Abstract
Compressible flow based image registration operates under the assumption that the mass of the imaged material is conserved from one image to the next. Depending on how the mass conservation assumption is modeled, the performance of existing compressible flow methods is limited by factors such as image quality, noise, large magnitude voxel displacements, and computational requirements. The Least Median of Squares Filtered Compressible Flow (LFC) method introduced here is based on a localized, nonlinear least squares, compressible flow model that describes the displacement of a single voxel that lends itself to a simple grid search (block matching) optimization strategy. Spatially inaccurate grid search point matches, corresponding to erroneous local minimizers of the nonlinear compressible flow model, are removed by a novel filtering approach based on least median of squares fitting and the forward search outlier detection method. The spatial accuracy of the method is measured using ten thoracic CT image sets and large samples of expert determined landmarks (available at www.dir-lab.com). The LFC method produces an average error within the intra-observer error on eight of the ten cases, indicating that the method is capable of achieving a high spatial accuracy for thoracic CT registration.
Keywords: Compressible Flow, Image Registration, Thoracic Computed Tomography, Moving Least Squares, Least Median of Squares, Point Cloud
1. Introduction
Deformable image registration (DIR) has many applications within the field of medical imaging, including (see [1] for a review) ventilation imaging [2,3], brain mapping [4], and cardiac motion estimation [5]. Regardless of the application, the goal of a DIR method is to produce an accurate spatial mapping that relates the position of the underlying anatomy represented by each voxel within a given reference image to its corresponding position within a target image. DIR methods were first developed for computer vision applications [6] where fluctuations in image intensities are primarily caused by lighting changes or shadows. As such, most DIR methods are based on the assumption that the imaged material will maintain its intensity throughout the deformation. For images depicting compressible fluid flow, or equivalently, a conserved mass deformation, the continuity (mass conservation) equation is a more appropriate model that allows for voxel intensity variations [5,7].
Within the context of thoracic CT registration, compressible flow DIR methods have been shown to produce spatially accurate deformations [7,8]. However, existing methods are limited by factors such as image quality, noise, large magnitude voxel displacements, and computational requirements. The compressible flow methods described in [5,7,9–12] are based on differential equation models that require approximations to spatial and temporal image derivatives. In the presence of image noise, the accuracy of spatial derivative approximations are known to suffer [13]. Moreover, temporal image derivatives, which are typically approximated by simply subtracting the reference image from the target image, are suspect in image pairs depicting large magnitude displacements. The methods described in [14,15] are based on a nonlinear compressible flow model that does not require temporal image derivatives. However, those approaches do require the application of a gradient-based optimization algorithm, such as Newton's Method, Steepest Descent, or Levenberg-Marquardt (see [16] for an over view), in order to obtain a numerical solution. Applying a gradient-based optimization algorithm to an image registration problem necessarily requires either finite difference approximations to spatial image derivatives, or a continuous (such as B-spline [17]) representation of the image data. Furthermore, each iteration of a gradient-based algorithm requires a large scale linear system solve (with exception of Steepest Descent). The compressible flow 4D trajectory modeling method introduced in [8] utilizes gradient based optimization. But, because of the trajectory formulation, does not require large scale linear system solves. However, the method still requires a B-spline representation of the image data. Optimal control-based registration methods have been shown to be effective on two dimensional image sets [18,19], but the computational cost associated with the 3D or 4D extensions of such methods would be prohibitively expensive.
The DIR method introduced here, referred to as Least Median of Squares Filtered Compressible Flow (LFC), is based on describing the motion of a single voxel with a locally defined, nonlinear least squares, compressible flow voxel motion model. The model easily lends itself to a simple grid search (block matching) optimization strategy, and therefore does not require the approximation of image derivatives to obtain a numerical solution. A well known drawback to this type of basic block matching search strategy is the possibility of computing an erroneous point match, corresponding to a spatially inaccurate local minimizer of the nonlinear voxel motion formulation. In order to provide robustness to the grid search solves, the method utilizes a novel point match filter, based on least median of squares fitting and the "forward search" [20–22] outlier detection method. A globally defined deformation map is generated from the filtered point matches using Moving Least Squares interpolation [23].
In accordance with the DIR validation framework presented in [24], the spatial accuracy of the LFC method is measured using ten thoracic CT inhale/exhale image pairs and large sets of expert-determined landmark point pairs. All validation data (including images and expert-determined landmarks) used in this study are publically available online at www.dir-lab.com.
2. Methods
The LFC method is composed of two main components: 1) a localized, nonlinear least squares, compressible flow voxel motion model, and 2) a point match filter based on least median of squares fitting and forward search outlier detection. After describing each component in section 2.1. and 2.2, we derive the full LFC method in section 2.3. Our numerical implementation of the method is discussed in section 2.4.
2.1 Localized Nonlinear Compressible Flow
Assuming the image pair to be registered, P0(x) and P1(x), are snapshots of an unknown density function ρ(x(t),t), t ∈[0,1], the compressible flow (mass conservation) continuity equation is defined in terms of the density function as:
| (1) |
where v represents the apparent velocity of the flow depicted in the images. Methods based on the differential equation model (1), such as those described in [5,7,9–12], are dependent on approximations to the partial derivatives of ρ. These approximations are typically computed by applying finite differences to the image data contained in P0(x) and P1(x). Consequently, most differential equation based methods are susceptible to poor image quality and noise. The integrated form of (1), derived in [14,15] for use in 2D image registration, does not require temporal image derivative information, and is defined as
| (2) |
where D(x) is the unknown displacement field. DIR methods based on (2) typically employ a gradient-based optimization routine to compute a numerical solution to a large-scale nonlinear optimization problem. Rather than utilize (2) within a standard global DIR framework, we propose adjusting the model to fit within the locally regularized DIR framework first introduced in [25] for standard optical flow.
Locally regularized DIR methods are based on using the image information contained in a local neighborhood to articulate a well-posed least squares problem describing the motion of a single voxel. A full DIR is recovered from the localized approach by either solving the least squares problem for every voxel in the image domain or by solving the problem for a subset of voxels and then interpolating the results to obtain a globally dense field. The nonlinear compressible flow voxel motion-intensity model can be adapted to fit within the localized DIR framework by first applying the natural log to formulation (2) and rearranging terms to obtain:
| (3) |
Compressible flow formulation (3) describes the motion of a single voxel x in terms of the unknown displacement vector d = D(x). However, formulation (3) only provides one equation for the three unknown components of d and can be viewed as an underdetermined system. An over-determined system can be obtained by incorporating the image information contained in a local voxel neighborhood Nε(x), centered on x with radius ε, into the model. The unknown displacement d is then described as a least squares fit:
| (4) |
The difficulty in solving the nonlinear least squares problem (4) is in computing the divergence of the displacement field D. Since D is unknown, a manner for approximating the divergence of D using only the image information contained in Nε(x) is required. Following the approach used in [7] for the differential equation form (1), the contribution of the div(D(x)) term can be approximated by an auxiliary variable ϕ:
| (5) |
In this way, both the displacement of x and the divergence of the displacement field evaluated at x are described as a nonlinear least squares fit over the image information contained in the voxel neighborhood Nε(x).
An examination of the objective function R for problem (5) reveals that for any fixed displacement value d̃, there is unique value ϕ* such that:
| (6) |
and,
| (7) |
This follows from the fact that (6) represents a simple linear least squares problem, the solution of which is given by (7). Thus, for any displacement vector d̃, the optimal value of ϕ that yields the smallest value in R is given by (7). Substituting this result into (5) yields a localized nonlinear compressible flow formulation that only depends on the unknown displacement d:
| (8) |
Considering that image data is inherently discretized into a voxel grid, an exhaustive grid search optimization scheme is an appropriate strategy for computing a numerical solution to problem (8). Assuming the unknown displacement vector to be integer valued: d = (d1,d2,d3)T ∈ ℕ3, the grid search solution is described as:
| (9) |
where B(s) = {d : −s ≤ di ≤ s}, represents a bounding box of length 2s centered at the origin. The solution to (9) is obtained by simply evaluating R̂ at each integer grid point contained in the bounding box. As such, the grid search approach does not require approximations to image derivatives or a continuous representation of the image data.
2.2 Least Median of Squares Filtering
Within the context of DIR, grid search optimization applied to a locally defined least squares problem, such as (8), can be classified algorithmically as a block matching registration approach (see [13,26] for basic overviews). Block matching methods approximate the underlying deformation by computing point matches representing optimal (according to a given image similarity metric) corresponding regions in the reference image and target image. Though robust to image noise and large magnitude voxel displacements [13], block matching methods are still susceptible to the issues associated with nonlinear, nonconvex optimization; namely, convergence to erroneous local minima.
The grid search (block matching) optimization procedure is not guaranteed to find the most spatially accurate point match for two main reasons. First, the grid search result depends on the search window, which may or may not include the correct minimizer. Second, due to the nature of deformable transformations, there is no guarantee that the strongest numerical minimizer corresponds to the most spatially accurate point match. Consequently, existing block matching methods typically compute a full DIR from the generated point match data by employing an interpolation scheme designed to dampen the contributions of erroneous point matches (as in [27]). Other approaches for alleviating the effects of inaccurate point matches (often referred to as outliers) include parameter sensitivity [28] analysis and local thresholding [29] which are more heuristic in nature and also difficult to generalize. In the case of rigid registration, least squares trimming [30] has been shown to be effective for obtaining the transformation while remaining robust to outliers [31]. This strategy has been incorporated into an iterative deformable registration framework [27], but the method relies on a global regularization strategy to smooth out a series of local affine approximations, in addition to multiple block matching runs. Rather than simply dampen the effects of outliers or apply a heuristic thresholding, we propose utilizing two robust statistical tools known as "least median of squares" (LMS) and the "forward search method" to identify and remove potentially inaccurate point matches altogether.
LMS is a data fitting formulation that determines the fit function by minimizing the median of the absolute residuals, whereas standard least squares fitting minimizes the sum of squared residuals. Specifically, the LMS fit of a function f parameterized by the coefficient vector q for the data sets , is defined as:
| (10) |
The LMS formulation can robustly estimate q for data sets containing up to 50% outliers, as opposed to standard least squares fitting which can be severely affected by just a few points with large magnitude errors [32]. The fitting function f is typically chosen to be a polynomial of degree n, with the choice of n depending on the nature of the data set X,Y. In this case, the data set X represents a set of voxel locations within the reference image domain, and Y is obtained by solving problem (8) for each voxel location contained in X using grid search optimization.
The least median of squares filtering approach presented here is based on adapting the LMS framework to classify and remove potential outliers in the grid search computed point match data. The remaining "outlier-free" points can then be confidently interpolated to obtain a full DIR. Therefore, unlike the previous methods based on least squares trimming [27,31], our interest is not in the actual solution q to problem (10), but rather the data points that most contribute to it. The forward-search statistical tool can be used to determine precisely this type of information.
The forward-search method (see [20–22] for a more detailed description) is an iterative procedure for outlier detection that has been shown to be an effective technique for computing LMS fits [33]. The method begins by first finding an initial subset of m outlier-free point matches from the full data set X,Y. This subset, denoted X(m),Y(m), is easily generated by the random sampling method described in [32], and requires that the value of m be greater than or equal to the number of parameters required by the fitting model. The least squares fit to the data contained in X(m),Y(m) serves as the initial estimate q(m) of the LMS solution. After initialization, the method moves forward to a larger subset by applying the estimated solution q(m) to the full data set X,Y and finding the m+1 points with the smallest squared residuals. These m+1 points comprise the new subset X(m+1),Y(m+1), and their least squares fit represents the corresponding LMS solution estimate q(m+1). This process is repeated resulting in a sequence of point match subsets X(k),Y(k), k = m,m+1,m+2,…,M, each with a corresponding LMS estimate q(k).
The proposed forward search LMS filter determines a filtered subset of point matches from the original data set X,Y using the sequence of parameter estimates and subsets generated by the forward search iteration. The filtered set is defined as Xk*,Yk*, where:
| (11) |
Put simply, the forward search LMS filter takes the original point match data X,Y as input, and returns the forward search generated subset that produces the best LMS parameter estimate. As described in [20–22], the forward search iteration systematically ranks the points in the data set X,Y according to their consistency with the parameterized fit model. Thus, points fitting to the model poorly are included in the estimation subsets towards the end of the iteration. Since the forward search LMS point filter (11) returns the subset that minimizes the LMS objective function, the filter retains the point matches fitting best to the model. Consequently, potential outliers are removed.
Naturally, the filter's performance depends in part on the choice of the parameterized function space. Considering that a polynomial of degree n defined on ℝ3 requires the identification of (n+1)(n+2)(n+3) / 6 parameters, the task of finding an outlier free data subset for initializing the forward search becomes increasingly difficult for higher-order fitting models. However, the goal of applying the filter is not to obtain an accurate approximation of the underlying deformation, but rather to determine the data points most consistent with one another. Thus, it is not necessary to incur the computational cost of filtering the entire input data set X,Y with a high order model suitable for a full DIR recovery. Instead, a strategy where the filter is applied to a series of smaller image subregions is a more practical alternative.
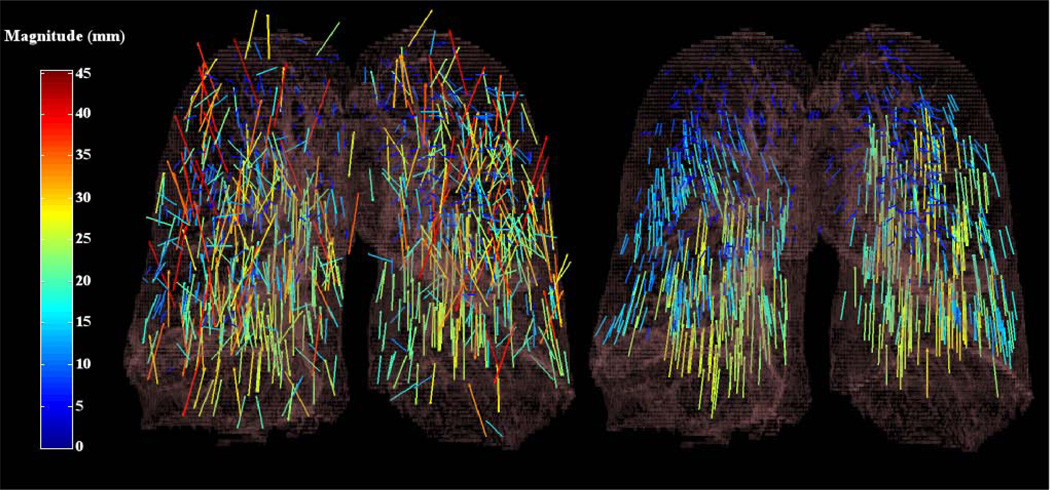
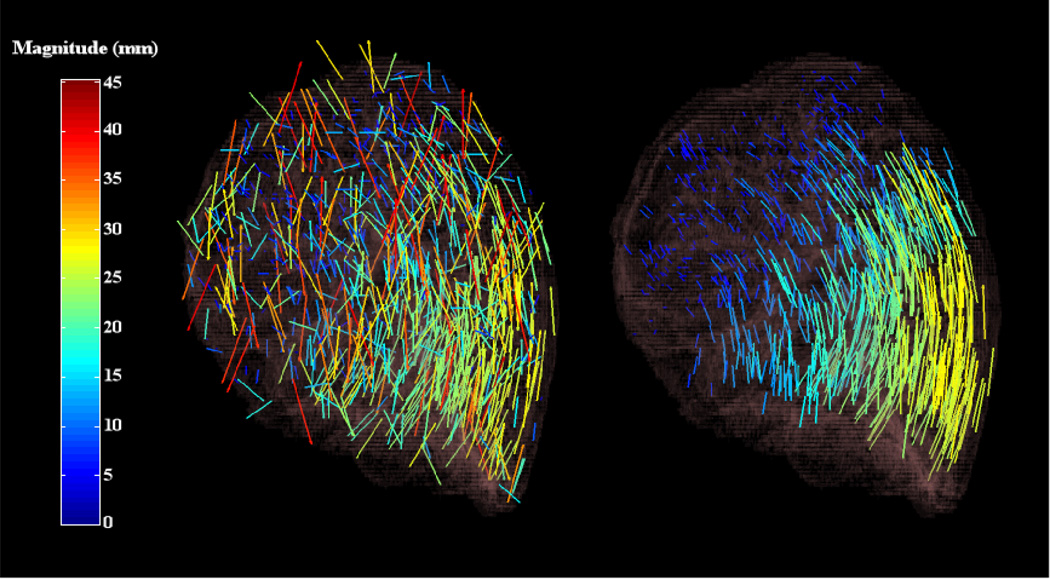
Under the mild assumption that the deformation field can be well approximated locally by a low order polynomial, the filter, utilizing a low order polynomial, can be applied to each subdomain contained in a decomposition of the full image domain. The union of the resulting filtered point sets obtained from each subdomain can then be interpolated confidently to recover the full DIR. This domain decomposition approach preserves the nonlinearity inherent to the underlying spatial transformation since the LMS filter only determines the points that are consistent with one another, leaving the data itself unaltered. Figures 1 and 2 illustrate the results of applying the filter to a data set of grid search computed point matches. The numerical implementation of the domain decomposition strategy is detailed in section 2.4.
Figure 1.
The effect of applying the forward search least median of squares filter to an input point match data set. The image on the left illustrates a coronal 3D rendering of the original grid search point matches. The image on the right illustrates the significantly improved filtered point match subset. The illustrated results correspond to test case 6 from (Results) section 3.
Figure 2.
The effect of applying the forward-search least median of squares filter to an input point match data set. The image on the left illustrates a sagittal 3D rendering of the original grid search point matches. The image on the right illustrates the significantly improved filtered point match subset. The illustrated results correspond to test case 6 from (Results) section 3.
2.3 Filtered Compressible Flow
The nonlinear least squares compressible flow formulation (8) provides a means for computing the displacement of a given voxel using simple grid-search optimization. Though robust to image noise and large magnitude deformations, the approach is susceptible to the effects of spatially inaccurate local minima. As the name suggests, the Least Median of Squares Filtered Compressible Flow (LFC) method described here utilizes the forward search least median of squares filter (11) to weed out the potentially erroneous point matches obtained by solving problem (8) with the grid search procedure (9).
LFC Algorithm
Given an image pair P0,P1, and a fitting function parameterized by the vector q, the LFC algorithm is as follows:
Define a set of uniformly spaced voxel locations on P0.
For i = 1, 2,3,…,M, compute di for the voxel location xi by solving problem (8) with the grid search procedure (9) and set yi = xi + di.
Determine the filtered data set X*,Y* by applying the forward-search LMS filter (11) to the data set .
Compute the fully dense displacement field by applying moving least squares interpolation to each voxel on the image grid using the filtered point match set X*,Y*.
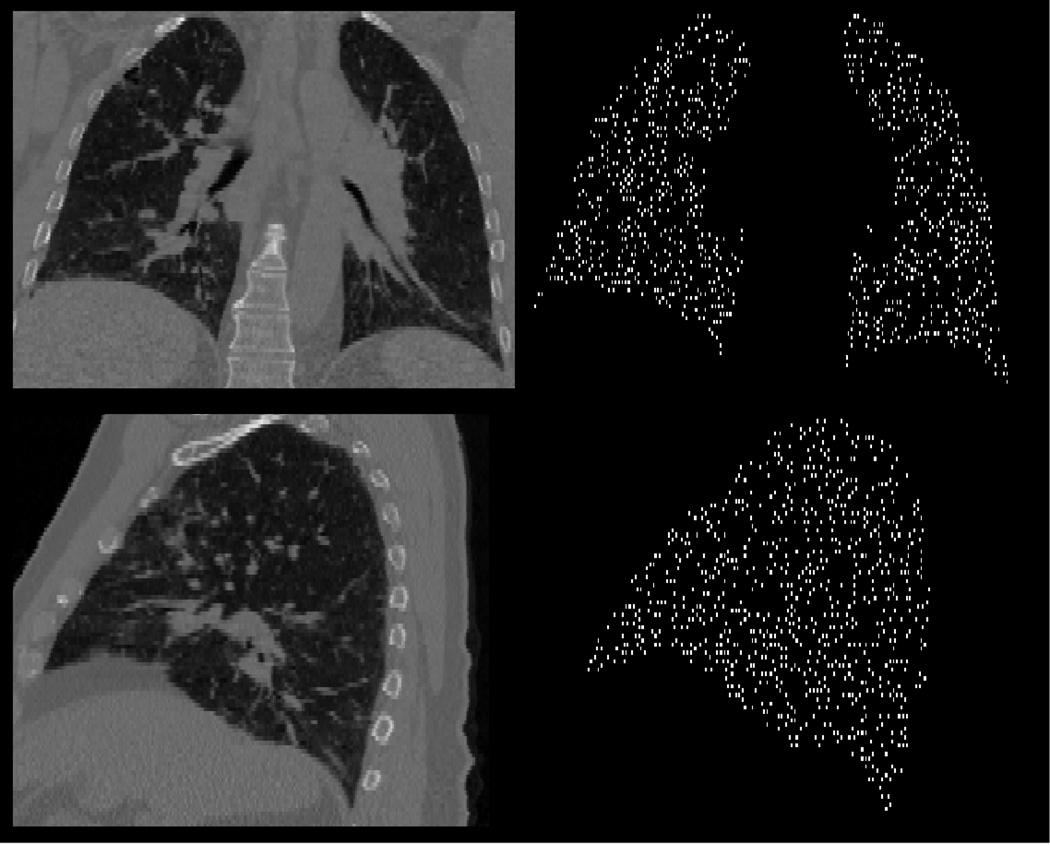
The LFC method is inherently point based, as such the first step of the algorithm simply determines the voxel locations in P0 on which to operate. For example, the set X might represent the voxels located on a coarse image grid. Our current implementation of the LFC method, as illustrated in Figure 3, generates the input source point set X using a simple dart throwing algorithm [34,35] applied to an input image mask. This approach ensures that X contains uniformly spaced voxel locations within a region of interest. However, despite the initial spatial structure placed on X, there is no a priori knowledge of which points will be filtered out by the forward search LMS filter. Consequently, the filtered source point set X* is not guaranteed to posses any spatial structure.
Figure 3.
An example source point cloud set generated by the dart throwing algorithm. (Top left) A 2D coronal slice of the inhale image for test case 6. (Top right) The corresponding 2D coronal slice of the point cloud generated by the dart throwing algorithm applied to the case 6 lung mask. (Bottom left) A 2D sagittal slice of the inhale image for test case 6. (Bottom right) The corresponding 2D sagittal slice of the point cloud generated by the dart throwing algorithm applied to the case 6 lung mask. All lung masks were computed using a simple histogram segmentation.
The filtered data X*,Y* represents an unstructured point cloud sampling of an underlying deformation. As such, interpolating the filtered sets X*,Y* to generate a full DIR requires an interpolation procedure that does not depend on structured input data. A similar problem arises in computer aided design where the goal is to recover closed shapes and surfaces from unstructured geometric point cloud data acquired by digital scanning devices. The moving least squares interpolation method is known to be robust and effective [23,36,37] for reconstructing surfaces from unorganized point clouds, and has also been shown to be effective for image registration applications [7,8,38]. Thus, the final step of the LFC method is to reconstruct a fully dense displacement field from the point cloud X*,Y* by applying moving least squares interpolation.
2.4 Numerical Implementation
The bulk of the computational workload associated with the LFC method is represented by the second step of the algorithm, namely computing the grid-search solution for a given set of voxel locations. However, due to the decoupled nature of block matching DIR, the grid-search step easily lends itself to a parallel implementation. Similarly, the moving least squares interpolation procedure required by the final step of the algorithm is also inherently parallelizable. Our current implementation of both the grid search procedure and the moving least squares interpolation utilizes the parallel computing power of Graphics Processing Units (GPUs). The software is written in the Compute Unified Device Architecture (CUDA) C programming language [39] for use with the NVIDIA Tesla 2070 GPU.
Our implementation of the forward-search LMS filter employs a simple affine function model:
| (12) |
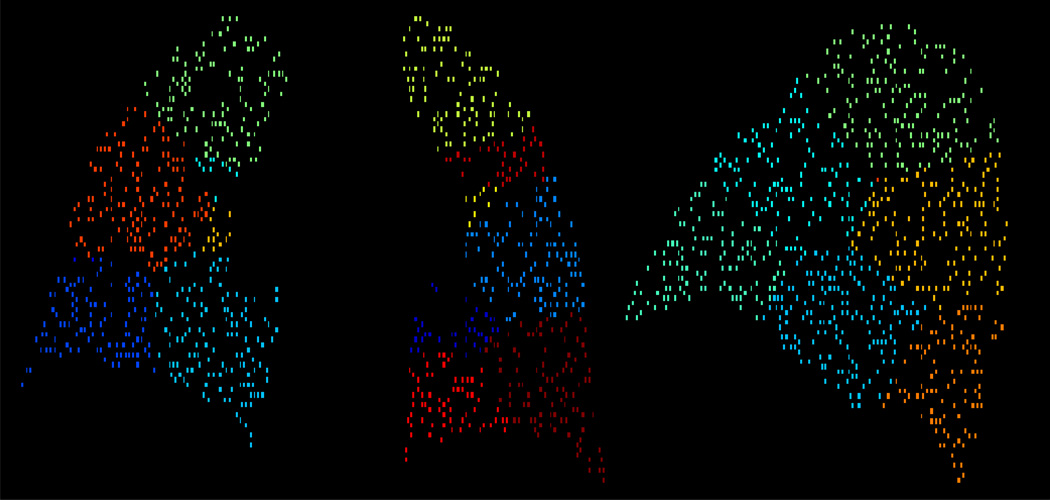
and, as discussed in section 2.2, is designed to operate on small image subdomains where the low-order affine model is an acceptable approximation. The K-means clustering algorithm described in [40] is used to divide the input point set X into K subsets, representing a spatial decomposition of the image domain. Figure 4 illustrates the results of a K-means subdomain decomposition. The filter, utilizing the affine function model (12), is then applied to each subdomain independently. The final filtered point match data set X*,Y* is then taken to be the union of the filtered sets determined on each subdomain. The dart throwing algorithm, subdomain generation, and the forward search LMS filter are all implemented in C++. All experiments listed in the Section 3 were conducted on a Silicon Mechanics Hyperform HPCg R2504.v2 workstation with two Intel Xeon X5675 Six-Core 3.06GHz processor and an NVIDIA Tesla 2070 GPU.
Figure 4.
2D coronal (left) and sagittal (right) slices of the K-means generated subdomain decomposition corresponding the point cloud shown in Figure 3. The different colors represent different subdomains. For all examples discussed in the Results (section 3), 32 subdomains were computed.
3. Results
Following the DIR validation framework presented in [24], the spatial accuracy of the LFC method is measured using large samples of expert-determined landmark point pairs and ten thoracic CT inhale/exhale image pairs obtained from the publically available database: http://www.dir-lab.com. A full treatment of the data acquisition, landmark generation, and statistical characterization of the reference feature pairs is detailed in our prior publications [8,24]. Tables 1 and 2 summarize the landmark data characteristics for all ten test cases. At each reference landmark position, the DIR spatial error is computed as the three-dimensional Euclidian distance (in millimeters) between the corresponding expert-determined target position and the estimated target position returned by the LFC method.
Table 1. Landmark Validation Data.
A summary of the expert determined landmarks for the ten validation inhale and exhale thoracic CT image pairs. All data is available publically at: www.dir-lab.com. The first column indicates the number of reference landmarks per case. The second column gives average (and standard deviation) reproducibility of feature registrations in units of mm, as described in [7,8,24]. The average (and standard deviation) displacement magnitudes for the set of reference landmark feature pairs are shown for the set of 10 CT image pairs utilized in this study are given in the last column. The distances are reported in units of mm.
| Case Number |
Number of Landmarks |
Intra-observer Error |
Avg. (Std.) Magnitude Displacement |
|---|---|---|---|
| 1 | 1280 | 0.85 (1.24) | 4.01 (2.91) |
| 2 | 1487 | 0.70 (0.99) | 4.65 (4.09) |
| 3 | 1561 | 0.77 (1.01) | 6.73 (4.21) |
| 4 | 1166 | 1.13 (1.27) | 9.42 (4.81) |
| 5 | 1268 | 0.92 (1.16) | 7.10 (5.15) |
| 6 | 419 | 0.97 (1.38) | 11.10 (6.98) |
| 7 | 398 | 0.81 (1.32) | 11.59 (7.87) |
| 8 | 476 | 1.03 (2.19) | 15.16 (9.11) |
| 9 | 342 | 0.75 (1.09) | 7.82 (3.99) |
| 10 | 435 | 0.86 (1.45) | 7.63 (6.54) |
Table 2. Sensitivity to Parameters.
The LFC method was applied to test case 8 using several different sets of input parameters.: M is the number of source points in the point cloud, the search time indicates (in seconds) the runtime required to compute the grid searches.
| M | Grid Search Time |
K | M* | Filter Time | Avg. (Std.) Error |
|---|---|---|---|---|---|
| 122657 | 174.90 | 16 | 63608 | 833.63 | 1.14 (1.22) |
| 32 | 64145 | 652.67 | 1.13 (1.11) | ||
| 64 | 66343 | 583.20 | 1.13 (1.23) | ||
| 68312 | 97.50 | 16 | 36477 | 331.57 | 1.13 (1.19) |
| 32 | 36645 | 290.63 | 1.11 (1.18) | ||
| 64 | 37127 | 261.58 | 1.14 (1.17) | ||
| 30647 | 44.51 | 1 | 14735 | 181.97 | 1.67 (2.16) |
| 2 | 15324 | 157.46 | 1.63 (2.05) | ||
| 4 | 16556 | 118.54 | 1.26 (1.35) | ||
| 8 | 16446 | 108.70 | 1.24 (1.42) | ||
| 16 | 15664 | 103.14 | 1.14 (1.19) | ||
| 32 | 15805 | 98.60 | 1.12 (1.16) | ||
| 64 | 16395 | 93.78 | 1.16 (1.25) | ||
| 128 | 16388 | 82.87 | 1.17 (1.32) | ||
| 22847 | 33.55 | 32 | 12065 | 65.07 | 1.18 (1.21) |
| 9704 | 15.29 | 32 | 4799 | 22.80 | 1.27 (1.26) |
K is the number of subdomains used by the LMS forward search filter and M* denotes the number of acceptable point matches returned by the filter. The filter time indicates (in seconds) the runtime required to compute the filtered points. The average error is given in millimeters. For all experiments in the table, the grid search bound s =17 and the local neighborhood match window is set to 9×9×5 voxels.
The LFC method requires the specification of four input parameters: 1) the radius ε of the local DIR neighborhood Nε(x), defined in formulation (5), 2) the bound for the grid search window s defined in (9), 3) the number of subdomains K used to decompose the input source locations (as described in 2.4 and illustrated in Figure 4), and finally the number of points M contained in the initial point cloud X resulting from the dart throwing algorithm. Tables 2 and 3 detail the results of applying the LFC method to test case 8 (the case containing the largest magnitude landmark displacements according to Table 1) using varying parameter values.
Table 3. Sensitivity to Match Window and Grid Search Bound.
The LFC method was applied to test case 8 using several different sets of input parameters.
| Match Window Dimensions |
S (voxels) | Grid Search Time |
M* | Avg. (Std.) Error |
|---|---|---|---|---|
| 9×9×5 | 7 | 5.64 | 17068 | 7.84 (9.97) |
| 12 | 18.62 | 16428 | 1.12 (1.23) | |
| 17 | 44.51 | 15805 | 1.12 (1.16) | |
| 22 | 89.36 | 15965 | 1.17 (1.30) | |
| 5×5×5 | 17 | 16.52 | 15162 | 2.63 (2.26) |
| 9×9×3 | 17 | 24.91 | 15659 | 1.24 (1.20) |
| 11×11×5 | 17 | 49.66 | 16431 | 1.14 (1.23) |
M* denotes the number of acceptable point matches returned by the filter and the average error is given in millimeters. For all experiments in the table, K =32 and M = 30647.
As demonstrated by the average millimeter errors reported in Table 2, the LFC method produces the best results when the value of K yields subdomains small enough for an affine function to adequately describe the underlying transformation on each subdomain. For instance, K=1 filters point matches based on an affine fit applied to the entire image domain. This produces the poorest spatially accurate result. However, the average error only fluctuates between 1.14 mm and 1.17 mm for K ≥16 (with the exception of the experiment where M=9704). Whereas for K ≤ 16, the fluctuations is more pronounced and the average error varies between 1.67 mm and 1.14 mm. Additionally, the computation time for the filter is sensitive to K. A larger value of K implies fewer points will be contained in each subdomain. Every iteration of the forward search method requires sorting the residuals obtained by applying the fit estimate from the previous iteration to all points matches in the subdomain. The sorting (our implementation utilizes the well know heap sort), becomes more expensive as the number of points in the subdomain increases. Thus, larger K (≥16 in this case) is beneficial for both accuracy and computational efficiency.
Table 2 also shows that the LFC results are not sensitive to size of the grid search bounding box, once the size is taken to be large enough to capture the displacement magnitude of the transformation. For test case 8, s = 7 is insufficient as evidenced by its associated high average error. This is because the voxel dimensions (as list on www.dir-lab.com) for the case 8 image data are 0.97×0.97×2.50, meaning that for s = 7 voxels, the grid search box can only capture displacements with a magnitude of 6.79 mm in the xy-plane and 17.5 mm in the z-direction. Considering that the average magnitude displacement for case 8 is 15.16 mm, the LFC method cannot accurately recover the transformation given s = 7. In comparison, the runs with s = 12,17, and 22 produce consistently accurate results.
Our implementation of the LFC method allow for anisotropic local neighborhoods to be used within the grid search. More specifically, the local DIR neighborhood Nε(x) is represented as a box of size l×l×w centered on x. We refer to this anisotropic local neighborhood as the match window. Table 3 also demonstrates the LFC's sensitivity to the match window dimensions. In general, too small a match window does not provide enough information to compute an accurate grid search point match. Larger match windows, though more robust, are more computationally expensive. Experimentation reveals that a match window of size 9×9×5 provides a compromise between computational expense and spatial accuracy as detailed by Table 3.
Though the bulk of the LFC method's computational workload is associated with the grid search, because of the GPU implementation, the majority of the method's actual run time is spent filtering the point matches. For comparison, we also implemented the grid search routine in C++ using the Open MP parallel library. Table 4 shows the runtimes for the grid search procedure using the GPU, a single CPU core, and 12 CPU cores in parallel. The GPU provides a speed up factor of approximately 300 over the single core run (with M=68312 and s = 17). Thus, without the GPU, the grid search time easily dominates the filtering time. In general, the LFC runtime depends on M and K. However, as demonstrated by Table 2, the spatial accuracy of the method does not greatly depend on either of these parameters. In fact, the average error achieved with M = 122657 and K =32 is only marginally better than the one achieved with M = 9704 and K = 32, despite the fact that the runtime for the former was 857.57 seconds, while the latter ran in 38.09 seconds.
Table 4. GPU vs. CPU Grid Search Run Time.
A comparison of the runtimes for the GPU based grid search and CPU based grid search is given. For these experiments, the match window was set to 9×9×5 voxels and s = 17. CPU sequential indicates that only one processor was used to compute the search whereas the CPU parallel run used 12 processors.
| M | GPU | CPU Sequential | CPU Parallel |
|---|---|---|---|
| 9704 | 15.29 | 3495.45 | 284.61 |
| 68312 | 97.50 | 2.94×104 | 2089.62 |
The mean errors and corresponding standard deviations produced by the LFC method for each individual case are detailed in Table 5. In addition, the parameter sets used to achieve the results are also given. For comparison, the previously reported spatial errors determined from the same ten validation test cases for the combined compressible local-global (CCLG) method [7], the four-dimensional trajectory modeling method (4DLTM) [8], and the GPU implementation of Therion's Demons algorithm [44], are presented in Table 6. Since measurements were made for each algorithm using the same reference images and landmark feature pairs, comparison can be made among the average spatial registration errors. Note that across all cases, the LFC method produced the smallest average spatial error.
Table 5. LFC Spatial Accuracy.
Average (and standard deviation) spatial registration errors are shown for each case, as well as combined for the set of reference feature pairs. All registration errors are given in units of mm. The parameters used by the LFC method to achieve the results are K=32, s=17, and a match window set to 9×9×5 voxels. The varying values of M reflect a constant spatial distribution of points across all cases, i.e., the point clouds for each case contained voxel positions uniformly spaced approximately 3mm apart. The p-values greater than 0.05 indicate there is no statistical difference between the distribution of errors produced by the LFC method and those represented by the intra-observer errors reported in Table 1.
| Case Number |
M | Grid Search Time |
M* | Filter Time |
Avg. (Std.) Error |
p-value |
|---|---|---|---|---|---|---|
| 1 | 29535 | 42.18 | 16552 | 90.86 | 0.85 (1.00) | 0.343 |
| 2 | 46391 | 66.33 | 26300 | 175.84 | 0.74 (0.99) | 0.147 |
| 3 | 36403 | 51.47 | 19558 | 123.90 | 0.93 (1.07) | 0.307 |
| 4 | 27592 | 39.19 | 13597 | 80.68 | 1.33 (1.51) | 0.029 |
| 5 | 32727 | 46.45 | 17203 | 102.40 | 1.14 (1.25) | 0.009 |
| 6 | 38181 | 54.16 | 20895 | 122.39 | 1.04 (1.05) | 0.002 |
| 7 | 46116 | 66.18 | 24912 | 165.66 | 1.03 (1.01) | 0.026 |
| 8 | 68312 | 97.50 | 36645 | 290.63 | 1.11 (1.18) | 0.193 |
| 9 | 24956 | 35.22 | 12425 | 70.92 | 1.04 (1.00) | 0.000 |
| 10 | 41475 | 59.55 | 21887 | 142.46 | 1.05 (1.10) | 0.010 |
Table 6.
Summary is shown of previously reported spatial accuracies derived using the publically available reference data set utilized in this study for spatial accuracy assessment of the LFC method. Since measurements were made for each algorithm using the same reference images and landmark feature pairs, comparison can be made among the average spatial registration errors. Results for the algorithms listed are reported in [7,8,44].
| Case Number |
LFC | CCLG | Demons | 4DLTM |
|---|---|---|---|---|
| 1 | 0.85 (1.00) | 1.02 (1.03) | 1.10 (1.09) | 0.97 (1.02) |
| 2 | 0.74 (0.99) | 1.29 (1.22) | 1.00 (1.15) | 0.86 (1.08) |
| 3 | 0.93 (1.07) | N/A | 1.32 (1.21) | 1.01 (1.17) |
| 4 | 1.33 (1.51) | N/A | 2.42 (2.48) | 1.40 (1.57) |
| 5 | 1.14 (1.25) | 2.50 (1.91) | 1.82 (1.87) | 1.67 (1.79) |
| 6 | 1.04 (1.05) | N/A | N/A | 1.58 (1.65) |
| 7 | 1.03 (1.01) | N/A | N/A | 1.46 (1.29) |
| 8 | 1.11 (1.18) | N/A | N/A | 1.77 (2.12) |
| 9 | 1.04 (1.00) | N/A | N/A | 1.19 (1.12) |
| 10 | 1.05 (1.10) | N/A | N/A | 1.59 (1.87) |
For each of the cases, the Wilcoxon rank sum test was performed to assess the statistical significance of differences between the LFC spatial accuracy and the intra-observer measurements, with p-values for each test shown in table 5. Under a null hypothesis that the distribution of LFC errors is equivalent to the pooled distribution of the observer errors, for a significance level of 0.05 the LFC errors are statistically equivalent to the observer errors for case 1,2,3 and 8 and statistically greater for the remaining cases. However, after applying the Bonferroni multiple-comparison correction to our significance level of 0.05 the significance level for each of the 10 cases becomes 0.005. Under this correction, the LFC errors are only statistically greater than the pooled observer errors in cases 6 and 9, with no significant statistical difference found between the two for any of the remaining cases.
4. Discussion
The LFC method is designed to compute accurate DIR for compressible flow image pairs utilizing 1) a localized nonlinear voxel motion model derived from the mass conservation equation, and 2) a novel point match filter based on least median of squares fitting and the forward search outlier detection method. The simplicity of the localized compressible flow formulation allows for the use of derivative free grid-search optimization, an approach known to be robust to image noise and large magnitude deformations. Considering that grid search generated point matches are not guaranteed to always be spatially accurate, we apply a novel point match filter to weed out erroneous point matches. A full DIR is computed from the resulting filtered point set using moving least squares interpolation. Our implementation of the LFC method utilizes GPU computing to mitigate the substantial computational workload represented by the grid search procedure. Given the emergence and growing popularity of GPU computing within the medical imaging community [41–43], this requirement is not a prohibitive limitation.
Examination of the average error summaries provided in Table 6 reveals that in general, the poorest performing methods were the two partial differential equation based methods: CCLG and Demon's. Though the CCLG method incorporates a local least squares fitting into the model to provide robustness to image noise, as discussed in [7], the method's performance tends to drop off for image pairs depicting large magnitude deformations, as does the performance of the Demon's algorithm. This result is not surprising since large magnitude deformations lead to poor approximations of the temporal image derivatives required by both methods, which in turn degrade the quality of the computed DIR. In the case of Demon's method, robustness to large displacements can be incorporated by adopting a multi-resolution scheme, and in fact, the results reported in Table 6 (from [44]) were obtained using a multi-resolution strategy. As such, Demon's produces better results than CCLG but does not achieve the accuracy of LFC. In contrast to LFC, the Demon's algorithm is based on the incompressible flow motion model and also does not guard against spatially inaccurate local minimizers. However, to what degree these properties contribute to the spatial error of the Demon's method is not known.
A nonlinear voxel motion model based on mass conservation and trajectory modeling is employed by the 4DLTM method [8]. As a result, the method does not require temporal image derivative approximations. However, 4DLTM does require gradient based optimization of a nonlinear objective function. Consequently, the method is susceptible to the errors introduced by the optimization solver's convergence to spatially inaccurate local minima. The forward search least median of squares filter used by the LFC method guards against spatially inaccurate grid search point matches, a feature the 4DLTM lacks. Consequently, the average errors for the 4DLTM method are smaller than the differential equation based methods, but larger than those returned by the LFC method, as reported in Table 6.
The LFC framework is based on a general point processing approach and is similar in nature to the types of algorithms used for point cloud processing. Though primarily known as statistical tools for robust regression analysis, both least median of squares fitting and the forward search iteration have been shown to be effective tools for reconstructing discontinuous surfaces, such as edges or corners, from unstructured point cloud data [45]. Within the surface reconstruction procedure, the forward search is used to determine which points are located along a geometric discontinuity. Similarly, the LFC method utilizes the forward search method to determine which point matches are erroneous. Because erroneous point matches are discarded, applying the filter results in "holes" or gaps in the uniformity of the source point distribution. The moving least squares method is therefore ideal for interpolating the set of filtered point matches, since the method does not require any spatial structure in the data. Considering that moving least squares interpolation, forward search, and least median of squares fitting have all been previously used to compute surface reconstructions from unstructured point clouds, the LFC method can be viewed as an adaptation of a surface reconstruction methodology for use within a DIR framework. Moreover, the point filter methodology introduced in this work is general in that it can be applied to any point match dataset.
Conclusions and Future Work
A method for computing compressible flow image registration using grid search determined point matches and a filtering scheme based on robust statistical tools is presented. The results of applying the method, referred to as Least Median of Squares Filtered Compressible Flow, to a validation data set consisting of ten thoracic CT image pairs and large sets of expert-determined landmark point pairs indicate that the method in general achieves a high spatial accuracy. Additionally, the LFC method produced spatial errors that are statistically indistinguishable from human observers on eight out of the ten test cases. An immediate area of future research is to apply the method to other more challenging CT and inter-modality data sets, and to compile new manual landmark validation sets for evaluating spatial accuracy.
Acknowledgements
This work was partially funded by the National Institutes of Health through a National Cancer Institute Grant R21CA141833 and through an NIH Director’s New Innovator Award DP2OD007044. Richard Castillo was partially supported by an NIH Training Grant T32CA119930.
References
- 1.Modersitzki J. Numerical Methods for Image Registration. Oxford: Oxford University Press; 2004. Numerical Mathematics and Scientific Computation. [Google Scholar]
- 2.Guerrero T, Sanders K, Castillo E, Zhang Y, Bidaut L, Pan T, Komaki R. Dynamic ventilation imaging from four-dimensional computed tomography. Phys. Med. Biol. 2006;51:777–791. doi: 10.1088/0031-9155/51/4/002. [DOI] [PubMed] [Google Scholar]
- 3.Castillo R, Castillo E, Martinez J, Guerrero T. Ventilation from four dimensional computed tomography: density versus Jacobian methods. Phys. Med. Biol. 2010;55:4661–4685. doi: 10.1088/0031-9155/55/16/004. [DOI] [PubMed] [Google Scholar]
- 4.Toga AW, Thompson PM. The role of image registration in brain mapping. Image Vision Comput. 2001;19:3–24. doi: 10.1016/S0262-8856(00)00055-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song SM, Leahy RM. Computation of 3-D velocity fields from 3-D cine CT images of a human heart. IEEE Trans. Med. Imaging. 1991;10:295–306. doi: 10.1109/42.97579. [DOI] [PubMed] [Google Scholar]
- 6.Horn BKP, Schunck BG. Determining optical flow. Artif. Intell. 1981;17:185–203. [Google Scholar]
- 7.Castillo E, Castillo R, Zhang Y, Guerrero T. Compressible image registration for thoracic computed tomography images. J. Med. Biol. Eng. 2009;29:222–233. [Google Scholar]
- 8.Castillo E, Castillo R, Martinez J, Shenoy M, Guerrero T. Four dimensional image registration using trajectory modeling. Phys. Med. Biol. 2010;55:305–327. doi: 10.1088/0031-9155/55/1/018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devlaminck V. A functional for compressible or incompressible elastic deformation estimation. IEEE Signal Proc. Let. 1999;6:162–164. [Google Scholar]
- 10.Fitzpatrick JM. A method for calculating velocity in time dependent images based on the continuity equation. (CVPR'95), San Francisco. 1985:78–81. [Google Scholar]
- 11.Qui M. Computing optical flow based on the mass conserving assumption. Proc. Int. Conf. on Pattern Recognition. 2000;3:7041–7045. [Google Scholar]
- 12.Wildes R, Amabile M, Lanzilloto A-M, Leu T-S. Recovering estimates of fluid flow from image sequence data. Comput. Vis. Image Underst. 2000;80:246–266. [Google Scholar]
- 13.Barron JL, Fleet DJ, Beauchemin SS. Performance of optical flow techniques. Int. J. Comput. Vis. 1994;12:43–77. [Google Scholar]
- 14.Corpetti T, Memin E, Perez P. Dense estimation of fluid flows. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24:365–380. [Google Scholar]
- 15.Corpetti T, Memin E, Perez P. Optical flow estimation in experimental fluid mechanics. Electron. Imaging. 1998;1:347–352. [Google Scholar]
- 16.Nocedal J, Wright SJ. Springer Series in Operations Research. In: Numerical Optimization. Glynn P, Robinson S, editors. New York, NY: Springer; 1999. p. 636. [Google Scholar]
- 17.Unser M. Splines: a perfect fit for medical imaging". Proc. SPIE. 2002;4684:225–236. [Google Scholar]
- 18.Papadakis E, Memin N. Variational assimilation of fluid motion from image sequence. SIAM J. Imaging Sciences. 2008;1:343–363. [Google Scholar]
- 19.Borzi A, Ito K, Kunisch K. Optimal control formulation for determining optical flow. SIAM J. Sci. Comput. 2002;24:818–847. [Google Scholar]
- 20.Atkinson AC, Riani M. Robust Diagnostic Regression Analysis. New York: Springer-Verlag; 2000. [Google Scholar]
- 21.Riani M, Atkinson AC, Cerioli A. Finding an unknown number of multivariate outliers. JR. Statist. Soc. B. 2009;71:447–466. [Google Scholar]
- 22.Atkinson AC, Riani M. Forward search added-variable t-tests and the effect of masked outliers on model selection. Biometrika. 2002;89:939–946. [Google Scholar]
- 23.Levin D. The approximation power of moving least squares. Math Comput. 1998;67:1517–1531. [Google Scholar]
- 24.Castillo R, Castillo E, Guerra R, Johnson V, McPhail T, Garg A, Guerrero T. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys. Med. Biol. 2009;54:1849–1870. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 25.Lucas B, Kanade T. An iterative image registration technique with an application to stereo vision; 7th International Joint Conference on Artificial Intelligence; Vancouver, BC, Canada. 1981. pp. 674–679. [Google Scholar]
- 26.Huang Y, Chen C, Tsai C, Shen C, Checn L. Survey on block matching motion estimation algorithms and architectures with new results. J. VLSI Signal Proc. 2006;42:297–320. [Google Scholar]
- 27.Commowick O, Isambert AVA, Costa J, Dhermain F, Bidault P-Y, Bondiau F, Ayache N, Malandain G. An Efficient Locally Affine Framework for the Smooth Registration of Anatomical Structures. Medical Image Analysis. 2008;12:427–441. doi: 10.1016/j.media.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Fedorov A, Billet E, Kikinis R, Chrisochoides N. Improved Block Matching for Non-Rigid Registration of Brain MRI. Image Processing and Data Visualization within SEECCM’09. 2009 [Google Scholar]
- 29.Garcia V, Commowick O, Malandain G. A Robust and Efficient Block Matching Framework for Non Linear Registration of Thoracic CT Images. A Grand Challenge on Pulmonary Image Registration (EMPIRE'10), held in conjunction with MICCAI'10. 2010 [Google Scholar]
- 30.Rousseeuw P, Leroy A. Robust Regression and Outlier Detection. New York, NY: John Wiley and Sons, Inc.; 1987. [Google Scholar]
- 31.Ourselin S, Roche A, Prima S, Ayache N. In: Block Matching: A General Framework to Improve Robustness of Rigid Registration of Medical Images Medical Image Computing and Computer-Assisted Intervention – MICCAI 2000. Delp S, DiGoia A, Jaramaz B, editors. Springer Berlin / Heidelberg: 2000. pp. CH373–CH373. [Google Scholar]
- 32.Rousseeuw PJ. Least median of squares regression. J. Am. Statist Assoc. 1984;79:871–880. [Google Scholar]
- 33.Atkinson AC. Fast Very Robust Methods for the Detection of Multiple Outliers. J Am Stat Assoc. 1994;89:1329–1339. [Google Scholar]
- 34.Cline D, Jeschke S, White K, Razdan A, Wonka P. Dart throwing on surfaces. Comput. Graph. Forum. 2009;28:1217–1226. [Google Scholar]
- 35.Cook R. Stochastic sampling in computer graphics. ACM Trans. Graph. 1986;5:51–72. [Google Scholar]
- 36.Dey TK, Sun J. An adaptive MLS surface for reconstruction with guarantees. Symposium on Geometry Processing. 2005:43–52. [Google Scholar]
- 37.Lipman Y, Or DC, Levin D. Data-dependent MLS for faithful surface approximation; SGP '07: Proceedings of the fifth Eurographics symposium on Geometry processing.2007. [Google Scholar]
- 38.Schaefer S, McPhail T, Warren J. Image deformation using moving least squares, in ACM SIGGRAPH 2006 Papers. Boston, MA: ACM Press; 2006. [Google Scholar]
- 39.Sanders J, Kandrot E. CUDA By Example: An Introduction to General-Purpose GPU Programming. Boston, MA: Addison-Wesley; 2011. [Google Scholar]
- 40.Arthur D, Vassilvitskii S. k-means++: the advantages of careful seeding. Symposium on Discrete Algorithms. 2007 [Google Scholar]
- 41.Dematté L, Prandi D. GPU computing for systems biology. Briefings in Bioinformatics. 2010;11:323–333. doi: 10.1093/bib/bbq006. [DOI] [PubMed] [Google Scholar]
- 42.Pratx G, Xing L. GPU Computing in Medical Physics: A Review. Medical Physics. 2011;38:2685–2697. doi: 10.1118/1.3578605. [DOI] [PubMed] [Google Scholar]
- 43.Samant SS, Xia J, Muyan-Ozcelik P, Owens JD. High performance computing for deformable image registration: Towards a new paradigm in adaptive radiotherapy. Medical Physics. 2008;35:3546–3553. doi: 10.1118/1.2948318. [DOI] [PubMed] [Google Scholar]
- 44.Gu X, Pan H, Liang Y, Castillo R, Yang D, Choi D, Castillo E, Majumdar A, Guerrero T, Jiang S. Implementation and evaluation of various demons deformable image registration algorithms on GPU. Phys. Med. Biol. 2010;55:207–219. doi: 10.1088/0031-9155/55/1/012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleishman S, Cohen-Or D, Silva CT. Robust moving least-squares fitting with sharp features. ACM SIGGRAPH. 2005;24:544–552. [Google Scholar]