marine cyanobacterium Prochlorococcus. Mol. Biol. Evol. 28:2751–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martiny AC, Coleman ML, Chisholm SW. 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U. S. A. 103:12552–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martin JH, et al. 1994. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature 371:123–129 [Google Scholar]
- 54. Smith DP, et al. 2010. Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus Pelagibacter ubique. PLoS ONE 5:e10487 http://dx.doi.org/10.1371/journal.pone.0010487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hamza I, Chauhan S, Hassett R, O’Brian MR. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669–21674 [DOI] [PubMed] [Google Scholar]
- 57. Paytan A, McLaughlin K. 2007. The oceanic phosphorus cycle. Chem. Rev. 107:563–576 [DOI] [PubMed] [Google Scholar]
- 58. Coleman ML, Chisholm SW. 2010. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc. Natl. Acad. Sci. U. S. A. 107:18634–18639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Temperton B, Gilbert JA, Quinn JP, McGrath JW. 2011. Novel analysis of oceanic surface water metagenomes suggests importance of polyphosphate metabolism in oligotrophic environments. PLoS ONE 6:e16499 http://dx.doi.org/10.1371/journal.pone.0016499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zaneveld JR, Lozupone C, Gordon JI, Knight R. 2010. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 38:3869–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vergin KL, et al. 2007. High intraspecific recombination rate in a native population of Candidatus Pelagibacter ubique (SAR11). Environ. Microbiol. 9:2430–2440 [DOI] [PubMed] [Google Scholar]
- 62. Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199–208 [DOI] [PubMed] [Google Scholar]
- 63. Rodríguez-Ezpeleta N, Embley TM. 2012. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7:e30520 http://dx.doi.org/10.1371/journal.pone.0030520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo H, Löytynoja A, Moran MA. 2012. Genome content of uncultivated marine Roseobacters in the surface ocean. Environ. Microbiol. 14:41–51 [DOI] [PubMed] [Google Scholar]
- 65. Fraser CM, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 66. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 67. Merhej V, Raoult D. 2011. Rickettsial evolution in the light of comparative genomics. Biol. Rev. Camb. Philos. Soc. 86:379–405 [DOI] [PubMed] [Google Scholar]
- 68. Dufresne A, et al. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U. S. A. 100:10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Giovannoni SJ, et al. 2008. The small genome of an abundant coastal ocean methylotroph. Environ. Microbiol. 10:1771–1782 [DOI] [PubMed] [Google Scholar]
- 70. Huggett MJ, Hayakawa DH, Rappé MS. 2012. Genome sequence of strain HIMB624, a cultured representative from the OM43 clade of marine Betaproteobacteria. Stand. Genomic Sci. 6:1 http://dx.doi.org/10.4056/sigs.2545743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596 [DOI] [PubMed] [Google Scholar]
- 72. Dufresne A, Garczarek L, Partensky F. 2005. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6:R14 http://dx.doi.org/10.1186/gb-2005-6-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Button DK, Robertson B. 2000. Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol. Oceanogr. 45:499–505 [Google Scholar]
- 74. Ivars-Martinez E, et al. 2008. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2:1194–1212 [DOI] [PubMed] [Google Scholar]
- 75. Grimes DJ, et al. 2009. What genomic sequence information has revealed about Vibrio ecology in the ocean—a review. Microb. Ecol. 58:447–460 [DOI] [PubMed] [Google Scholar]
- 76. Lauro FM, et al. 2009. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:15527–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302:1401–1404 [DOI] [PubMed] [Google Scholar]
- 78. Newton RJ, et al. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798 [DOI] [PubMed] [Google Scholar]
- 79. Andrews JH, Harris RF. 1986. r- and K-selection and microbial ecology. Adv. Microb. Ecol. 9:99–147 [Google Scholar]
- 80. Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A. 97:10231–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Markowitz VM, et al. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278 [DOI] [PubMed] [Google Scholar]
- 83. Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. 2011. Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr. 3:RRN1213 http://dx.doi.org/10.1371/currents.RRN1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Salichos L, Rokas A. 2011. Evaluating ortholog prediction algorithms in a yeast model clade. PLoS ONE 6:e18755 http://dx.doi.org/10.1371/journal.pone.0018755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee K-B, et al. 2005. The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55:1907–1919 [DOI] [PubMed] [Google Scholar]
- 86. Williams KP, Sobral BW, Dickerman AW. 2007. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 189:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. DeSantis TZ, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 89. Liu W, et al. 2012. Comparative genomics of Mycoplasma: analysis of conserved essential genes and diversity of the Pan-genome. PLoS ONE 7:e35698 http://dx.doi.org/10.1371/journal.pone.0035698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Starkenburg SR, et al. 2008. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl. Environ. Microbiol. 74:2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene content in SAR11 genomes as a function of distance from dnaA. Gene categories were plotted for 10-kbp blocks for each genome. y axes, numbers of genes; x axes, distances from dnaA in bp. Blue, core; green, additional subclade 1a core; orange, shared non-core; red, unique; black, rRNA. *, since HTCC9565 was not a single contig, the histogram assumes a length of 1.46 Mbp to match data in Fig. 5. Download Figure S1, PDF file, 0.3 MB.
Circos plot showing paralogs according to genomes. The genomes are arranged in order from the outermost to the innermost as follows: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, and HIMB59. Organisms are aligned with 0 at dnaA, sequences going clockwise to dnaN and continuing in the order in which they are presented at IMG. Blue, core SAR11 paralogs; bright green, additional subclade Ia core paralogs; orange, shared non-core paralogs; red, unique paralogs; black, rRNA genes; grey, single-copy genes. The outer scale is measured in units of 10-kbp increments. HVR2 is highlighted in black. Download Figure S2, PDF file, 1.3 MB.
Box plot of paralogs by COG category. Boxes show 25th to 75th interquartile range, with the median indicated by a dark line; whiskers indicate adjacent values to quartile ranges, and circles indicate outside values. Download Figure S3, PDF file, 0.1 MB.
Distribution of genes for C1 and methylated compound oxidation by strain. Figure modified with data from Fig. 2 in the work of Sun et al. (17). Download Figure S4, PDF file, 0.2 MB.