Abstract
The P2Y1 purinoceptor cloned from chick brain (Webb, T. et al. (1993) FEBS Lett., 324, 219–225) is a 362 amino acid, 41 kDa protein. To locate residues tentatively involved in ligand recognition a molecular model of the P2Y purinoceptor has been constructed. The model was based on the primary sequence and structural homology with the G-protein coupled photoreceptor rhodopsin, in analogy to the method proposed by Ballesteros and Weinstein ((1995) Meth. Neurosci. 25, 366–428). Transmembrane helices were constructed from the amino acid sequence, minimized individually, and positioned in a helical bundle. The helical bundle was then minimized using the Amber forcefield in Discover (BIOSYM Technologies) to obtain the final model. Several residues that have been shown to be critical in ligand binding in other GPCRs are conserved in the P2Y1 purinoceptor. According to our model the side chains of these conserved residues are facing the internal cleft in which ligand binding likely occurs. The model also suggests four basic residues (H121 in TM3, H266 and K269 in TM6 and R299 in TM7) near the extracellular surface that might be involved in ligand binding. These basic residues might be essential in coordinating the triphosphate chain of the endogenous ligand adenosine 5′-triphosphate (ATP). Potential binding sites for agonists have been explored by docking several derivatives (including newly synthesized N6-derivatives) into the model. The N6-phenylethyl substituent is tolerated pharmacologically, and in our model this substituent occupies a region predominantly defined by aromatic residues such as F51 (TM1), Y100 (TM2) and F120 (TM3). The dimethylated analogue of ATP, N6,N6-dimethyl-adenosine 5′-triphosphate, is less well tolerated pharmacologically, and our model predicts that the attenuated activity is due to interference with hydrogen bonding capacity to Q296 (TM7).
Keywords: Molecular modelling, sequence analysis, ATP receptor(s), purinoceptor, nucleotides, G-protein coupled receptors
INTRODUCTION
About twenty years ago Burnstock proposed the ‘purinergic hypothesis’ which suggested that extracellular ATP is responsible for a variety of pharmacological effects mediated via membrane bound receptors. These purinoceptors were divided into P1 and P2 subclasses that differentiate receptors for adenosine and ATP, respectively.1 Pharmacological effects of extracellular ATP are exerted in peripheral tissues,4 where ATP acts as a neurotransmitter at autonomic neuromuscular junctions,2 as well as within the central nervous system.3
The family of P2 receptors consists of no fewer than five (and likely many more) subtypes. P2X (excitatory ion channel), P2Y (inhibitory G-protein coupled), P2U (G-protein coupled and responding to UTP), P2Z (opens membrane pore in mast cells), and P2T subtypes (ADP receptor on platelets) have been defined and characterized.4–8 This classification was made on the basis of functional responses and agonist specificity. Recently, a reorganization of the nomenclature has been proposed,8 in which all of the G-protein coupled purinoceptors are to be considered subtypes within a ‘P2Y superfamily”, and all of the ion channel purinoceptors are to be considered subtypes within a “P2X superfamily”. The most well-established second messenger system for the metabotropic P2Y and P2U receptors is activation of phospholipase C via coupling to G-proteins.9–11 ATP acting at P2X receptors activates Ca2+ channels in smooth muscle.12,13
The complete elucidation of P2 subtypes and mechanisms of action have been impeded by the limited number of stable and selective agonists and antagonists available. We have recently reported the synthesis of a variety of ATP analogues modified at either the ribose or the purine, including a series of functionalized congeners based on the potent P2Y purinoceptor agonist 2-methylthioadenosine 5′-triphosphate (2MeSATP).14,15 The 2-thio-ether derivatives were shown to be potent P2Y purinergic ligands stimulating the production of inositol phosphates in turkey erythrocyte membranes with K0.5-values ranging from 1.5 to 770 nM. Moreover, N6-methyl-2-(5-hexenylthio)-ATP was shown to be a potent and selective P2Y agonist (K0.5 = 26 ± 7 nM). Since substitution at the N6 position of the adenine moiety seemed to convey selectivity for a specific P2Y receptor subtype,14,15 we now synthesized the 5′-mono- and triphosphates of N6,N6-dimethyl-adenosine and N6-(2-phenylethyl)-adenosine. Progress in the development of antagonists has also been made recently.16–18
A very characteristic feature of G-protein coupled receptors (GPCRs) is their overall topology. They all have similar hydrophobicity profiles19 with seven stretches of increased hydrophobicity, most probably corresponding to α-helical regions spanning the cell membrane.20 The new potent and, in some cases, selective P2 agonists, together with the recent elucidation of the ammo acid sequences of the chick and turkey P2Y purinoceptor21,22 and the related mouse, rat and human P2U purinoceptor,23–25 encouraged us to proceed in our research effort towards understanding of the 3D structure of P2 purinoceptors, as well as towards identification of the ligand binding site. Unfortunately, isolation, purification and crystallization of P2 purinoceptors, as for other GPCRs, remains to be demonstrated. Not only is a crystallographic structure as yet unobtainable, but the very limited quantities of receptor protein present in the membrane, and isolation problems, prohibit the use of modern techniques, such as nuclear magnetic resonance spectroscopy, towards structure elucidation. Chemical modification of the ligand binding site and site-directed mutagenesis have been shown to be useful techniques in understanding receptor-ligand interactions,26 but is in the case of P2 purinoceptors restricted by the lack of suitable radioligands. A possible further approach is to construct models of the receptors and their ligand complexes using molecular modelling, as was first proposed by Hibert et al.27 In this paper we present a model of the chick P2Y purinoceptor based on the electron density map of rhodopsin,28 rather than on the bacteriorhodopsin template29 used by Hibert et al.27 The model is verified by incorporation of site-directed mutagenesis 41,50,51 and ligand affinity data.14,15
MATERIALS AND METHODS
Computational Methods
Software and hardware
Calculations and manipulations were performed using the Quanta (Molecular Simulations Inc., Sunnyvale, CA, releases 3.1.1 and 4.0) and Insight II (BIOSYM Technologies, San Diego, CA, version 2.2.0) modelling packages. All minimization calculations, on either proteins or the ligand-receptor complex, were performed using the Amber forcefield in Discover (BIOSYM Technologies, San Diego, CA, version 2.90). Single ligands were minimized using the MNDO hamiltonian in the MOPAC program (Quantum Chemistry Program Exchange, version 6.0). The Sequence Analysis Software Package of the Genetics Computer Group (University of Wisconsin, Madison, WI, version 7.3.1-UNIX) was used for producing the Kyte-Doolittle, surface probability, helical wheel and dendrogram analyses. The MOPAC program was run on a Convex C3830 system (Convex Computer Corp., Richardson, TX) and GCG was run on an SGI Challenge XL (Silicon Graphics Inc., Mountain View, CA, MIPS R4400 CPU). All other programs were run on an Iris Indigo XZ4000 (Silicon Graphics Inc., Mountain View, CA, MIPS R4000 CPU) workstation.
Model building
The sequences for the chick and turkey P2Y purinoceptor were taken from Webb et al.21 and Filtz et al.,22 respectively, and the sequences for the mouse, rat, and human P2U receptor were taken from Lutstig et al.,23 Rice et al.,25 and Parr et al.,24 respectively. Kyte-Doolittle hydrophobicity and Emini surface probability parameters were calculated using a 7 amino acid window. With these parameters, the putative transmembrane domains were identified. A sequence homology search was performed on the third transmembrane domain of the chick P2Y sequence (see discussion), using the World Wide Web version of the Basic Logical Alignment Search Tool (National Center for Biotechnology Information (NCBI), Bethesda, MD). Sequences were obtained from the Swiss, GenPept, EMBL, or PIR databases maintained by the NCBI. Sequences of the putative transmembrane domains (TMs) were aligned manually using the P2Y, P2U, and rhodopsin sequences based on common patterns, rather than on amino acid homology alone. The alignment for bacteriorhodopsin was based on the procedure developed by Oliveira et al30 The identifier for the first TM was either GX3N or GN where X represents any occurring amino acid. The identifiers for TM2 through TM7 were LX3DX7P or LX3DX8P (TM2), SX3LX2IX2DR (TM3) or SX3LX2IX2Hr, WX8P or WX9P (TM4), FX2PX7Y (TM5), FX2CX2P (TM6) and LX3NX3DPX2Y or LX3NX3NPX2Y (TM7), respectively.
Helices were built from the full length of the TM, using the Biopolymer module in the Insight II program. The N terminus was capped with an acetamido group, and the C terminus was capped with a carboxamido group. The secondary structure was assumed to be a right-handed alpha helix (default ϕ and Ψ angles from the Biopolymer module). After generation of the Amber atom type parameters and template charges, the helices were energy minimized as described in the Energy Minimization section.
The energy minimized helices were converted to Protein Database Brookhaven (PDB) format files and imported in the Quanta program. Coils (an alpha carbon backbone representation of the helix) of the minimized helices were drawn, that resemble the helical wheel representation to aid in orienting the individual helices towards each other.
The following constraints were used constructing the helical bundle:
The helical axes of the first, third, fifth and seventh helices were quasi-antiparallel to those of the second, fourth and sixth helices.
The hydrophobic side of each helix was facing the lipid phase and the hydrophilic side of each helix was facing either another helix or the pore formed by the helical bundle.
Conserved residues, either identical throughout a subset or highly homologous, determined the orientation of each helix relative to the other helices.
Distances in the electron density map of rhodopsin28 correlated to atomic coordinates in the model. This was obtained through triangulation of the electron density map and conversion of the coordinates to a grid on the screen.
The assembly of helices maintained a clockwise order, when seen from the intracellular side, as argued by Baldwin.31
None of the helices were intersecting.
The helical bundle was then energy minimized as described in the Energy minimization section.
Energy minimization
The individual helices generated with Insight II’s Biopolymer module were energy minimized in Discover using a stepwise process. Initially, 200 steps of Steepest Descent were performed, followed by minimization using the Conjugate Gradient (CG) method until the gradient reached a value below 0.1 kcal/mol/Å. Use of the Amber forcefield in Discover required that in all calculations the 1–4 nonbonded interactions were scaled by a factor 0.5, and the dielectric constant was assumed to be distance independent with a magnitude of 4. The bond, theta, phi, out-of-plane, nonbonded and Coulombic interactions were all used to obtain the final energy, but were not expressly scaled.
The helical bundles generated with Quanta were energy minimized in Discover in a stepwise process. Initially, 500 steps CG were performed with the backbone of the helices tethered with a force constant of 100 kcal/Å. In consecutive runs (500 steps CG each), the force constant was reduced to 50 kcal/Å, then 25 kcal/Å and, finally to 0 (no tethering).
Ligand docking
Six ligands were used to probe the receptor for possible ligand binding domains (BDs). Adenosine 5′-triphosphate (ATP) was constructed from guanosine 5′-(3-thiotriphosphate) co-crystallized with transducine38 and modified using the Builder module of Insight II (see discussion). 2-Methylthio-adenosine 5′-triphosphate (2MeSATP), 2-(2-(4-aminophenyl)ethyl)thio-adenosine 5′-triphosphate (APSATP), N6-(2-phenylethyl)-adenosine 5′-triphosphate (N6PEATP), N6,N6-dimethyl-adenosine 5′-triphosphate (N6diMeATP), and triphosphate were constructed from ATP using the Builder module of Insight II. The ligands were then fully minimized using the MNDO hamiltonian of MOPAC. All ligands were rigidly docked into the helical bundle using graphical manipulation coupled to continuous energy monitoring, i.e. the ligand was manually docked into the binding site without relaxing the atomic coordinates of either ligand or protein while, continuously, calculating the energy of the whole. When a final position was reached, consistent with a low local energy and known pharmacological data, the complex of receptor and ligand was subjected to a minimization run of 4000 steps CG (or until the gradient was < 0.1 kcal/mol/Å). Charges for the ligands were imported from the MOPAC output files.
Empirical Methods
Synthesis
New compounds were characterized by proton nuclear magnetic resonance using a Varian GEMINI-300, a Bruker AM-300 or a Bruker AC-200 NMR spectrometer. Spectra were taken in D2O. In all cases H-2′ was associated with the water peak. Nucleotides were also characterized by 31P NMR in D2O using H3PO4 as an external reference. Samples were treated with CHELEX-100 (BioRad, Richmond, CA) prior to measurement. Nucleotides were desorbed from a glycerol matrix under fast atom bombardment (FAB) conditions using 6 kV Xe atoms on a Joel SX102 spectrometer. Preparation of tri-n-butylammonium pyrophosphate for the triphosphate synthesis, as well as the preparation of triethylammonium bicarbonate (TEAB) buffer was done as published.14,15 Purification of nucleotides was achieved on an Isco UA-6 LC system using DEAE A-25 Sephadex columns and a linear gradient of 0–0.4 or 0.6 M NH4HCO3. Peaks were detected by UV absorption at 280 nm. The final purification was done on a Hewlett-Packard 1090 HPLC system using a semipreparative SynChropak RPP-100 column (10 × 250 mm, SynChrom, Inc., Lafayette, IN) with a flow rate of 3 ml/min. For analytical purposes, a nucleoside/nucleotide 7U column (4.6 × 250 mm, Alltech Associates, Inc., Deerfield, IL) was used with a flow rate of 1 ml/min. Separation was obtained using either a linear gradient of 0.1 M triethylammonium acetate buffer (TEAA, pH = 8.3) and acetonitrile (20% to 60% in 20 min; solvent system I) or 60 mM ammonium phosphate and 5 mM tetrabutylammonium phosphate (TBAP) in 90% water/10% methanol and 5 mM TBAP in methanol (25% to 75% in 8 min; solvent system II). Peaks were detected by UV absorption at 260 nm, and spectra collected with a diode array detector. Nucleotides were generally > 90 % pure. N6,N6-dimethyladenosine was purchased from Sigma (St. Louis, MO), and N6-(2-phenylethyl)adenosine was from Research Biochemical International (Natick, MA).
N6-(2-phenylethyl)-adenosine 5′-monophosphate bis-triethylammonium) salt (1, N6PEAMP) and N6-(2-phenylethyl)-adenosine 5′-triphosphate tetrakis-(triethylammonium) salt (2, N6PEATP): The procedure for nucleoside triphosphate synthesis was adapted from Kovács and Ötvös32 and Moffat.33 The reaction was carried out on 25 mg (0.067 mmol) of N6-2-phenylethyl-adenosine. The reaction mixture was then lyophilized and separated on a Sephadex DEAE A-25 column using a 0–0.5 M NH4HCO3 linear gradient. The monophosphate product 1 was obtained in 76% yield (25 mg). HPLC chromatography (nucleoside/nucleotide 7U column; solvent system II) showed > 98% purity. Retention time: 6.36 min. 1H NMR (D2O) δ 8.50 (1H, s, H-8), 8.13 (1H, s, H-2), 7.26 (5H, m, Ph), 6.06 (1H, d, J = 6 Hz, H-1′), 4.46 (1H, t, J = 6 Hz, H-3′), 4.31 (1H, m, H-4′), 3.95 (2H, t, J = 4 Hz, H-5′), 3.85 (2H, m, CH2NH), 2.98 (2H, t, J = 7.2, Hz, CH2Ph) ppm. High resolution FAB: calculated for C18H19N5O7P1 450.1173, found 450.1179.
The triphosphate product 2 was obtained in 21% yield (10 mg). This product was further purified on a semipreparative SynChropak RPP-100 column (solvent system I), and obtained in > 90% purity. Retention time (nucleoside/nucleotide 7U column; solvent system I) 4.13 min. 1H NMR (D2O) δ 8.50 (1H, s, H-8), 8.15 (1H, s, H-2), 7.28 (5H, m, Ph), 6.08 (1H, br.s, H-1′), 4.52 (1H, t, J = 6 Hz, H-3′), 4.38 (1H, m, H-4′), 3.22 (2H, m, H-5′), 3.87 (2H, m, CH2NH), 2.95 (2H, t, J = 7 Hz, CH2Ph) ppm. High resolution FAB: calculated for C18H23N5O13P3, 610.0525, found 610.0505.
N6,N6-dimethyl-adenosine 5′-monophosphate bis-(triethylammonium) salt (3, N6diMeATP) and N6N6-dimethyl-adenosine 5′-triphosphate tetrakis-(triethylammonium) salt (4, N6diMeATP): The reaction was carried out on 25 mg (0.054 mmol) of N6,N6-dimethyl-adenosine. The reaction mixture was lyophilized and separated on a Sephadex DEAE A-25 column using a 0–0.5M NH4HCO3 linear gradient. The monophosphate product 3 was obtained in 62% yield (19.3 mg). This product was obtained upon purification on a semipreparative SynChropak RPP-100 column (solvent system I) resulting in > 98% purity. Retention time (nucleoside/nucleotide 7U column; solvent system I) 2.55 min. 1H NMR (D2O) δ 8.51 (1H, s, H-8), 8.18 (1H, s, H-2), 6.10 (1H, d, J = 5.6 Hz, H-1′), 4.45 (1H, m, H-3′), 4.32 (1H, m, H-4′), 3.95 (2H, t, J = 4 Hz, H-5′), 3.42 (s, 6H, Me) ppm. High resolution FAB: calculated for C12H17N5O7P1 374.0888, found 374.0866.
The triphosphate product 4 was obtained in 5% yield (2.5 mg). This product was obtained upon purification on a semipreparative SynChropak RPP-100 column (solvent system I) resulting in > 98% purity. Retention time (nucleoside/nucleotide 7U column; solvent system I) 2.8 min. 1H NMR (D2O) δ 8.46 (1H, s, H-8), 8.20 (1H, s, H-2), 6.12 (1H, d, J = 5.7Hz, H-1′), 4.60 (1H, m, H-3′), 4.39 (1H, m, H-4′), 4.20 (2H, t, J = 4 Hz, H-5′), 3.44 (s, 6H, Me) ppm. High resolution FAB: calculated for C12H19N5O13P3 534.0167, found 534.0192.
Pharmacology
The biological characteristics of the novel compounds, determined using standard methodology,10,14 will be published separately.56
RESULTS AND DISCUSSION
The P2Y1, purinoceptor cloned from chick brain21 is a 362 amino acid, 41 kDA protein that shares < 20% identity with adenosine receptors. We built a model of this receptor in order to locate residues tentatively involved in ligand recognition and signalling. The steps of approach consisted of 1) Defining approximate boundaries of the helical regions from Kyte-Doolittle plots19 and from determination of sequence homology to other receptors (Figures 1 and 2). This also established which residues of P2 receptors correspond to those that have been postulated to be involved in ligand recognition in other receptors,41,46,47,48,50,51 and are, therefore, directed toward the binding site. This was useful for proper rotation of the helices.; 2) Building the receptor model using rhodopsin,31 with which it shares sequential and structural homology, as a template.34 Transmembrane helices were constructed from the amino acid sequence, minimized individually, and positioned in a helical bundle; and 3) Docking of ATP and synthetic analogues.14,15 For this purpose we have prepared, in addition to the already published compounds, the mono- and triphosphate analogues of N6-(2-phenylethyl)- and N6,N6-dimethyl adenosine, as probes of the N6 region in the P2Y receptor binding site.
Figure 1.
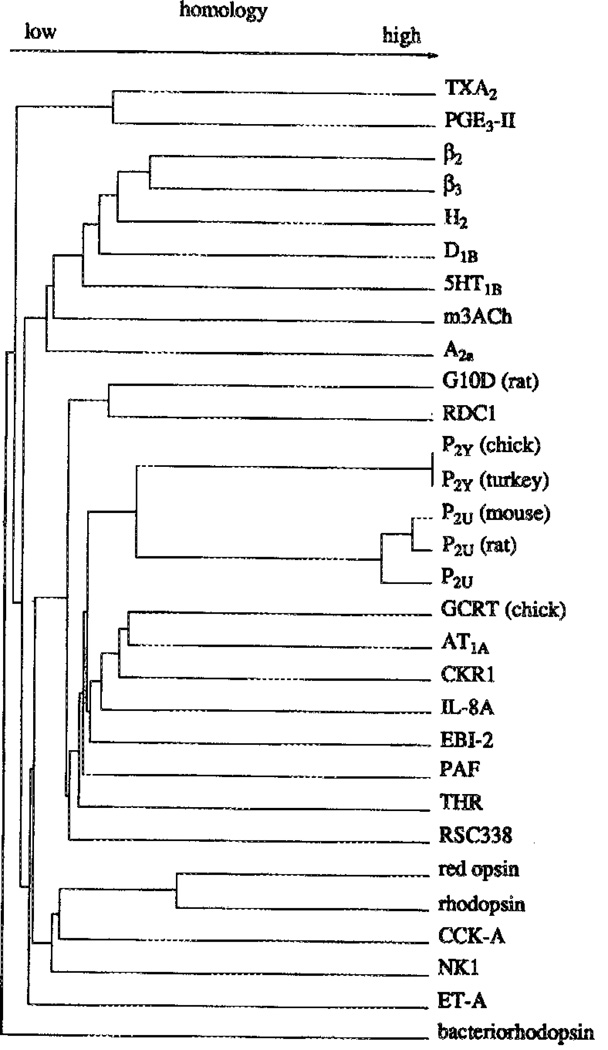
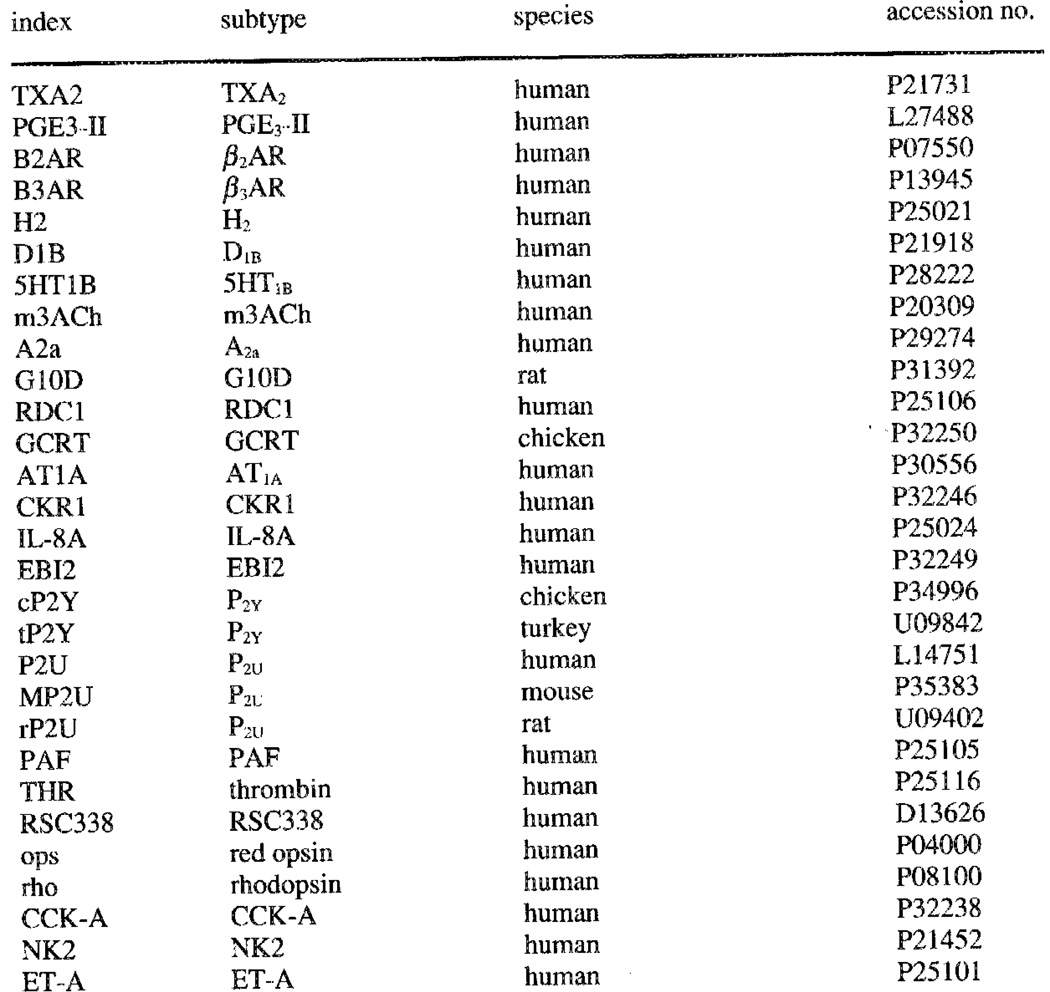
Dendrogram for selected G-protein coupled receptors, including the Known P2 receptor sequences.
Dendrogram and included sequences.
Figure 2.
Alignment of putative transmembrane domains, constructed as described in Materials and Methods and allowing no gaps in the sequence. Human sequences, where available, were used. In addition to the receptors shown: TXA2, PGE3-II, β2AR, β3AR, H2, D1B, 5HT1B, G10D, RDC1, GCRT, CKR1, IL-8A, EBI2, RSC338, red opsin, CCK-A, NK2, ET-A, and bacteriorhodopsin were also used in the alignment (see Figure 1). Unless specified, sequences are from the human, c = chick, t = turkey, m = mouse, r = rat. The residues of the alignment motifs are underlined, and the discussed in this paper are printed in bold type.
Alignment with other G protein-coupled receptors and dendrogram
A dendrogram (Figure 1) composed for the sequences of 29 G protein-coupled receptors (including orphan receptors) and bacteriorhodopsin shows that the known metabotropic P2 receptor sequences (P2Y and P2U) are more closely related to each other than to any other receptor. Comparison of the P2Y (chick) and P2U (rat) receptor subtypes with other GPCR sequences showed only low percentages of sequence identity (e.g., angiotensin II – P2Y 27%, angiotensin II – P2U 22%, thrombin – P2Y 25%, thrombin – P2U 25%; interleukin 8 – P2Y 22%; interleukin 8 – P2u; 23%). Both P2Y and P2U receptors bear a marginal sequence identity with the A1 adenosine (21% for P2Y and less than 12% for P2U) or cAMP receptors (17% for P2Y and less than 12% for P2U). A comparison of P2Y vs. P2U receptors revealed 38.8% identity and 58.6% similarity. In the transmembrane regions the identity percentage was even higher - 52%. Also, the P2 receptors are only distantly related to the biogenic amine receptors. An alignment of selected sequences (Figure 2) was constructed. The patterns on which this alignment is based differ markedly between the biogenic amine subclass and the P2Y and P2u type receptors.
In TM1, the motif GXXXN (P2Y) or GXXGN (P2U) occurs rather than the GN motif in the biogenic amine receptors. The alternate motifs, however, are not exclusively used by P2 receptors, but are shared with, e.g., the PGE3-II (GXXGN) and the PAF (GXXXN) receptors. In all sequences the last 5 C-terminal residues of this helix are frequently occupied by lysine or arginine residues, indicating the end of the transmembrane domain. Such basic residues may serve as ‘membrane anchors’,34,32 and are useful in determining the position of the helix in the lipid bilayer.
In TM2, the residue preceding the conserved leucine in the LXXXD motif is a conserved serine for the biogenic amine receptors, but is either an asparagine or a histidine in the P2Y and P2U receptors, respectively. The asparagine is conserved among the P2Y, AT1A, CKR1, IL-8A, PAF, NK2 receptors, the opsins and rhodopsin. The histidine is shared between P2U and thrombin receptors. Another difference between biogenic amine receptors and P2 receptors is the position of the conserved proline relative to the conserved aspartate. Although absent in the muscarinic receptor, the proline is consistently spaced by 8 residues from the aspartate in the other biogenic amine receptors, whereas in many other receptors, the P2 receptors included, only 7 residues separate these two residues. This last difference is possibly significant since the proline is located near the luminal side of the receptor protein, which is thought to be important for ligand binding. The only exceptions, observed so far, to the conservation of the aspartate occur in the substance P receptor where it is substituted with a glutamate, and in the gonadotropin-releasing hormone receptor where an asparagine replaces this residue. It has been argued by Zhou et al.52, that the conserved aspartate in TM2 is in close proximity to the conserved asparagine in TM7, and that the concurrent change of D to N in TM2 and N to D in TM7 in the gonadotropin-releasing hormone receptor is consistent with this finding. However, all P2Y and P2U receptor sequences identified so far, and several other receptors such as the TXA2, thrombin, PAF, and PGE3-II receptors, contain an aspartate in both TM2 and TM7. This argues against the hypothesis of close proximity of the two residues, which is indeed found in our model of the chick P2Y1 receptor. The conserved aspartate in TM2 was shown by Horstman et al. 53 to be a sodium-dependent allosteric regulatory site in the α2-adrenergic receptor.
The DRY motif, characteristic of the third transmembrane domain in biogenic amine receptors, is replaced by a HRY motif in the P2 GPCRs. This motif has not been found in other receptors. The only substitution for the conserved aspartate, seems to be the D to E in e.g., the TXA2, PGE3-II, opsin and rhodopsin sequences. The significance of this deviation is not clear, but is supposedly important for coupling of the P2 receptors to G-proteins and not for ligand binding, the subject of this study.
One or more of the first 6 positions in the N-terminal sequence of TM4 are generally (37 out of 40 aligned sequences of family A GPCRs) occupied by lysine or arginine residues. Again these residues could well serve as ‘membrane anchors’. They seem to be occurring more frequently towards the cytosolic side of helices than on the luminal side, thus reflecting the polarity of the membrane. These ‘membrane anchors’ also occur much less frequently in the C-terminal sequences of helices, than in the N-terminal sequences as is demonstrated by the alignment for TM5. The reason for this disparity is believed to be the direction in which the side-chains are pointing, i.e. opposite to the propagation direction of the helix, allowing more rotational freedom for lysine and arginine residues near the cytosolic N-terminus than for the cytosolic C-terminus. There are no other marked differences between the various receptor subfamilies for either TM4 or TM5.
The CXXP motif used for the alignment of TM6 could be substituted by WXP for most receptors, but the P2 receptors deviate at this position. The P2Y receptor has a tyrosine at the position of the conserved tryptophan, and this seems to be rather unique. The phenylalanine in the P2U receptor at the same position occurs more frequently, such as in the thrombin, and various orphan receptors. Characteristic of the P2 receptors is the presence of a lysine (P2Y: K269) or an arginine (P2U: R265) at an otherwise non-conserved position. It shares this feature only with the endothelin receptors (both ET-A and ET-B subtypes) and the orphan receptor RSC338. The proposed ‘membrane anchors’ occur quite frequently at the first position in the alignment of this transmembrane domain.
TM7 is best aligned by means of the NPXXY motif. However, the P2 receptors, the gonadotropin-releasing hormone receptor, the TXA2, PGE3-II, and several orphan receptors constitute an exception to this rule. The conserved asparagine is replaced by an aspartate residue in the latter cases. A non-conserved aspartate (D352) in the 5HT1B receptor aligns perfectly with aspartates in the IL-8A receptor (D288), and in the D1B receptor (D342), a glutamine in the P2Y receptor (Q296) and a lysine in the P2U receptor (K289). Arginines 299 (P2Y) and 292 (P2U) align with Y530 in the m3, E291 in the IL-8A and E287 in the CKR1 receptor, respectively. Both positions are near the luminal side of the receptor and are likely involved in ligand binding. The K and R residues occurring near the C-terminus are probably better characterized as ‘membrane anchors’.
According to Abbracchio et al.,8 the chick P2Y and turkey P2Y receptors should be classified as P2Y1 and P2Y4, respectively, based on their pharmacological profile. On the basis of the currently available sequences however, this subclassification is redundant since the two sequences differ only at position 28 in the N-terminus of the sequences. The chick threonine residue is replaced by the highly homologous serine residue in the turkey sequence. Any pharmacological difference would be methodological rather than sequence related. The mouse, rat, and human P2U (P2Y2) receptors are much more divergent, as is illustrated by the dendrogram (Figure 1). The P2 receptors, regardless of subtype, whether GPCR or ligand-gated ion channel, are often regarded as related to the better characterized and more widely known adenosine receptors [Burnstock4 and most subsequent reviews]. From the dendrogram (Figure 1), however, it must be concluded that, e.g., the A2a adenosine receptor is more closely related to the biogenic amine receptors, than to any of the GPCR P2 receptors. The P2 receptors are more closely related to the PAF, AT1A, and IL-8A receptors, and various orphan receptors, than to any other GPCR subfamily. Bacteriorhodopsin, included to facilitate comparison with other modelling studies, [e.g., 27,35] is clearly not related to any of the GPCR subfamilies. The degree of relatedness between bacteriorhodopsin and GPCRs shown in the dendrogram is probably an overestimate of the actual distance, caused by the residue alignment procedure of the program.
Kyte-Doolittle hydrophobicity and Emini surface probability analysis
At the portion of the sequence where proteins are supposed to cross the lipid bilaycr membrane, the hydrophobicity of that segment is usually increased relative to the cytosolic and luminal portions. The procedure developed by Kyte and Doolittle19 uses this phenomenon to identify transmembrane domains in protein sequences with unknown structure. In the case of GPCRs, these transmembrane domains are thought to be alpha helical, and more importantly amphiphilic. The amphiphilicity of the TMs supposedly reflects the way a GPCR is built out of seven of these helical transmembrane domains. Consequentially, the hydrophobicity profile of these GPCRs is not as clear as one would wish. An example of this effect is the poor separation of the sixth and seventh TM in both the P2Y receptor profile and the bacteriorhodopsin profile (data not shown). To facilitate identification of TMs, the Kyte-Doolittle hydrophobicity method is often supplemented with methods describing other sequence dependent vectorial parameters, such as the ‘conservation moment’ and ‘hydrophilic moment’,36 for each helix or other computational methods43. Use of the ‘conservation moment’ method requires a particularly well defined alignment of highly homologous sequences, and can not be applied to the P2 GPCRs because of the low sequence similarity of these receptors with any other GPCR subfamily. The ‘hydrophilic moment’ method relies on a database of partial hydrophilic factors for any given amino acid and is dependent on the method with which these factors were measured. Furthermore, the high incidence of basic residues in the P2Y receptor sequence greatly influences the results obtained with this method. In contrast, the Emini surface probability can be calculated from a given sequence, indicating the propensity of a stretch of amino acids (the window) to be at the surface of a protein. The combination of an increase in the Kyte-Doolittle hydrophobicity index and a decrease in the Emini surface probability index was successfully applied to predict the TMs in the P2Y receptor and in the reference protein bacteriorhodopsin. The start and end residue of each helix was usually predicted within 3 residues of the TMs used by Henderson et al.29 The only deviation from ideality that could not easily be explained by invoking the actual sequence, is the propagation of TM2 in bacteriorhodopsin 9 residues beyond its established terminating residue. Three other deviations could be explained by a closer examination of the profiles with regard to the sequence. The only major deviation in the P2Y receptor prediction occurred at the N-terminus of TM7. This particular sequence contains 3 hydrophilic residues (K, Q and R) that are possibly involved in ligand binding, thus delaying the start of the predicted TM.
Building the receptor model
Modelling of G protein-coupled receptors has become an important tool in understanding drug-receptor interactions and in the development of new ligands for these receptors. The first widely accepted method was the homology modeling method by Hibert et al.27 This method involved the alignment of the receptor sequence with the sequence of bacteriorhodopsin, and the subsequent mapping of the sequence onto the structure of bacteriorhodopsin that was determined by Henderson et al.29 Bacteriorhodopsin is a proton pump in the outer membrane of Halobacterium halobium, and lacks any functional or sequence homology with GPCRs. Nevertheless, the procedure was based on the assumption that there would be considerable structural homology. This structural homology was inferred by the extraordinary similarity in the hydrophobicity plots, or Kyte-Doolittle plots, of the biogenic amine subfamily of GPCRs and bacteriorhodopsin. Recently, a low resolution electron density map of rhodopsin, a true member of the GPCR superfamily, was published.28 The low sequence homology with bacteriorhodopsin, the structural differences that must arise from the different placement of proline residues (causing bends in helices) in bacteriorhodopsin- and GPCR-sequences, and the availability of an electron density map of a true member of the GPCR superfamily prompted us to adapt a new method to build models of GPCRs.34 This novel method is based on a computational approach rather than strict compliance with the atomic coordinates of a distantly related protein, albeit with higher resolution.
To ascertain the viability of modelling transmembrane proteins (based on the structure of rhodopsin34) by the methods described in this paper, we built models of bacteriorhodopsin (data not shown) based on the electron density map of bacteriorhodopsin,29 and rhodopsin (data not shown) and the P2Y receptor based on the electron density map of bovine rhodopsin as published by Schertler et al.28 root mean square (r.m.s.) distance calculations on superimposed structures were performed to establish how well the models fitted the experimental data. The bacteriorhodopsin model that was constructed, compared with the one deposited in the PDB,29 had an r.m.s. value of 2.15 Å when measured on all backbone atoms and 2.06 Å on all Cα atoms.
Both values are lower than the resolution of the model, i.e. 3.5 Å. When the rhodopsin model was compared with the bacteriorhodopsin model by Henderson et al.,29 however the r.m.s. value increased to 16.96 Å when measured on all backbone atoms and 16.86 Å on all Cα atoms. These rather high values indicate that there are more structural differences between bacteriorhodopsin and rhodopsin than have been assumed thus far 44 In our opinion, the position of the proline residues in the helices is a major determinant in this mater. As can be seen from the alignment for the helices, proline residues occur less frequently and at different positions in the sequence of bacteriorhodopsin than in the sequence of rhodopsin. The influence of proline residues in α-helices was extensively studied by Sankararamakrishnan and Vishveshwara37 and von Heijne.45 We have found that applying the Amber forcefield to our calculations yields results in agreement with Sankararamakrishnan and Vishveshwara37 and von Heijne,45 and that those results are consistent with data obtained from a crystallographic study of the transmembrane protein photosynthetic reaction center (PDB: 1prc) and globular proteins such as phosphoglycerate kinase (PDB: 3pgk), lysozyme (PDB: 1127) and alcohol dehydrogenase (PDB: 5adh). The differences between the structures derived from the electron density maps of bacteriorhodopsin and rhodopsin also illustrate why helical wheel models, widely used by molecular biologists, are highly imprecise when applied to GPCRs.31
Ligand docking
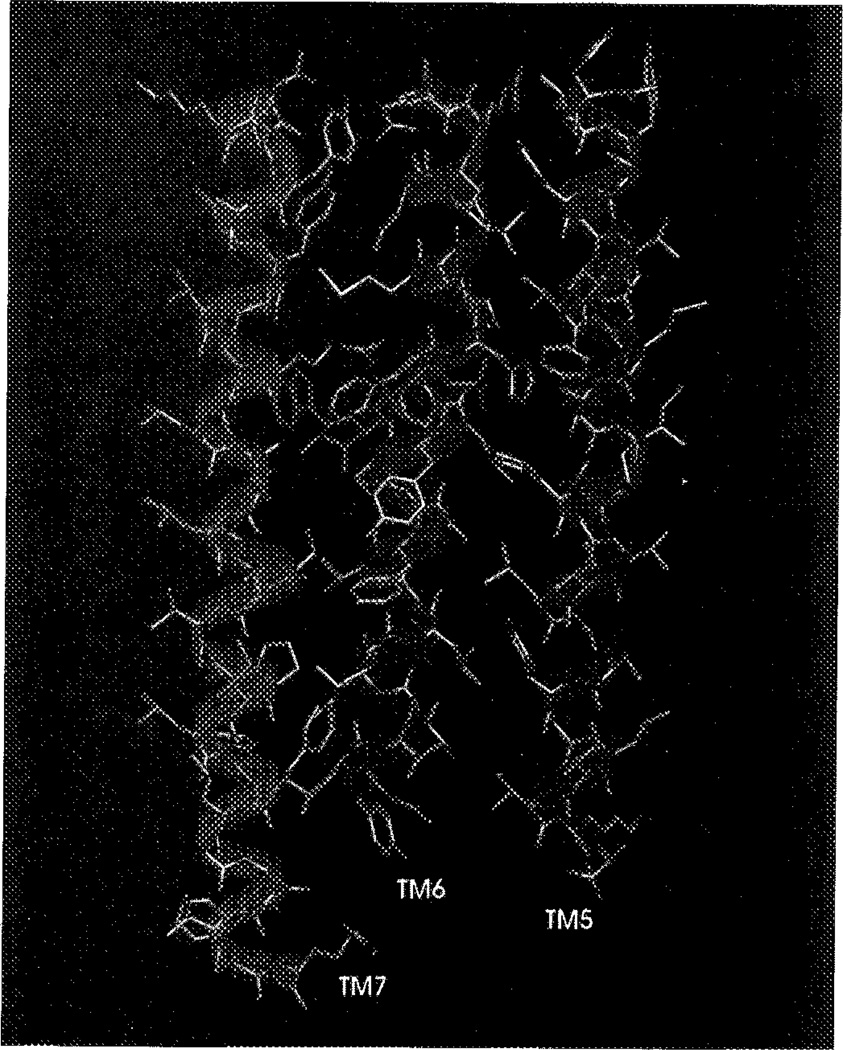
Figure 3 represents the final helical bundle with ATP docked into the purported ligand binding cavity. It is viewed from the luminal or extracellular side, as opposed to the rhodopsin template that was determined from the intradiscal or intracellular side. The backbones of the helices are accentuated with ribbons to emphasize the tilt of the helices towards each other and the effect of prolines in the sequence. Marked kinks are visible in, e.g., TM2 and TM4. ATP, which is significantly larger than the biogenic amine neurotransmitters, occupies the binding cleft formed by helices 2, 3, 5, 6, and 7. TM4, in particular, seems to be located too far outward to participate in ligand binding. Figure 4 is an cut-away drawing of TMs 5–7, shown to contain multiple ligand binding residues in the A2a receptor.51 The binding of the endogenous ligand ATP seems to occur around the upper third to upper half of the helical bundle. ATP is oriented in the plane of the lipid bilayer, almost perpendicular to the TMs. Figure 4 also demonstrates the use of ‘membrane anchors’ near the bottom of TM5 and TM7, and the top of TM6. Figure 5 focuses on the ligand binding domain (BD) formed by TMs 5–7. Although not directly involved in ligand binding, Pro218 in TM5 and Pro264 in TM6 have a great impact on receptor structure, and therefore, the BD. They are both located at the same distance from the membrane surface as the ligand and, more importantly, Phe215 and Phe219, located at opposite sides of the discontinuity formed by Pro218, are in close proximity of the terminal phosphate of ATP. This particular geometry is highly suggestive of the much heralded, but as yet unproven conformational change induced by agonists.
Figure 3.
View of the helical bundle of the chick P2Y1 receptor from the luminal side. Displayed (and coloured by atom type) are the helix backbone atoms, the side chains of TMs 5–7, and His121 in TM3. The adenosine moiety of ATP is displayed in dark green, and the triphosphate chain in purple. (See Color Plate IV at back of this issue).
Figure 4.
TMs 5–7 and ATP, as viewed in the plane of the membrane along the short axis. The adenosine moiety of ATP is displayed in dark green, and the triphosphate chain in purple. (See Color Plate V at back of this issue).
Figure 5.
Display of TMs 5–7 and ATP, in the vicinity of the binding site. The adenosine moiety of ATP is displayed in dark green, and the triphosphate chain in purple. (See Color Plate VI at back of this issue).
Ligand docking was initially performed with ATP, using a typical conformation based on crystallographic data for protein-bound nucleotides. To avoid the characteristically curled conformation of the triphosphate chain found in several phosphate, transferases, we opted to use the nucleotide bound to the phosphate hydrolase transducin,38 even though it involved substituting the purine guanine with the purine adenine. The orientation of the adenine moiety relative to the ribose ring was anti (i.e., the dihedral angle χ C9-N9-C1′-O4′ was 30.18°). The ring puckering, defined by the dihedral angle C1′-C2′-C3′-C4′ was, −3.85°, resulting in a 2′-exo, 3′-endo conformation for the two hydroxyl groups. After energy minimization the receptor was probed for possible hydrogen bonds (up to 5 Å between heavy atoms), electrostatic interactions (up to 10 Å between heavy atoms), and aromatic interactions (up to 10 Å between heavy atoms) There appeared to be only one favourable interaction between the adenine moiety of ATP and the receptor. The side chain of Gln296 (TM7) was within hydrogen bonding distance of the N6 atom at 4.51 Å. More residues were tentatively involved in coordinating the ribose moiety. The side chain of Ser306 (TM7) and the O2′ are separated by only 3.03 Å. The side chain of Ser306 (TM7) is within hydrogen bonding distance of 02′ at 2.95 Å and O3′ at 2.84 Å and the backbone carbonyl of Ala302 (TM7) is positioned at 4.53 Å of both hydroxyl groups. Arg299 (TM7) is within range of several heavy atoms, including O5′ (2.83 Å to Nε), O1α (3.30 Å to Nε), O1β (3.69 Å to NH), 02β (3.12 Å to NH), and O3α (2.90 Å to NH). Other interactions with the triphosphate chain seem to be constituted by His121 (TM3; 3.85 Å, Nε to O1β), Tyr125 (TM3; 2.73 Å, OH to O1β), Lys269 (TM6; 2.71 Å O3γ, 4.16 Å to O1β, and 4.49 Å, to O3β; all from Nε), and His266 (TM6; 4.91Å Nε to O1γ, 2.97 Å, Nε to O2γ and 3.77 Å, Nε to O3γ, 3.50 Å Nδ to O2γ, 3.78 Å Nδ to O3γ, and 4.64 Å Nδ to O3β). Aromatic residues can be found at 3.62 Å (O3γ to m-position of Phe219 in TM6),5.06 Å (03γ to o-position of Phe215 in TM6), 4.30 Å (C2 to m-position of Phe51 in TM1), 5.57 Å (C2 to m-position of Phe55 in TM1), and 9.98 Å (C2 to m-position of Tyr47 in TM1). Of special interest are four basic residues (His121 in TM3, His266 and Lys269 in TM6, and Arg299 in TM7) near the extracellular side of the helical bundle. In our model these residues are essential to coordinate the triphosphate moiety of the natural ligand ATP. Experimental support for this hypothesis was not only granted by experiments by Erb et al.,41 but also derived from the alignment and sequence analysis of over 40 GPCRs. Erb et al.41 showed that residues His262 (TM6), Arg265 (TM6) and Arg292 (TM7) in the human P2U, receptor (corresponding to His266 (TM6), Lys269 (TM6) and Arg 299 (TM7) in the P2Y purinoceptor, respectively) are directly involved in receptor activity. This was also the case for Lys289 (TM7; Gln296 in the P2Y receptor), but the difference in character between a lysine (P2U: UTP) and a glutamine (P2Y: ATP) residue suggests different functions in the mode of binding of the respective ligands. Tyr114 (TM3; Hisl21 in the P2Y receptor) was not investigated in the study, but the importance of this residue emerged from the modelling work. The central position of TM3 in the rhodopsin template, the distance from the extracellular surface, and the hydrogen bonding capacity of both histidine and tyrosine are all consistent with a role in ligand binding. H262 in the P2U receptor,41 H250 in the A2a receptor,51 and H265 in the NK1 receptor,50 the equivalents of H266 (TM6) in the P2Y receptor, were all shown to be important for ligand binding. The same holds true for R265 (P2U), N253 (A2a) and F268 (NK1) [the equivalents of K269 (TM6)], K289 (P2U) and Y271 (A2a) [the equivalents of Q296 (TM7)], R292 (P2U) and I274 (A2a) [the equivalents of R299 (TM7)], H278 (A2a) corresponding to S303 (TM7), and S281 (A2a) corresponding to S306 (TM7). His121 (TM3; P2Y) aligns, e.g., with Tyr529 in the rat m3,46 Asn412 in the human β2,47 and Thr355 in the human 5HT1B receptor,48 and all residues were shown to be essential for agonist binding. In contrast, Lys114 (TM3) in the P2Y and Lys107 (TM3) in the P2U receptor were not implicated in ligand binding41, whereas mutation of the equivalent residue, Asp99, in the rat ml receptor resulted in loss of affinity.49 This residue is located at the fringe of the transmembrane domain or even in the first extracellular loop in our model. It is therefore likely that the residue is involved in accessibility of the ligand binding domain or in maintaining a specific structure in the loop.
In addition to ATP, we also docked N6-(2-phenylethyl)-adenosine 5′-triphosphate (N6PEATP), 2-(2-(4-aminophenyl)ethyl)thio-adenosine 5′-triphosphate (APSATP), N6,N6-dimethyl-adenosine 5′-triphosphate (N6diMeATP), 2-methylthioadenosine 5′-triphosphate (2MeSATP) and triphosphate into the helical bundle. After minimization, the energy of all complexes was between −2500 and −2600 kcal/mol and the complexes were 400 to 500 kcal/mol more stable than the sum of the components. Since triphosphate yielded values similar to the adenosine derivatives, the energy contribution of the triphosphate moiety by far exceeds the combined contribution of the ribose and the adenine moiety. Energy differences between compounds must therefore be regarded as qualitative rather than quantitative indications of ligand affinity.
The role of divalent cations such as Mg2+ in P2 purinergic transmission has been described throughout the literature.1–11 This is not addressed in our model, because current pharmacological data are not sufficient to hypothesize the position and mechanism of such divalent cations in receptor structure. The impact of the divalent cation Zn2+ on binding of antagonists to the tachykinin NK-1 receptor and its mutants was recently demonstrated and described in detail by Elling et a1.50
Correlation between modelling and pharmacology
It appears that the binding of the triphosphate moiety is a major determinant in binding of ligands to the P2Y purinergic receptor. Since this part of the molecule contains multiple negative charges, one would expect to find counterions in the BD. Indeed, the P2Y1 receptor sequence contains several positively charged residues. Our modelling study reveals that, of these, Lys269 (TM6) and Arg299 (TM7) are likely candidates for this function and are appropriately positioned within the helical bundle to exert this function. We propose that these two basic residues are assisted by two histidine residues, His121 (TM3) and His266 (TM6), and one tyrosine residue, Tyr125 (TM3). These residues tentatively coordinate the α-phosphate (His121 and R299) the β-phosphate (Y125, K269 and R299), and the γ-phosphate (His266, and K269). Although adenosine 5′-monophosphate analogues are widely regarded as inactive at P2 purinergic receptors,2,5,6 Fischer et al.14 recently demonstrated that one monophosphate analogue in particular, 2-(5-hexenyl)thio-adenosine 5′-monophosphate, is a more potent (K0.5 =328 ± 43 nM; 8.5-fold over ATP) agonist at P2Y receptors on turkey erythrocytes than ATP (K0.5 = 2800 ± 700 nM). In our model there is sufficient coordination of the α-phosphate to warrant such an interaction, although the number of stabilizing interactions, and hence the interaction energy and affinity, will be lower than in the case of the corresponding triphosphate. This is pointedly illustrated by the K0.5 values of 2-(5-hexenylthio)-ATP (10 ± 4 nM) and 2-(5-hexenylthio)-AMP (328 ± 43 nM).14 Since the interaction between the receptor and a ligand monophosphate is much weaker than with a triphosphate, the effect of substituents at distal sites, such as in N6PEAMP no effect ≤ 10−4 M) and N6diMeAMP (no effect ≤ 10−4 M), increases and the combined effect of deleting two phosphates and adding N6-substituents proved detrimental to activity. Interestingly, Erb et al.41 showed that Lys289 (TM7) in the mouse P2U receptor, corresponding to Gln296 (TM7) in the chick P2Y receptor, when mutated to an arginine, reversed the selectivity of the triphosphates ATP and UTP to the corresponding diphosphates. In our proposed model Gln296 (TM7) is not in the vicinity of the phosphate BD, and this suggests that P2Y and P2U receptors display significantly different modes of binding of ligands, as implied by the pharmacology-derived nomenclature of the receptors.
Gln296 (TM7), in our model, is positioned in the vicinity of the N6 amine of the adenine moiety. Substitution of the hydrogens on this amine with methyl groups, thus reducing the extent of the interaction between the N6 region and Gln296 (the distance between the two increases), greatly decreases the activity of the analogues: N6MeATP (K0.5 ≈ 19 µM) is 6.8 times less potent than ATP (K0.5 = 2.8 ± 0.7 µM.) and N6diMeATP (K0.5 ≈ 64 µM) is over 20-fold potent less than ATP in stimulating phospholipase C in turkey erythrocyte membranes.10,14 The loss of affinity by the methyl substitution may, however, be compensated for by introducing an aromatic side chain on this substituent, as in N6PEATP (K0.5 = 7.1 ± 0.3 µM; only 2.5-fold less active than ATP). Such an aromatic substituent may be accommodated by residues Phe51 (TM1), Tyr100 (TM2), and Phe120 (TM3), which are located at distances ranging between 3.5 and 10 Å from the N6 amine. Certain substituents on the 2-position of the adenine ring enhance activity, such as in 2MeSATP (K0.5 = 8 ± 2 nM; 350-fold more potent than ATP) and APSATP (K0.5 = 1.53 ± 0.21 nM; 1830-fold more potent than ATP).14 Substituents in the 2-position may occupy the same region of the receptor as the N6-substituents, as was proposed for the rat A2a receptor,54 or an entirely different region of the receptor as was proposed for the m3 muscarinic receptor.55 In the latter case, the side chain would extend through a largely hydrophobia region accommodating the phenyl ring, into a more hydrophilic region occupied by the conserved aspartate in TM2 that confers allosteric regulation of α2-adrenergic receptors by sodium,53 where it could form a salt bridge with the amine. In our current model the phenyl ring of N6PEATP occupies the same region as the phenyl ring of APSATP, thus adhering to the model proposed for the A2a adenosine receptor.54
CONCLUSIONS
We have sought to identify positively charged amino acid residues (Arg or Lys) as anchoring points which could contribute major electrostatic interactions with the phosphates of ATP. Such residues should be conserved within the P2 GPCR family and should also be pointing towards the center of the receptor cavity. Likewise, they should probably be located around the middle or upper third of the transmembrane regions, where most of the non-peptide GPCRs are thought to bind ligands. These requirements only yield two possible anchoring points: Lys269 in TM6 and Arg299 in TM7.
As a result of the above conclusions, we identified six more residues, Gln296 in TM7 in the adenine BD, Ser303 and Ser306 in TM7 in the ribose BD, His121 in TM3, His266 in TM6, and Tyr125 in TM3 in the triphosphate BD, that are involved in ligand binding according to this model. Furthermore, we have shown that our model is consistent with the current pharmacological data.
It will be interesting to construct models of the P2x receptor, now cloned,39–40 and compare the binding sites as a means of getting insights for achieving ligand selectivity.
Acknowledgements
The authors wish to thank Prof. H. Weinstein, Dr. J.A. Ballesteros and Ms. L. Laakonen of Mount Sinai Medical School, NY, and Dr. R. Pearlstein of the Division of Computer Research and Technology, NIH, MD, for their helpful suggestions towards this project. AMvR should like to thank Prof. G. Burnstock of University College London and the Cystic Fibrosis Foundation for financial support. The authors also wish to thank Dr. J.L. Boyer and Prof. T.K. Harden of the Univ. of North Carolina for the pharmacological determinations, which will be published in a separate study.
List of Abbreviations
all single and three letter notations for amino acids.
- 2MeSATP
2-methylthioadenosine 5'-triphosphate
- APSATP
2-(2-(4-aminophenyl)ethyl)thio-adenosine 5′-triphosphate
- ATP
adenosine 5′-triphosphate
- BD
binding domain
- cAMP
adenosine 3′,5′-cycle monophosphate
- CG
Conjugate Gradient
- GPCR
G-protein coupled receptor
- MNDO
Medium Neglect of Differential Overlap
- N6diMeATP/N6diMeAMP
N6,N6-dimethyl-adenosine 5′-triphosphate/-monophosphate
- N6PEATP/N6PEAMP
N6-(2-phenylethyl)-adenosine 5′-triphosphate/-monophosphate
- PDB
Protein Database Brookhaven
- r.m.s
root mean square
- TBAP
tetrabutylammonium phosphate
- TEAB
triethylammonium bicarbonate
- TM
transmembrane domain
- UTP
uridine 5′-triphosphate
Footnotes
The coordinate files for the molecular models described in this article will be included in the CD-ROM edition of the journal. The files are also available from the Editor.
References
- 1.Burnstock G. A basis for distinguishing two types of purinergic receptor. In: Straub PW, Bolis L, editors. Cell Membrane Receptors for Drugs and Hormones: a Multidisciplinary Approach. New York: Raven Press; 1978. pp. 106–118. [Google Scholar]
- 2.Hoyle CHV. Transmission: purines. In: Burnstock G, Hoyle CHV, editors. Autonomic neuroeffector mechanisms. Chun Harwood: Academic Press; 1992. pp. 367–407. [Google Scholar]
- 3.Edwards FA, Gibb AJ, Colquhoun D. ATP receptor mediated synaptic currents in the central nervous system. Nature. 1992;359:144–147. doi: 10.1038/359144a0. [DOI] [PubMed] [Google Scholar]
- 4.Hoyle CHV, Burnstock G. ATP receptors and their physiological roles. In: Stone TW, editor. Adenosine in the nervous system. London: Academic Press Ltd.; 1991. pp. 43–76. [Google Scholar]
- 5.Cusack NJ. P2, receptors: subclassification and structure-activity relationships. Drug Dev. Res. 1993;28:244–252. [Google Scholar]
- 6.Gordon JL. Extracellular ATP: effects, sources and fate. J. Biochem. 1986;223:309–319. doi: 10.1042/bj2330309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden TK, Jacobson KA, Leff P, Williams M. Nomenclature and classification of purinoceptors. Pharmacol. Rev. 1994;46:143–156. [PMC free article] [PubMed] [Google Scholar]
- 8.Abbracchio MP, Burnstock G. Purinoceptors: are there families of P2X and P2Y purinoceptors? Pharmac. Ther. 1994;64:445–475. doi: 10.1016/0163-7258(94)00048-4. [DOI] [PubMed] [Google Scholar]
- 9.Dubyak GR. Signal transduction by P2-purinergic receptor for extracellular ATP. Am. J. Respir. cell. Mol. Biol. 1991;4:295–300. doi: 10.1165/ajrcmb/4.4.295. [DOI] [PubMed] [Google Scholar]
- 10.Harden TK, Hawkins PT, Stephens L, Boyer JL, Downes P. Phosphoinositide hydrolysis by guanosine 5′([γ-thio]triphosphate)-activated phospholipase C of turkey erythrocyte membranes. Biochem. J. 1988;252:583–593. doi: 10.1042/bj2520583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirotton S, Bocynaems JM. Transduction mechanism of P2 purinergic receptor: Role of phospholipase C and Calcium. Nucleos. Nucleot. 1991;10:1009–1017. [Google Scholar]
- 12.Benham CD, Tsien RW. A novel receptor-operated Ca2+-permeable channel activated by ATP in smooth muscle. Nature. 1987;328:275–278. doi: 10.1038/328275a0. [DOI] [PubMed] [Google Scholar]
- 13.Bean BP. Pharmacology and electrophysiology of ATP activated ion channels. Trends Pharmacol. Sci. 1992;13:87–90. doi: 10.1016/0165-6147(92)90032-2. [DOI] [PubMed] [Google Scholar]
- 14.Fischer B, Boyer JL, Hoyle CHV, Ziganshin AU, Brizzolara AL, Knight GE, Zimmet J, Burnstock G, Harden TK, Jacobson KA. Identification of potent, selective P2Y-purinoceptor agonist structure-activity relationship for 2-thioether derivatives of adenosine 5′-triphosphate. J. Med. Chem. 1994;36:3937–3946. doi: 10.1021/jm00076a023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burnstock G, Fischer B, Hoyle CHV, Maillard M, Zinganshin AU, Brizzolara AL, Von Isakovics A, Boyer JL, Harden TK, Jacobson KA. Structure activity relationship for derivatives of adenosine 5′-triphosphate as agonists at P2 purinoceptors; heterogeneity within P2X and P2Y subtypes. Durg Dev. Res. 1994;31:206–219. doi: 10.1002/ddr.430310308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windscheif U, Ralevic V, Bäurnert HG, Mutschler E, Lambrecht G, Burnstock G. Vasoconstrictor and vasodilator by responses to various agonists in the rat perfused mesenteric arterial bed – Selective-inhibition by PPADS of contractions mediated Via P2X-purinoceptor. Br. J. Pharmacol. 1994;113:1015–1021. doi: 10.1111/j.1476-5381.1994.tb17094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphries RG, Tomlinson W, Ingall AH, Cage PA, Leff P. FPL-66096 – A Novel, highly potent and selective antagonist at human platelet P2T-purinoceptors. Br J. Pharmacol. 1994;113:1057–1063. doi: 10.1111/j.1476-5381.1994.tb17100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Rhee AM, van der Heijden MPA, Beukers MPA, IJzerman AP, Soudijin W, Nickel P. Novel competitive antagonists for P2 purinoceptor. Eur. J. Pharmacol. Mol. Pharmacol. Sect. 1994;268:1–7. doi: 10.1016/0922-4106(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 19.Kyte J, Doolittle RF. A Simple method for displaying the hydrophobic character of protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Kubo T, Fukuda K, Mikama A, Maeda A, Takahashi H, Mishina M, Haga Y, Ichiyama A, Kangawa K, Kajima M, Matsuo H, Hirose T, Numa T. Cloning, sequencing and expressing of complementary DNA coding the muscarinic acetylcholine receptor. Nature. 1986;323:411–416. doi: 10.1038/323411a0. [DOI] [PubMed] [Google Scholar]
- 21.Webb TE, Simon J, Krishek BJ, Bateson AN, Smart TG, King BF, Burnstock G, Barnard EA. Cloning and functional expression of a brain G-protein-coupled ATP receptor. FEBS Lett. 1993;324:219–225. doi: 10.1016/0014-5793(93)81397-i. [DOI] [PubMed] [Google Scholar]
- 22.Filtz TM, Li Q, Boyer JL, Nicholas RA, Harden TK. Expression of a P2Y purinergic receptor that couples to phospholipase C. Mol. Pharmacol. 1994;40:8–14. [PubMed] [Google Scholar]
- 23.Lustig KD, Shiau AK, Brake AJ, Julius D. Expression cloning of an ATP receptor from mouse neuroblastoma cells. Proc. Natl. Acad. Sci. USA. 1993;91:5113–5117. doi: 10.1073/pnas.90.11.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parr CE, Sullivan DM, Paradiso AM, Lazarowski ER, Burch LH, Olsen JC, Erb L, Weisman GA, Boucher RC, Tuner JT. Cloning and expression of a human P2U nucleotide receptor, a target for cystic fibrosis pharmacotherapy. Proc. Natl. Acad. Sci. USA. 1994;91:3275–3279. doi: 10.1073/pnas.91.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice WR, Burton FM, Fiedeldey DT. Cloning and expression of the alveolar type II cell P2U-purinergic receptor. Am. J. Respir. Cell Mol. Biol. 1995;12:27–32. doi: 10.1165/ajrcmb.12.1.7811468. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson KA, Stiles GL, Ji XD. Chemical modification and irreversible inhibition of striatal A2a adenosine receptors. Mol. Pharmacol. 1992;42:123–133. [PMC free article] [PubMed] [Google Scholar]
- 27.Hibert MF, Trumpp-kallmeyer S, Bruinvels A, Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol. Pharmacol. 1991;40:8–15. [PubMed] [Google Scholar]
- 28.Schertler GF, Villa C, Henderson R. Projection structure of rhodopsin. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 29.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the Structure of bacteriorhodopsin based on high-resolution electron crymicroscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira L, Paiva ACM, Vriend G. A common motif in G-protein-coupled seven transmembrane helix receptors. J. Comp. Aided Mol. Design. 1993;7:649–658. [Google Scholar]
- 31.Baldwin JM. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovács T, Ötvös L. Simple synthesis of 5-vinly and 5-ethynyl-2'-deoxyuridine-5'-triphosphates. Tetrahedron Lett. 1988;29:4525–4528. [Google Scholar]
- 33.Moffat JG. A general synthesis of nucleoside-5′-triphosphates. Can. J. Chem. 1964;42:599. [Google Scholar]
- 34.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Method Neurosci. 1995;25:366–428. [Google Scholar]
- 35.IJzerman AP, van Galen PJM, Jacobson KA. Molecular modeling of adenosine receptors. I. The ligand binding site on the A1 receptor. Drug Des. Discov. 1992;9:49–67. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Weinstein H. Polarity conserved positions in transmembrane domains of G-protein coupled receptors and bacteriorhodopsin. FEBS Lett. 1994;337:207–212. doi: 10.1016/0014-5793(94)80274-2. [DOI] [PubMed] [Google Scholar]
- 37.Sankararamakrishnan R, Vishveshwara S. Geometry of proline-containing alpha-helices in proteins. Int. J. Peptide Protein Res. 1992;39:356–363. doi: 10.1111/j.1399-3011.1992.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 38.Noel JP, Hamm HE, Sigler PB. The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature. 1993;366:654. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 39.Brake AJ, Wagenbach MJ, Julius D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature. 1994;371:519–523. doi: 10.1038/371519a0. [DOI] [PubMed] [Google Scholar]
- 40.Valera S, Hussy N, Evans RJ, Adami N, North RA, Surprenant A, Buell G. New class of ligand-gated ion-channel defined by P2X receptor or extracellular ATP. Nature. 1994;371:516–519. doi: 10.1038/371516a0. [DOI] [PubMed] [Google Scholar]
- 41.Erb L, Garrad R, Wang Y, Quinn T, Turner JT, Weisman GA. Site-directed mutagenesis of P2U purinoceptors. J. Biol. Chem. 1995;270:4185–4188. doi: 10.1074/jbc.270.9.4185. [DOI] [PubMed] [Google Scholar]
- 42.von Heijne G, Manoil C. Membrane proteins: from sequence to structure. Protein Eng. 1990;4:109–112. doi: 10.1093/protein/4.2.109. [DOI] [PubMed] [Google Scholar]
- 43.Sander C, Schneider R. Database of homology-derived protein structures. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 44.Hoflack J, Trumpp-Kallmeyer S, Hibert M. Re-evaluation of bacteriorhodopsin as a model for G protein-coupled receptors. Trends Pharmacol. Sci. 1994;43:348–350. doi: 10.1016/0165-6147(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 45.von Heijne G. Proline kinks in transmembrane α-helices. J. Mol. Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 46.Wess J, Gdula D, Brann MR. Site-directed mutagenesis of the m3 muscarinic receptor: identification of a series of threonine and tyrosine residues involved in agonist but not antagonist binding. EMBO J. 1991;10:3729–3734. doi: 10.1002/j.1460-2075.1991.tb04941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suryanarayana S, Daunt DA, von Zastrow M, Kobilka BK. A point mutation in the seventh hydrophobic domain of the α2 adrenergic receptor increases its affinity for a family of βreceptor antagonists. J. Biol. Chem. 1991;266:15488–15492. [PubMed] [Google Scholar]
- 48.Oksenberg D, Marsters SA, O’Dowd BF, Jin H, Havlik S, Peroutka SJ, Ashkenazi A. A single amino-acid difference confers major pharmacological variation between human and rodent 5-HT1B receptors. Nature. 1992;360:161–163. doi: 10.1038/360161a0. [DOI] [PubMed] [Google Scholar]
- 49.Fraser CM, Wang CD, Robinson DA, Gocayne JD, Venter JC. Site-directed mutagenesis of ml muscarinic acetylcholine receptors: conserved aspartic acids play important roles in receptor function. Mol. Pharmacol. 1989;36:840–847. [PubMed] [Google Scholar]
- 50.Elling CE, Møller Nielsen S, Schwartz TW. Conversion of antagonist binding site to metal-ion site in the tachykinin NK-1 receptor. Nature. 1995;374:74–77. doi: 10.1038/374074a0. [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Wess J, van Rhee AM, Schöneberg T, Jacobson KA. Site-directed mutagenesis identifies residues involved in ligand recognition in the human A2a adenosine receptor. J. Biol. Chem. 1995;270:13987–13997. doi: 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W, Flanagan C, Ballesteros JA, Konvicka K, Davidson JS, Weinstein H, Millar RP, Sealfon SC. A reciprocal mutation supports helix 2 and helix 7 proximity in the gonadotropin-releasing hormone receptor. Mol. Pharmacol. 1994;2:165–170. [PubMed] [Google Scholar]
- 53.Horstman DA, Brandon S, Wilson AL, Guyer CA, Cragoe EJ, Limbird LE. An aspartate conserved among G-protein receptors confers allosteric regulation of α2-adrenergic receptors by sodium. J. Biol. Chem. 1990;265:21590–21595. [PubMed] [Google Scholar]
- 54.IJzerman AP, van der Wenden EM, van Galen PJM, Jacobson KA. Molecular modeling of adenosine receptors – the ligand-binding site on the rat adenosine A2a receptor. Eur. J. Pharmacol. Mol. Pharmacol Sect. 1994;268:95–104. doi: 10.1016/0922-4106(94)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobson KA, Fischer B, van Rhee AM. Molecular probes for muscarinic receptors: functionalized congeners of selective muscarinic antagonists. Life Sci. 1995;56:823–830. doi: 10.1016/0024-3205(95)00016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyer JL, Harden TK, Fischer B, Jacobson KA. in preparation. [Google Scholar]