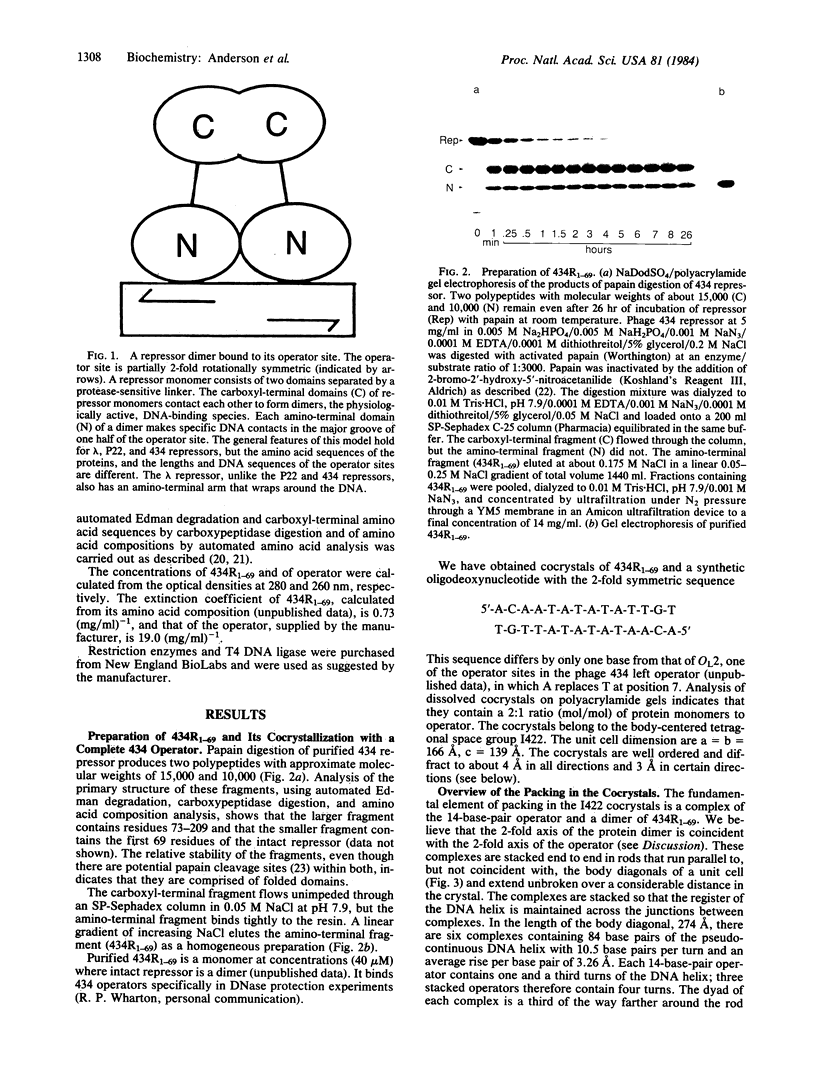
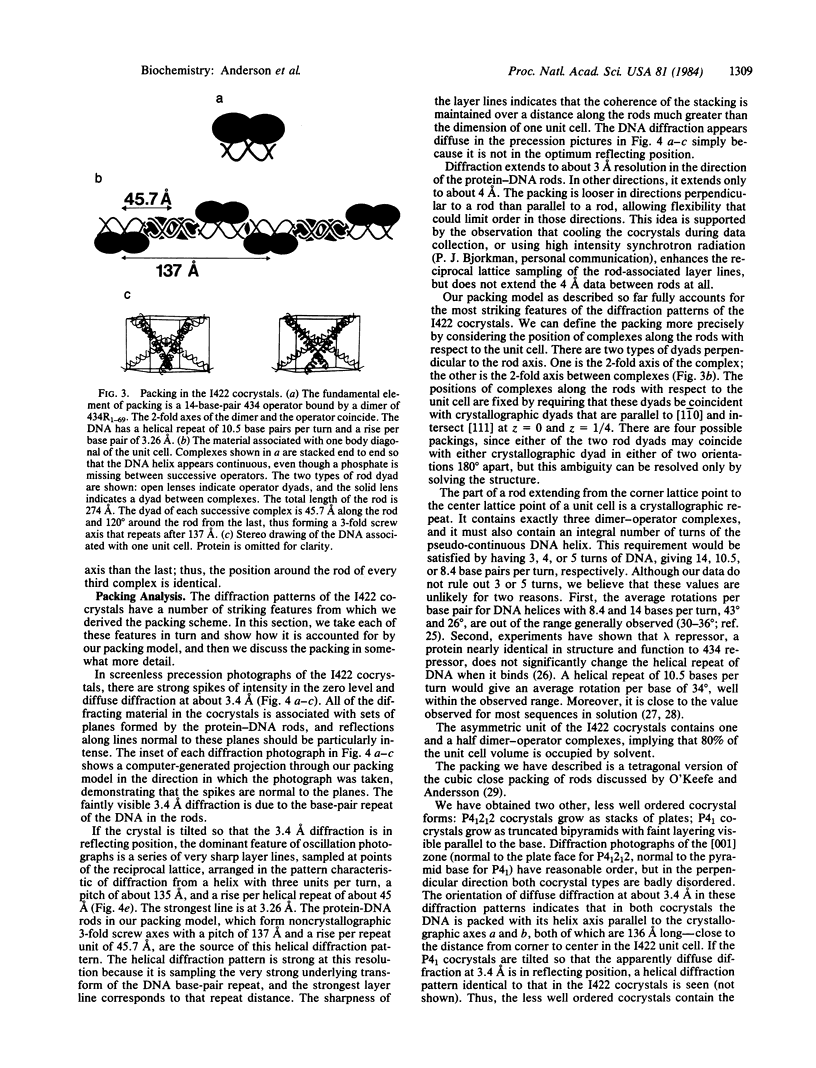
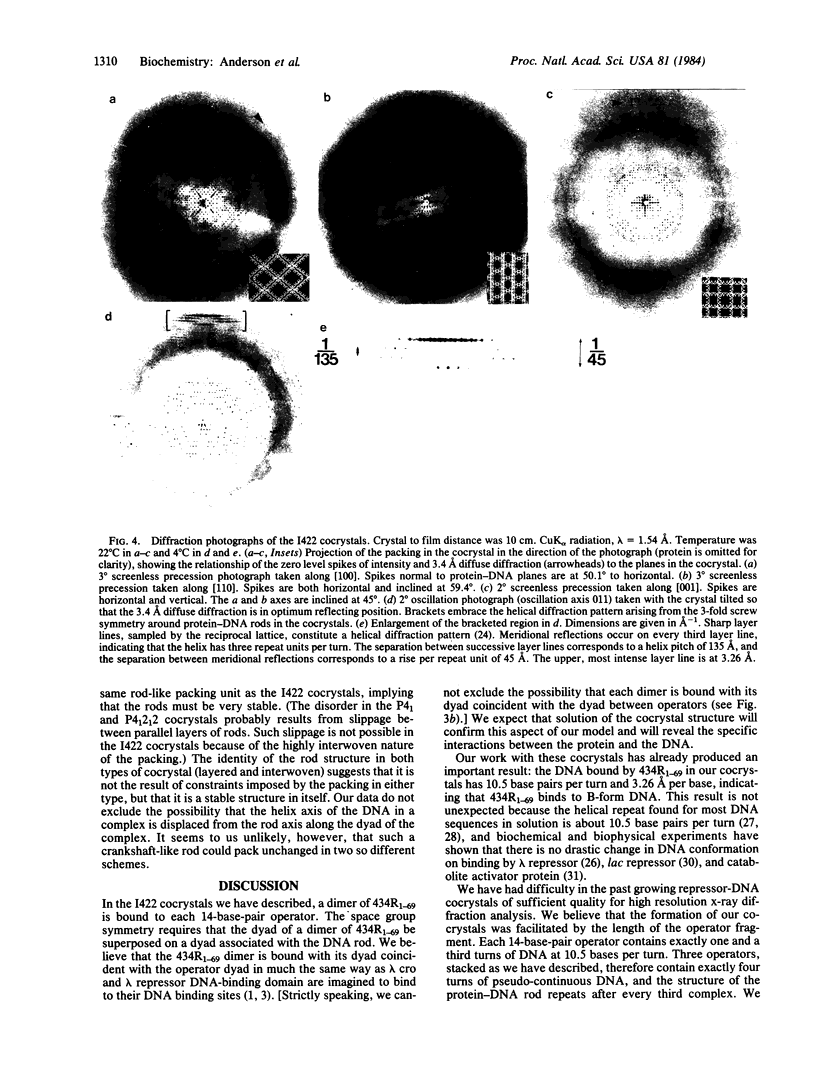
Abstract
The amino-terminal domain of the phage 434 repressor forms cocrystals with a synthetic phage 434 operator. The cocrystals diffract to at least 4 A, and x-ray crystallographic analysis of them is in progress. An analysis of the packing in the cocrystals shows that complexes consisting of dimers of amino-terminal domain bound specifically to operators are stacked end to end in longer protein-DNA rods parallel to the unit cell body diagonals. The DNA in the complexes has 10.5 base pairs per turn and a rise per base of 3.26 A--values consistent with B-form DNA--indicating that DNA is neither unwound nor overwound by bound repressor. The packing analysis suggests an approach that might facilitate the cocrystallization of other DNA-binding proteins with the DNA they recognize.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Cygler M., Vandonselaar M., Ohlendorf D. H., Matthews B. W., Kim J., Takeda Y. Crystallographic data for complexes of the Cro repressor with DNA. J Mol Biol. 1983 Aug 25;168(4):903–906. doi: 10.1016/s0022-2836(83)80082-5. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F. Proposed alpha-helical super-secondary structure associated with protein-dna recognition. J Mol Biol. 1982 Aug 25;159(4):745–751. doi: 10.1016/0022-2836(82)90111-5. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lauer G., Pastrana R., Sherley J., Ptashne M. Construction of overproducers of the bacteriophage 434 repressor and cro proteins. J Mol Appl Genet. 1981;1(2):139–147. [PubMed] [Google Scholar]
- Maniatis T., Ptashne M. Multiple repressor binding at the operators in bacteriophage lambda. Proc Natl Acad Sci U S A. 1973 May;70(5):1531–1535. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Anderegg R. Primary structure of the lambda repressor. Biochemistry. 1978 Mar 21;17(6):1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Saxe S. A., Revzin A. Cooperative binding to DNA of catabolite activator protein of Escherichia coli. Biochemistry. 1979 Jan 23;18(2):255–263. doi: 10.1021/bi00569a003. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Barkley M. D., Bourgeois S. Measurements of unwinding of lac operator by repressor. Nature. 1974 Sep 20;251(5472):247–249. doi: 10.1038/251247a0. [DOI] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Cleavage of a COOH-terminal hydrophobic region from D-alanine carboxypeptidase, a penicillin-sensitive bacterial membrane enzyme. Characterization of active, water-soluble fragments. J Biol Chem. 1979 Jun 10;254(11):4863–4875. [PubMed] [Google Scholar]
- Yocum R. R., Rasmussen J. R., Strominger J. L. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilus D-alanine carboxypeptidase. J Biol Chem. 1980 May 10;255(9):3977–3986. [PubMed] [Google Scholar]





