Abstract
N-hydroxy amides can be found in many naturally occurring and synthetic compounds and are known to act as both strong proton donors and chelators of metal cations. We have initiated studies of peptoids, or N-substituted glycines, that contain N-hydroxy amide side chains to investigate the potential effects of these functional groups on peptoid backbone amide rotamer equilibria and local conformations. We reasoned that the propensity of these functional groups to participate in hydrogen bonding could be exploited to enforce intramolecular or intermolecular interactions that yield new peptoid structures. Here, we report the design, synthesis, and detailed conformational analysis of a series of model N-hydroxy peptoids. These peptoids were readily synthesized, and their structures were analyzed in solution by 1D and 2D NMR and in the solid-state by X-ray crystallography. The N-hydroxy amides were found to strongly favor trans conformations with respect to the peptoid backbone in chloroform. More notably, unique sheet-like structures held together via intermolecular hydrogen bonds were observed in the X-ray crystal structures of an N-hydroxy amide peptoid dimer, which to our knowledge represent the first structure of this type reported for peptoids. These results suggest that the N-hydroxy amide can be utilized to control both local backbone geometries and longer-range intermolecular interactions in peptoids, and represents a new functional group in the peptoid design toolbox.
Introduction
Peptoids, or oligomers of N-substituted glycines, represent a versatile class of biologically relevant foldamers.1,2 These non-native oligomers have been found to recognize and bind to proteins,3–10 mimic peptidic lung surfactants,11–13 serve as molecular transporters across cell membranes,14–18 and act as antimicrobial agents.19–25 In addition, peptoids are resistant to proteolytic degradation,26,27 and are easily synthesized in a modular fashion by solid-phase techniques with access to a nearly limitless diversity of amide side chains.28,29 These promising features have catalyzed significant recent research in the peptoid field.2,30
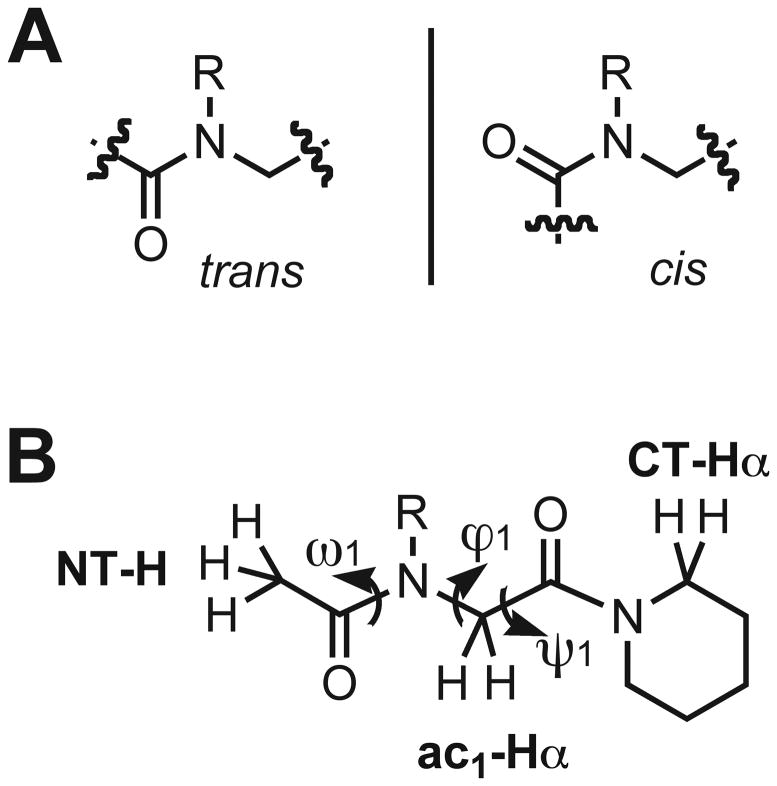
However, relative to other foldamer systems, predicting the secondary structure of peptoid oligomers a priori remains a major challenge.31–34 This problem originates in the peptoid backbone. First, there are no hydrogen bond donors or C-α substituents within the peptoid backbone, which are two major contributors to the folding of α-peptides.35–37 Second, there is often a negligible energetic difference between the cis and trans rotamers of peptoid backbone tertiary amides with N-alkyl substituents (Figure 1A).38–41 Therefore, extensive conformational heterogeneity is possible within peptoids. Indeed, the limited number of distinct and conformationally stable peptoid secondary structures, and primary sequences known to engender them, is in stark contrast with the boundless variety of side chain functionality that can be incorporated into peptoids.42–48
Figure 1.
(A) Depiction of trans- versus cis-amide peptoid rotamers. (B) Chemical structure of our peptoid monomer model system that was designed to isolate local non-covalent interactions between amide side chains and the peptoid backbone. Backbone atom and dihedral angle nomenclature is indicated as previously described for peptoids.42,49
Peptoids capable of adopting well-defined secondary and tertiary structures clearly would be valuable for a range of fundamental and applied studies. Within this broad context, there is significant interest in identifying peptoid side chains capable of restricting the rotation of local backbone dihedral angles and thereby enforcing specific conformational preferences in peptoid oligomers. The strategic installation of peptoid side chains capable of participating in distinct non-covalent interactions that influence peptoid amide rotameric equilibria (i.e., ω angles, Figure 1B) represents one approach toward this end, and has been a focus of several recent studies in our laboratory and others.38,39,45,46,48,50–53 In 2010, we demonstrated that N-aryl side chains with hydrogen bond donors at the ortho position of the aryl ring can form discrete intramolecular hydrogen bonds with backbone amides in peptoids, enforce exclusively trans backbone amides, and potentially influence the phi (ϕ) and psi (ψ) bond angles (Figure 1B).47 This recent study inspired us to examine the ability of other amide side chains containing hydrogen bond donors to influence peptoid backbone conformations and the formation of higher order structures. Conceptually, one of the simplest hydrogen bond donating peptoid side chain is an N-hydroxy amide (Figure 2A), and we selected this functional group for analysis in the current study.
Figure 2.
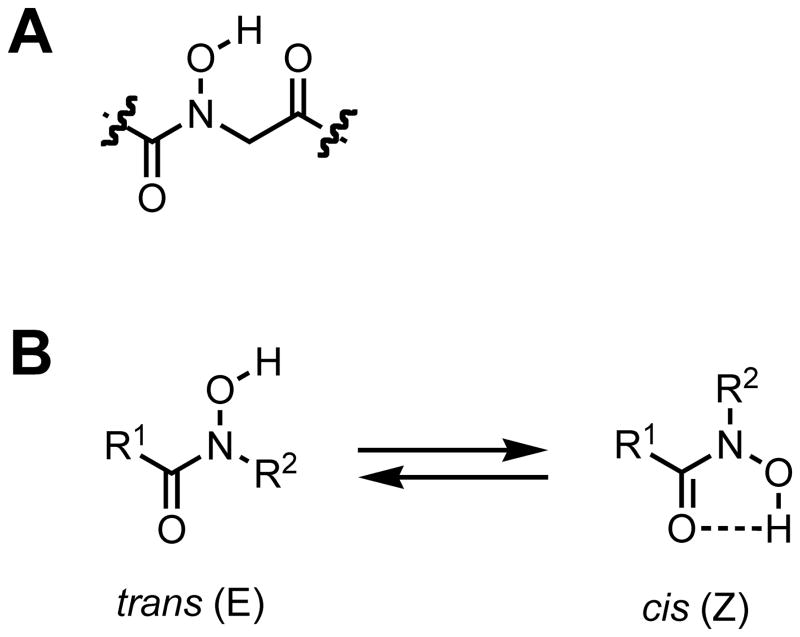
(A) An N-hydroxy amide peptoid monomer unit. (B) Two hydroxamic acid isomers defined as trans (E) and cis (Z).
N-hydroxy amides, or hydroxamic acids, were discovered in 1869 by Lossen.54 Since then, both naturally occurring and synthetic hydroxamic acid derivatives have been studied extensively, largely due to their ability to chelate a variety of metals.55 Structural studies of these complexes have established that the typical hydroxamate binding mode to most transition metals is by O, O′ bidentate coordination to the metal, requiring the cis (Z) geometry of the amide bond.56–58 The conformational preferences of hydroxamic acids in the absence of metal ions depends largely on the nature of N-substituents, acyl groups, and solvent.59 For example, if either the acyl group or the nitrogen is unsubstituted (Figure 2B; R1 or R2 = H), then the cis hydroxamic acid is predominately favored.59–63 This conformation is stabilized through a weak intramolecular hydrogen bond that forms a 5-membered ring. However, substitution on both the acyl group and nitrogen can cause the trans (E) isomer to be favored, especially when sterically bulky substituents or competing hydrogen bond acceptors are present (Figure 2B).59,62 The first of such structures were reported by Smith and Raymond in 1980, who found that the two hydroxamic acids in N,N′-dihydroxy-N,N′-diisopropylhexanediamide were trans in the solid state.64 Subsequent experimental and computational methods have shown that N-hydroxylated peptides exist almost exclusively as trans isomers.65–67 In addition, N-hydroxy peptides are known to be strong hydrogen bond donors, and the O-H bonds in these systems have been found to extend perpendicular to the amide plane.64–66,68 Collectively, these findings suggested that the installation of N-hydroxy amide side chains into peptoids could impact their local backbone conformations by both enforcing trans amides and engaging in unique hydrogen bonds. We envisaged that the latter interactions could allow for the formation of novel peptoid secondary and tertiary structures through intramolecular and/or intermolecular associations.
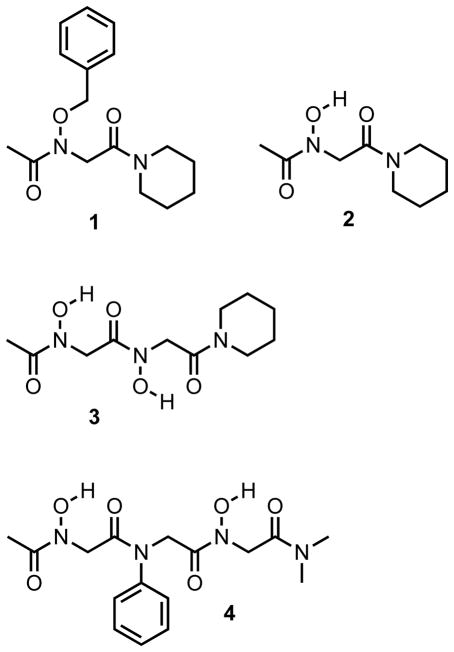
Herein, we report our initial investigations of peptoids containing N-hydroxy amides. A series of simple peptoid model systems (1–4) were designed and synthesized in solution (Figure 3). We were specifically interested in identifying the cis/trans preferences of the N-hydroxy amides in these model systems, as well as determining if discrete intramolecular or intermolecular hydrogen bonds would be formed. The solution-phase conformations of peptoids 1–4 were examined in organic solvent using 1D 1H and 2D magnetization exchange NMR experiments. The solid-state conformations of peptoids 2–4 were examined by X-ray crystallography. Overall, our studies of N-hydroxy amide model systems indicate that this side chain can enforce trans amide geometries in peptoids in chloroform and in the solid state. More notably, unique sheet-like structures were observed in the X-ray crystal structures of an N-hydroxy amide peptoid dimer (3), which to our knowledge represents the first structure of this type reported for peptoids.
Figure 3.
Structures of the N-hydroxy amide containing peptoids (1–4) evaluated in this study.
Results and Discussion
Peptoid Design and Synthesis
We sought to isolate and understand the conformational preferences of N-hydroxy amides in simple peptoid model systems. Previous studies in our laboratory have shown that a peptoid monomer with appropriate N-terminal (acetyl) and C-terminal (piperidinyl) capping groups is a useful tool for the study of local non-covalent interactions of individual side chains (shown in Figure 1B).38,39 Therefore, we utilized this monomeric scaffold and longer homologs in the present study.
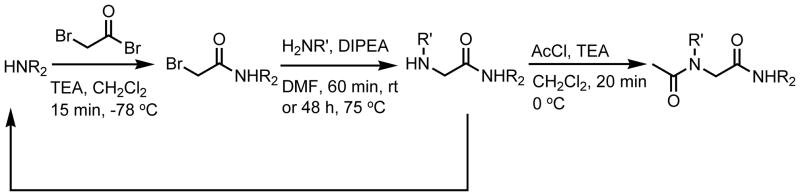
The O-benzyl protected monomer 1 is the synthetic precursor of the N-hydroxy monomer of interest (2; Figure 3), and was designed as a non-hydrogen bonding control compound for 2. Monomers 1 and 2 were synthesized using solution phase techniques that were analogous to the standard solid-phase submonomer synthesis of peptoids.28 Our general synthetic route is shown in Scheme 1. Briefly, the desired C-terminal capping secondary amine (piperidine) was acylated using bromoacetyl bromide, followed by displacement of the α-bromide with O-benzyl hydroxylamine. This afforded the uncapped O-benzyl protected monomer, which was acetylated to yield target monomer 1. Hydrogenation of 1 using Pd/C cleanly removed the benzyl group and gave monomer 2. Coupling of protected hydroxylamine was necessary in order to avoid disproportionation of the resulting secondary hydroxylamine, or subsequent acylation of the oxygen.69–73 We note that the reactivity of O-benzylhydroxylamine was substantially diminished compared to other primary amines typically used in the synthesis of peptoids, requiring ~48 h reaction times at 75 °C. This observation is consistent with other reports in the literature (see Experimental Section for further synthetic details).73,74
Scheme 1.
General route for the synthesis of peptoids 1–4.a
a Reagents and conditions: HNR2 = piperidine or dimethylamine hydrochloride, TEA = triethylamine, CH2Cl2 = dichloromethane, H2NR′ = O-benzyl hydroxylamine hydrochloride or aniline, DIPEA = diisopropylethylamine, DMF = N,N-dimethylformamide, rt = room temperature, AcCl = acetyl chloride.
Peptoids 3 and 4 (Figure 3) were designed as model compounds for homo and hetero N-hydroxy amide peptoid oligomers, respectively. Homodimer 3 is the dimeric analog of the N-hydroxy amide monomer 2. Heterotrimer 4 consists of two terminal N-hydroxy amide residues (i and i+2) flanking an N-phenyl residue at position two (i+1). As N-aryl side chains predominantly enforce trans amide isomers in peptoids,46,47 we reasoned that heterotrimer 4 would have reduced conformational heterogeneity, and this would facilitate its conformational analysis. The N-dimethyl amide was used as a C-terminal capping group in trimer 4 instead of a piperidinyl amide to simplify its 1H NMR spectrum. Model peptoids 3 and 4 were synthesized according to similar procedures as described for monomers 1 and 2 above (Scheme 1). The uncapped O-benzyl protected monomer was resubmitted to acylation with bromoacetyl bromide to generate the α-bromoacetamide monomer. Subsequent nucleophilic displacement/acylation steps provided the O-benzyl protected derivatives of dimer 3 and trimer 4. These intermediates were then hydrogenated to yield the target peptoids (see Experimental Section). The deprotected peptoids 2–4 were found to be relatively polar and soluble in both polar organic solvents and water, with dimer 3 exhibiting the highest degree of water solubility relative to monomer 2 and heterotrimer 4. This solubility trend indicates that the N-hydroxy amide side chain can heighten the polarity of peptoids.
Conformational Analysis of Peptoids 1 and 2 in Solution
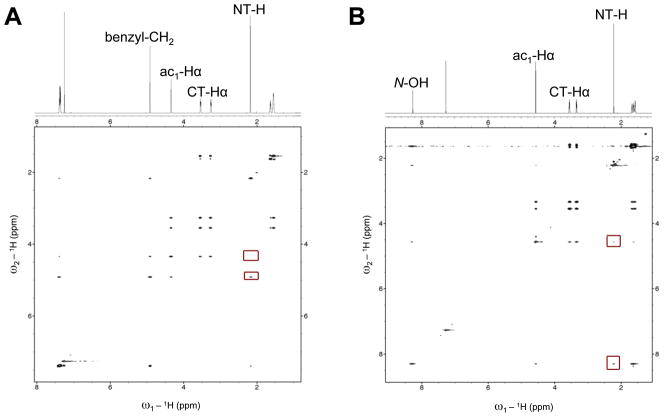
We analyzed the conformations of the N-O-benzyl and N-hydroxy peptoid monomers (1 and 2) using standard NMR techniques at room temperature. As is routinely done in peptoid analyses, the presence or absence of multiple conformations was determined by 1D 1H NMR spectroscopy, and the geometry of the N-OR (R = benzyl or H) amide was established via 1H–1H nuclear Overhauser effect spectroscopy (NOESY).49 Specifically for our monomer systems, a cis N-OR amide isomer was predicted to display an NOE between the N-terminal methyl (NT-H) and the backbone methylene (ac1-Hα) protons, while an NOE involving the N-terminal methyl (NT-H) and side chain protons was expected for the trans N-OR amide isomer (see Figure 1B for nomenclature).
The 1H NMR spectrum of monomer 1 in CDCl3 showed that one N-O-benzyl amide rotamer was favored exclusively (Figure 4A). Two-dimensional NOESY experiments of monomer 1 firmly established that the N-O-benzyl amide was in the trans conformation. Figure 4A illustrates the NOE between the NT-H methyl and the side chain benzyl methylene protons, and correspondingly, the lack of an apparent NOE between the NT-H methyl and ac1-Hα methylene protons. (For additional investigations of the conformational preferences of related N-alkoxy polypeptoids, we direct the reader to the report of Kirshenbaum and co-workers published in this issue.[ref to be added later])
Figure 4.
(A) Partial 1H NMR and NOESY spectra of peptoid 1 (600 MHz, CDCl3, 5 mM, 24 °C). Red boxes highlight the presence of an NOE between the NT-H—side chain methylene protons and the lack of an observed NOE between NT-H—ac1-Hα. (B) Partial 1H and NOESY spectra of peptoid 2 (600 MHz, CDCl3, 5 mM, 24 °C). Red boxes highlight the presence of an NOE between NT-H—N-OH and a weak NOE between NT-H—ac1-Hα.
Similar 1D and 2D NMR studies revealed that monomer 2, analogous to 1, was exclusively the trans isomer in CDCl3 at room temperature (Figure 4B). An NOE was observed in 2 involving the NT-H and the side chain N-hydroxyl protons, consistent with predicted expectations for the trans isomer. Interestingly though, a small cross peak was also observed between the NT-H methyl and backbone ac1-Hα methylene group, which was originally expected only for the cis isomer (as described above). The strength of the weak NOE was compared by integration to the NOE between the ac1-Hα protons and the α-methylene protons of the piperidinyl group at the C-terminus (CT-Hα), as these two groups are necessarily cis to one another. The small NT-H—ac1-Hα cross peak was 22 times less intense by integration than the cis ac1-Hα—CT-Hα cross peak. These data suggested that weak NOEs were observable between the N-terminal methyl and backbone methylene groups in the trans isomer of 2. We recognize that our NOESY experiments were conducted with relatively lengthy mixing times and d1 values, due to the long T1 relaxation times of the protons of interest; therefore, the appearance of such weak NOEs could be expected. We return to this issue below upon evaluating the structure of 2 in the solid state.
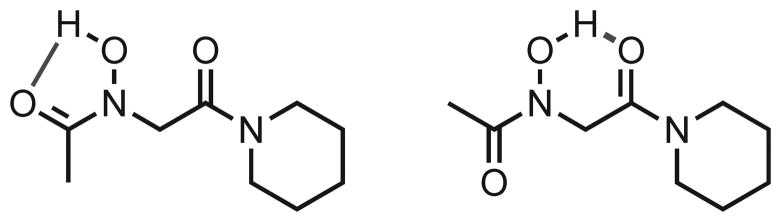
The N-hydroxyl proton in monomer 2 was assigned to the singlet at 8.26 ppm (Figure 4B); such a downfield chemical shift is indicative of hydrogen bonding. Moreover, this chemical shift was independent of sample concentration between 0.1–100 mM in CDCl3 (data not shown), suggesting that monomer 2 engages in an intramolecular hydrogen bonding interaction between the side chain N-hydroxyl proton and the C-terminal carbonyl oxygen in CDCl3 (Figure 5). We note that 2 can potentially form two distinct intramolecular hydrogen bonds (depicted in Figure 5). A hydrogen bond to the N-terminal carbonyl oxygen would require the N-hydroxy amide to be cis and would form a 5-membered ring. The other possible hydrogen bond would form a 6-membered ring to the C-terminal carbonyl oxygen, permitting the N-hydroxy amide to adopt either the cis or trans conformation. As peptoid 2 was found to adopt exclusively the trans rotamer in CDCl3, the proposed intramolecular hydrogen bond must exist between the side chain N-hydroxyl proton and the C-terminal carbonyl oxygen forming a 6-membered ring.
Figure 5.
Two hypothetical intramolecular hydrogen bonds (green lines) that could form in peptoid 2. (Left) Hydrogen bond to the N-terminal carbonyl. (Right) Hydrogen bond to the C-terminal carbonyl.
Conformational Analysis of Peptoid 2 in the Solid State
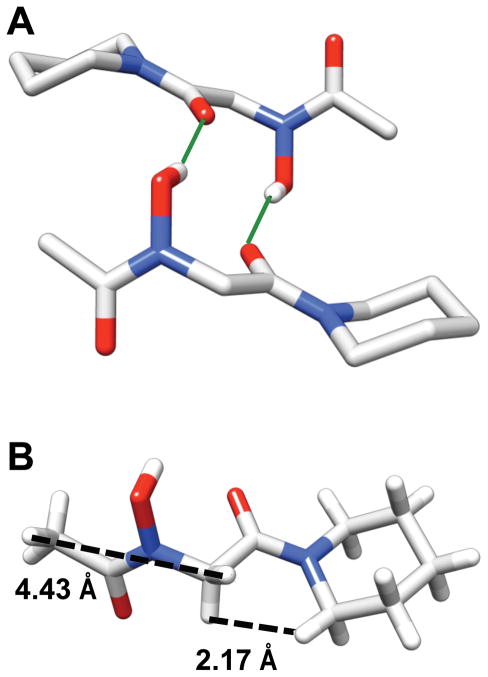
We examined the solid-state conformation of peptoid monomer 2 using single crystal X-ray crystallography. Slow dissolution of 2 from CHCl3/n-hexanes afforded large plate-like crystals for solid-state structure determination. Figure 6A depicts a view of the X-ray crystal structure of 2, and Table 1 lists key backbone dihedral angles. The structure revealed the N-hydroxy amide in monomer 2 to adopt a trans conformation, analogous to our observations in CDCl3 outlined above. Also, similar to previous reports of hydroxamic acid structures, the N-hydroxyl oxygen atom had significant sp3 character (N-O-H bond angle = 105°), with the O-H bond oriented perpendicular to the amide plane.66,68
Figure 6.
Views of an X-ray crystal structure of peptoid 2. (A) Two molecules of 2 participating in an intermolecular hydrogen bond [solid green lines indicate the H-bonds; both intermolecular O • • • O distances = 2.071Å]. Hydrogens are omitted for clarity except for the amide side chain N-OH. (B) A single molecule of 2 illustrating the intramolecular distances between NT-H—ac1Hα and ac1Hα—CT-Hα protons [dashed black lines].
Table 1.
Dihedral angles observed in the X-ray crystal structures of peptoids 2–4
| Peptoid | Residue | ω (°) | ϕ (°) | ψ (°) |
|---|---|---|---|---|
| 2 | N-OH (i) | −178.71 | 96.92 | 175.44 |
| 3 | N-OH (i) | −169.90 | −117.79 | 168.58 |
| N-OH (i+1) | −176.03 | 110.28 | 178.23 | |
| 4 | N-OH (i) | −172.44 | 95.65 | 160.73 |
| N-ph (i+1) | 170.49 | −95.90 | 174.14 | |
| N-OH (i+2) | 164.29 | −89.42 | 173.04 |
Interestingly, monomer 2 dimerized in a head-to-tail pattern in the solid state through two identical intermolecular hydrogen bonds between the N-hydroxyl proton of one monomer and the C-terminal carbonyl oxygen of the other (Figure 6A and Supporting Information, Table S-2). This dimerization complicates our initial interpretations of the NMR data for 2, which suggested that the hydrogen bonding was intramolecular in solution. We note that peptoid could potentially be dimeric over the concentration range investigated in our solution-phase NMR studies described above (0.1 mM – 100 mM). The fact that the same atoms are involved in intermolecular hydrogen-bonding interactions in the solid state that we predict are involved in intramolecular interactions in solution further complicates our understanding of the nature of the hydrogen bonding pattern of 2. Diffusion ordered NMR spectroscopy (DOSY) experiments represent a possible method for further interrogation of this phenomenon in solution, and represent a current focus of investigation in our laboratory.
Additional scrutiny of the solid-state structure of monomer 2 helped to clarify our interpretation of its NOESY data (see above). Despite the trans N-hydroxy amide in 2, the distance between the ac1-Hα methylene protons and the nearest proton of the NT-H methyl group in the crystal structure is ~4.4 Å due to the dihedral angle of ϕ1 (Figure 6B; Table 1). This distance falls within the range expected for a weak NOE, and supports our assertion that peptoid 2 is the trans isomer in chloroform. The distance between ac1Hα—CT-Hα, which are cis to one another across the C-terminal amide, was ~2.2 Å, and substantiates the strong NOE observed between these groups in the NOESY data discussed above. Collectively, these NMR and X-ray crystallographic data strongly suggest that peptoid 2 is exclusively the trans isomer in both chloroform and the solid state.
Conformational Analysis of Peptoid 3 in Solution
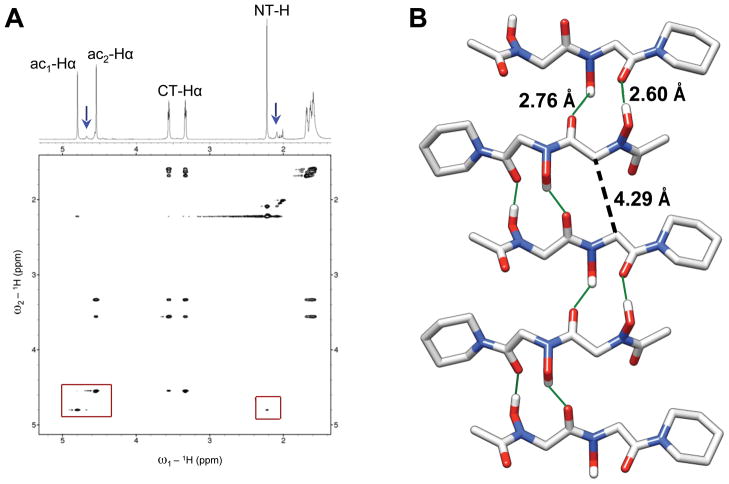
Homodimeric peptoid 3 was designed to examine the effects of N-hydroxy amides on the backbone conformations of homooligomer systems. NMR analyses of 3 were performed in CDCl3; however, we note that the solubility of 3 was limited to less than 8 mM in this solvent. Close inspection of the 1H NMR spectrum revealed a set of minor conformational peaks (blue arrows, Figure 7A). Although these peaks were poorly dispersed from the major conformational peaks, the ratio of the major:minor conformer was determined to be 6:1 by integration (see Supporting Information).
Figure 7.
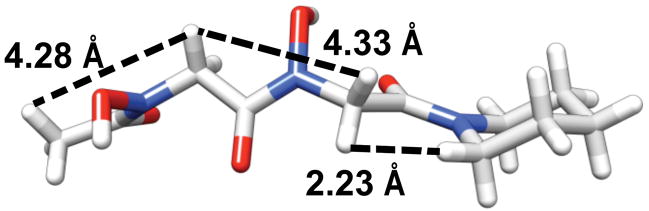
(A) Partial 1H NMR and ROESY spectra of peptoid 3 (600 MHz, CDCl3, 4 mM, 24 °C). The blue arrows point to the observable minor conformational peaks and the red boxes highlight a weak ROE between the NT-H and ac1-Hα backbone protons (right box), as well as the absence of an observed ROE between the ac1-Hα and ac2-Hα backbone protons (left box). (B) X-ray crystal structure depicting five molecules of peptoid 3 in an accordion-like hydrogen bonded network. The solid green lines indicate H-bonds with the intermolecular O • • • O distances provided. The black dashed line indicates the intermolecular ac1-Cα—ac2-Cα distance. All hydrogens except for the amide side chain N-OHs are omitted for clarity.
A 2D NOESY experiment in CDCl3 was initially conducted in order to facilitate assignments of peaks and determine whether the N-hydroxy amides were trans or cis in the major conformation of 3 in solution. However, characteristic NOEs between methylene protons of the C-terminal piperidinyl ring were absent in the resulting spectrum, which lead us to pursue rotating frame Overhauser effect spectroscopy (ROESY) for this analysis.75,76 The molecular weight of peptoid 3 (273.29 g/mol) is well below the weight of molecules typically considered for ROESY experiments.77 Nevertheless, as evidenced by the severely reduced solubility of peptoid 3 in CDCl3, it is reasonable to assume that 3 could be aggregating in this solvent even at low concentration, thereby affecting the molecular tumbling of this peptoid in solution and causing it to behave like a higher molecular weight compound.
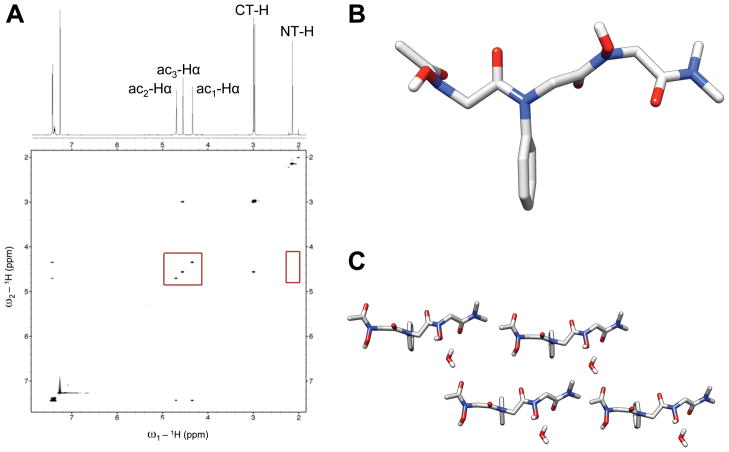
Partial 1H NMR and ROESY data of homodimer 3 in CDCl3 at 4 mM are shown in Figure 7A, with the major conformational peaks labeled on the 1H spectrum. The two backbone methylene groups were distinguishable from one another based on ROE correlations. The ac2-Hα group was assigned to the singlet at 4.50 ppm due to the strong cross peaks involving this singlet and the CT-Hα multiplets. The ac1-Hα group was then designated as the singlet at 4.67 ppm. The side chain N-hydroxyl protons of 3 were determined to be participating in hydrogen bonds in CDCl3, from their downfield chemical shifts of 7.55 and 8.59 ppm (see Supporting Information for full 1H spectrum of 3). Variable concentration NMR studies were not permissible due to poor solubility in this solvent, again highlighting the intermolecular component to the nature of the hydrogen bonding in 3.
Similar to our analyses of monomers 1 and 2 above, ROESY data were used to determine cis/trans preferences of the N-hydroxy amides in homodimer 3. ROEs were not expected between minor conformational peaks due to conformational exchange during the ROESY pulse sequence, and only correlations between major peaks were observed. Given our knowledge of the conformational bias of peptoid 2, we expected that trans N-hydroxy amides would be favored in the homologous dimer 3. No ROE was detected between the ac1-Hα—ac2-Hα backbone methylenes. This observation suggests that, within our limits of detection, the i+1 N-hydroxy amide in peptoid 3 is not cis. A weak correlation was observed between NT-H—ac1-Hα, which was 6.3 times less intense by integration than the ROE between ac2-Hα—CT-Hα of 3 (a necessarily cis relationship in this molecule), and may represent a weak “trans-indicating ROE” across the N-terminal N-hydroxy amide of peptoid 3. However, ROEs were not observed between the side chain N-hydroxyl protons to either the NT-H or backbone ac1-Hα protons (side chain protons are not included within the partial view of the ROESY data in Figure 7A). This apparent lack of backbone to side chain correlation is difficult to rationalize, considering an NOE between the NT-H and the side chain N-hydroxyl proton was observed in monomer 2 (see above).
Conformational Analysis of Peptoid 3 in the Solid State
Crystallization of peptoid dimer 3 by slow evaporation from methanol afforded single crystals that were suitable for structure determination. X-ray crystallographic analysis revealed 3 to adopt a structure analogous to the solid-state structure of monomer 2, in that both amides are trans. Perhaps more interestingly, however, dimer 3 was found to pack into one-dimensional, accordion-like pleated strands in the crystal through intermolecular hydrogen bonds extending in opposite directions (Figure 7B). Individual peptoid backbones were arranged antiparallel to one another and neighboring backbones were held in close proximity with an intermolecular distance between adjacent ac1-Cα—ac2-Cα equal to 4.29 Å. The ψ1 and ψ2 dihedral angles of dimer 3 are nearly identical to ψ1 of monomer 2 (Table 1). However, the ϕ1 (−117.79 °) and ϕ2 (110.28 °) angles of dimer 3 are not rotated out of planarity (180 °) to the same degree as the ϕ1 angle of monomer 2 (96.92 °), and the ϕ angles of 3 are opposite to one another in sign, thereby causing both N-hydroxyl side chains of individual dimers to be on the same face of the resulting sheets. The novel structure adopted by peptoid 3 in the solid state is noteworthy, because to our knowledge it represents the first hydrogen bonded, sheet-like structure to be reported for peptoids. This structure, although generated only from a peptoid dimer, is complementary to the recent development of two-dimensional, crystalline peptoid nano-sheets reported by Zuckermann and co-workers, which were held together in one dimension by electrostatic interactions, and in the second dimension via hydrophobic/aromatic interactions.78 Our structural data indicate that the N-hydroxy amide side chain may be an additional new tool for the generation of discrete peptoid structures coordinated through intermolecular noncovalent interactions.
We further analyzed the X-ray crystal structure of peptoid 3 to gain insight into its ROESY data discussed above. In the solid-state structure, the N-hydroxy amide separating the NT-H—ac1Hα protons in 3 was trans, and yet the distance between these groups was ~4.3 Å due to the ϕ1 angle (Figure 8; Table 1). As mentioned above, a weak ROE was observed involving these protons and was compared against the strong ROE observed between the ac2Hα—CT-Hα protons, which were ~2.2 Å apart in the crystal structure. These distances obtained from the X-ray crystal data are in agreement with our interpretation of the major conformation of 3 in solution. We note that the intramolecular distance in the crystal structure between backbone ac1Hα—ac2Hα protons (~4.3 Å) was also within the distance one would expect to observe the buildup of weak ROE cross peaks, (Figure 8). However, no ROE was present between these backbone protons in the ROESY data of 3.
Figure 8.
A view of a single molecule of peptoid 3 illustrating the intramolecular distances between the NT-H—ac1Hα, ac1Hα—ac2Hα, and ac2Hα—CT-Hα protons in the X-ray crystal structure [dashed black lines].
Conformational Analysis of Peptoid 4 in Solution
Peptoid heterotrimer 4 was designed as a model to examine the effects of N-hydroxy amides in heterooligomeric systems. N-hydroxy amides were placed at the i and i + 2 residues and an N-phenyl amide at the i + 1 residue. Peptoid 4 was predicted at the outset to have relatively low conformational heterogeneity, as N-aryl peptoid residues are known to exist as trans amides.43,46,47 1H NMR analyses of 4 in CDCl3 revealed it to adopt a single conformation (Figure 9A). Two-dimensional NOESY data were analyzed in order to facilitate assignments of peaks and to determine the rotameric states of the three unsymmetrical amides in peptoid 4 (Figure 9A). The ac3-Hα group was assigned as the singlet at 4.56 ppm due to its strong NOE to the CT-Hα groups. Distinguishing between ac1-Hα and ac2-Hα, however, was more challenging. Both singlets at 4.35 and 4.71 ppm exhibited NOEs to the ortho protons of the N-phenyl side chain. Furthermore, no NOEs were observed to either of the N-hydroxyl protons (not included within the partial view of the NOESY data in Figure 9A; see Supporting Information for the full 1H spectrum of 4 in CDCl3). Backbone ac1-Hα and ac2-Hα were labeled in Figure 9A solely based on their chemical shift. Side chain N-hydroxyl protons were assigned to the broad singlets at 8.10 and 8.67 ppm; their downfield shifts indicate that these protons are participating in hydrogen bonds in CDCl3. We note that peptoid 4 was highly soluble in CDCl3, analogous to monomer 2, and that there was no change in the chemical shifts of the N-hydroxyl protons over concentrations of 0.1–100 mM (data not shown).
Figure 9.
(A) Partial 1H NMR and NOESY spectra of peptoid 4 in (600 MHz, CDCl3, 5 mM, 24 °C). The red boxes highlight the lack of an NOE between the NT-H and ac1-Hα backbone protons (right box), as well as the lack of an NOE between any of the ac-Hα backbone protons (left box). (B) View of the X-ray crystal structure of 4 without the co-crystallized water molecule. (C) A packing diagram of four molecules of 4 from the X-ray crystal data including water molecules. All hydrogens are omitted for clarity except for water molecules and amide side chain N-OHs.
Considering our analyses of model peptoids 2 and 3 above and the reported rotameric preferences of N-aryl peptoids, the three unsymmetrical amides in trimer 4 were predicted to be trans. Figure 9A shows that no NOEs were observed between any of the backbone methylenes, and that no NOE was observed to the NT-Hα methyl peak. An NOE between ac2-Hα and the side chain phenyl protons unambiguously identified the N-phenyl residue as trans; however, no backbone—side chain NOEs were observed in the N-hydroxy residues i and i + 2. The lack of NOEs between the side chain hydroxyl protons and backbone protons of peptoid 4 is similar to the absence of analogous ROEs in homodimer 3.
Interestingly, a weak NOE was not observed between NT-H—ac1-Hα in 4. This correlation was present in both 2 and 3. We postulate that strong intermolecular hydrogen bonding interactions in o2 and 3 in organic solvents could enforce conformations in which the NT-H and ac1-Hα protons are close enough in space to generate NOE cross peaks. In contrast, peptoid 4 may not be aggregating in the same fashion, which could allow intramolecular hydrogen bonds to form and potentially enforcing a conformation that prevents the buildup of an NOE between the NT-H and ac1-Hα protons.
Conformational Analysis of Peptoid 4 in the Solid State
Crystals of trimer 4 were obtained by slow evaporation from 1-propanol under atmospheric conditions and analyzed by X-ray crystallography. Figure 9B shows the solid-state structure determined for trimer 4. Under our crystallization conditions, one molecule of water surprisingly co-crystallized with each peptoid molecule. We had initially expected trimer 4 to adopt a dimerized structure reminiscent of 2 that would be held together via interactions between the two N-hydroxy amide residues. All of the amides in 4 were trans and the backbone dihedral angles maintained a linear conformation; however, trimer 4 was not dimeric and there were no intermolecular hydrogen bonds between N-hydroxy amides. Instead, the intermolecular hydrogen bonding nature of 4 in the solid state appeared to be significantly influenced by the presence of water molecules. The water was hydrogen bonded to the C-terminal carbonyl oxygen of one trimer and to the N-terminal acetyl oxygen of another timer. We note that the ϕ angles of the N- and C-terminal N-hydroxy amide residues are opposite in sign (see Table 1). If the C-terminus was rotated by 180° about ϕ3, this may facilitate antiparallel dimerization of 4 by aligning the carbonyl oxygens of the i and i+3 residues; however, the co-crystallized water molecules appear to be preventing this from occurring in this solid-state structure of 4. Ongoing work is directed at crystallization of trimer 4 from alternate solvents to address this interesting hypothesis, and explore its capacity to form sheet-like structures in the solid state.
Summary and Outlook
The ability to synthesize peptoids capable of adopting predetermined and discretely folded structures could enhance their utility for a multitude of applications. In this study, we have demonstrated that N-hydroxy amides have a strong preference for trans amide rotamers in peptoid model systems in both organic solvents and in the solid state. We also note that N-alkoxy amides reside in the trans conformation, as reveled by our study of O-benzyl monomer 1. Additional studies supporting and expanding upon these data are reported by Kirshenbaum and co-workers in this issue.[to be addedref]
One of the most significant outcomes of this study was the discovery that N-hydroxy amide side chains are capable of promoting novel sheet-like structures in peptoids. We present X-ray crystallographic data for dimer 3 that provide a view of these extended peptoid strands. Prior to this study, there have been no reports to our knowledge of peptoids that can mimic the β-sheet-type structures prevalent in peptides and proteins. This absence is perhaps not surprising because β-sheet structures are dependent on both the hydrogen bonding capabilities of secondary amides and the trans nature of peptide bonds, features that are either absent or poorly populated, respectively, in peptoids.36 Our results indicate that incorporating N-hydroxy amides into polypeptoids may be a potential design strategy for generating peptoid-sheets.
We recognize that peptoids 2–4 are short model peptoids, and further studies of longer oligomers are essential to thoroughly define and characterize such novel peptoid sheets. Current research efforts in our laboratory seek to determine the isomeric equilibria and spectral properties of the N-hydroxyl side chain in a variety of solvents. As mentioned, peptoids containing N-hydroxy amides are highly polar, and can be soluble in polar organic solvents and water. Additional investigations are focused on exploring the ability of N-hydroxy amides to stabilize peptoid secondary structures, sheet-like and beyond, in longer peptoid oligomers. The results of these ongoing studies will be reported in due course.
Experimental Methods
Materials
All reagents were purchased from Aldrich and used without further purification. Solvents were purchased from commercial sources (Aldrich and Acros) and used as is, with the following exceptions: dichloromethane (CH2Cl2) and hexanes were distilled immediately prior to use, and N,N-dimethylformamide (DMF) was dried/stored over 3 Å molecular sieves for at least 24 h prior to use. All glassware was flame-dried and cooled under N2 prior to reactions. Thin layer chromatography (TLC) was performed on silica gel 60 F254 plates (E-5715-7, EMD). SiliaFlash® P60 silica gel (40–63 μm, Silicycle) was used for manual flash column chromatography.79
Spectroscopy
NMR spectra were recorded on a Varian Inova 600 (for final peptoid products: 1H, NOESY/ROESY, 600 MHz), a Bruker AC-300 (for all intermediates: 1H, 300 MHz), or a Varian MercuryPlus 300 (for all intermediates and products: 13C, 75 MHz) spectrometer. Data for final peptoid products were recorded at known concentrations (5–10 mM) in deuterated solvents at 297K. Chemical shifts are reported in parts per million (ppm, δ) and referenced to solvent or tetramethyl silane (TMS, 0.0 ppm).
2D NOESY or 2D ROESY spectra were used to identify cis or trans amide rotameric peaks, as reported in previous studies.38,39 All 2D NMR experiments were recorded in phase transfer mode with the following parameters. The number of points collected was 2048, and 192 points were obtained by linear prediction in the f1 dimensions. Square cosine window functions were applied in both dimensions. The spectra were zero-filled to yield f1f2 matrices of 4096 × 4096 before Fourier transformation. Table 2 lists parameters that were specific for each experiment. Mix times were calculated to be 0.4–0.6 x T1 for the protons of interest, and d1 values were set to 2.5–3.0 x T1 for the protons of interest. The data were processed using the Varian VNMR software package (v. 6.1C) and visualized using SPARKY software.80
Table 2.
Selected parameters used for NOESY and ROESY experiments
| Peptoid | Solvent | Spectral width (Hz) | nt | ni (s) | Mix time (s) | d1 (s) |
|---|---|---|---|---|---|---|
| 1 | CDCl3 | 5040 | 8 | 200 | 2.0 | 19.0 |
| 2 | CDCl3 | 5610 | 8 | 220 | 2.0 | 19.0 |
| 3 | CDCl3 | 3540 | 16 | 150 | 2.0 | 14.0 |
| 4 | CDCl3 | 6100 | 8 | 300 | 2.0 | 13.0 |
Mass spectrometry was performed with a Waters (Micromass) LCT™ instrument equipped with a time-of-flight analyzer (electrospray ionization, ESI). Samples were dissolved in methanol and sprayed with a sample cone voltage of 20. Perfluorokerosene (PFK) was used for calibration. Attenuated total reflectance (ATR)-IR spectra were recorded with a Bruker Tensor 27 spectrometer outfitted with a single reflection ATR containing a Ge crystal with a refractive index of 4.0 at 1000 cm−1 and a long wave cutoff of 575 cm−1.
Synthesis and Characterization Data for Peptoids 1–4
Peptoid synthesis was performed iteratively according to Scheme 1. Representative synthetic protocols for each of the three steps that yielded peptoid 1, along with the O-benzyl deprotection method for peptoid 2, are provided below. Additional details of the intermediates isolated in the syntheses of peptoids 1, 3 and 4 can be found in the Supporting Information.
Representative Acylation Protocol Using Bromoacetyl Bromide
Bromoacteyl bromide (3.26 mL, 37.5 mmol) was added to CH2Cl2 (25 mL) and cooled to −78 °C by stirring in a dry ice/acetone bath. Piperidine (2.47 mL, 25.0 mmol) and triethylamine (5.56 mL, 40.0 mmol) were dissolved in CH2Cl2 (40 mL) and added drop-wise via an addition funnel to the stirred solution at −78 °C under N2. After the addition was complete, the reaction mixture was stirred for 10 min at room temperature (rt). CH2Cl2 (75 mL) was added, and the mixture was washed with cold aq. NaHCO3 (2x), 10% w/v aq. citric acid (2x), and brine (1x). The organic phase was isolated, dried over MgSO4, and filtered. The solvent was removed in vacuo to yield 2-bromo-1-(piperidin-1-yl) ethanone as a faintly tan crude oil that was carried directly on to the amination step without purification.
Representative Amination Protocol
O′-benzyl hydroxylamine (1.08 mg, 6.77 mmol) was dissolved in DMF (5 mL) containing diisopropylethylamine (1.2 mL, 6.89 mmol), and warmed to 75 °C in an oil bath while stirring. Crude 2-bromo-1-(piperidin-1-yl) ethanone (from above, 465 mg, 2.26 mmol) was dissolved in DMF (10 mL) and then added to the stirring solution drop-wise. After the addition was complete, the reaction mixture was stirred for 48 h at 75 °C. DMF was removed in vacuo, and ~75 mL ethyl acetate (EtOAc) was added. The reaction mixture was washed with aq. NaHCO3 (3x) and brine (1x). The organic phase was isolated, dried over MgSO4, filtered, and the solvent was removed in vacuo. The crude product was purified to homogeneity by flash silica gel chromatography (Rf = 0.29, 1:1 EtOAc/hexanes + 1% TEA) to yield intermediate H-NOBn-Pip as a clear oil (60% yield over two steps).
Representative Acetylation Protocol
H-NOBn-Pip (from above, 323 mg, 1.30 mmol) was dissolved in CH2Cl2 (5 mL) containing triethylamine (363 μL, 2.60 mmol), and stirred at 0 °C in an ice bath. Acetyl chloride (185 μL, 2.60 mmol) in CH2Cl2 (10 mL) was added drop-wise to the stirring solution. After the addition was complete, the reaction mixture was stirred for 30 min at rt. The solvent was removed in vacuo, and the crude product was purified by flash silica gel chromatography (TLC: Rf = 0.23, 4:1 EtOAc/hexanes) to yield peptoid 1 as a white crystalline solid (95% yield). Prior to NMR studies, peptoid 1 was lyophilized overnight (2x) to a white powder (first from 1:1 CH3CN:H2O, then from CH3CN).
Representative Benzyl Deprotection Protocol
Peptoid 1 (85 mg) was dissolved in MeOH (10 mL) and Pd/C (10 mg, 10% mol/wt) was added to the solution. The reaction flask was sealed with a rubber septum and purged with H2 gas for 30 sec. Thereafter, the reaction was stirred at rt under a balloon of H2 for 10 h. The reaction mixture was then filtered through celite, and the solvent was removed in vacuo. Recrystallization from EtOAc afforded peptoid 2 as a colorless crystalline solid (90% yield).
Peptoid 1: Ac-NOBn-Pip
TLC: Rf = 0.23 (4:1 EtOAc/hexanes); 1H NMR (600 MHz, CDCl3, 10 mM): δ 7.39 (m, 5H), 4.91 (s, 2H), 4.34 (s, 2H), 3.55 (m, 2H), 3.26 (m, 2H), 2.17 (s, 3H), 1.67-1.51 (m, 6H); 13C NMR (75 MHz, CDCl3, 1H broadband-decoupled): δ 164.8, 135.1, 129.7, 129.0, 128.8, 50.1, 46.1, 43.4, 26.4, 25.5, 24.6, 20.6; IR (ATR, cm−1): 3087, 3065, 3036, 3004, 2930, 2852, 1672, 1659, 1445, 1393, 1266, 1231, 1139, 1128, 1078, 1018, 972, 854, 844, 800, 756, 740, 701, 667,; HR-MS (ESI): [M+H]+ calcd m/z = 313.1523, observed m/z = 313.1520.
Peptoid 2: Ac-NOH-Pip
TLC: Rf = 0.41 (5% MeOH/EtOAc); 1H NMR (600 MHz, CDCl3, 10 mM): δ 4.49 (s, 2H), 3.82 (s, 3H), 3.55 (m, 2H), 3.43 (m, 2H), 2.17 (s, 3H), 1.74-1.50 (m, 6H); 13C NMR (CDCl3,75 MHz, 1H broadband-decoupled): δ 175.2, 167.3, 51.5, 47.2, 44.5, 27.4, 26.8, 25.5, 20.2; IR (ATR,, cm−1): 3271, 2996, 2949, 2903, 2871, 1657, 1629, 1473, 1421, 1391, 1286, 1261, 1251, 1231, 1193, 1141, 1123, 1084, 1018, 990, 950,899, 843, 811, 752, 702,; HR-MS (ESI): [M+Na]+ calcd m/z = 223.1054, observed m/z = 223.1063.
Peptoid 3: Ac-(NOH)2-Pip
Synthesized by acetylation of H-NOBn-NOBn-Pip (see Supporting Information), followed immediately by benzyl deprotection according to the general procedures described above. Purified by recrystallization from MeOH:CH3CN (3:1). Isolated yield over two steps = 26%. TLC: Rf = 0.32 (1% AcOH in 5%MeOH/EtOAc); 1H NMR (600 MHz, CDCl3, 4 mM; major conformational peaks are reported): δ 8.59 (s, 1H), 7.55 (s, 1H), 4.80 (s, 2H), 4.55 (s, 2H), 3.56 (m, 2H), 3.33 (m, 2H), 2.22 (s, 3H), 1.69 (m 2H) 1.63 (m, 2H), 1.59 (m, 2H); 13C NMR (75 MHz, CDCl3, 1H broadband-decoupled): δ 170.3, 169.2, 166.4, 48.7, 48.2, 46.2, 43.4, 26.3,25.5, 24.3, 20.4; IR (ATR, cm−1): 3305, 3163, 2999, 2960, 2922, 2896, 1675, 1655, 1634, 1474, 1452, 1419, 1394, 1385, 1347, 1252, 1185, 1174, 1086, 1037, 1017, 985, 855, 801, 723, 668, 618; HR-MS (ESI): [M+Na]+ calcd m/z = 296.1217, observed m/z = 296.1208.
Peptoid 4: Ac-NOH-Nph-NOH-Dma
Synthesized from dimethylamine by iterative acylation/amination steps according to Scheme 1. Deprotection of both benzyl groups was accomplished according to the general hydrogenation procedure described above, and was followed by flash column chromatography to yield a white solid. Isolated yield = 74%. Prior to NMR experiments, 4 was lyophilized overnight (3x) to a white powder (first from 1:1 CH3CN:H2O, then from CH3CN, and finally from 2% t-butanol/CH3CN). TLC: Rf = 0.18 (1% AcOH in 5% MeOH/EtOAc); 1H NMR (600 MHz, CDCl3, 5 mM): δ 8.67 (brd s, 1H), 8.10 (brd s, 1H), 7.45-7.41 (m, 4H), 7.38 (m, 1H), 4.71 (s, 2H), 4.56 (s, 2H), 4.35 (s, 2H), 2.99 (s, 3H), 2.97 (s, 3H), 2.15 (s, 3H); 13C NMR (CDCl3, 75 MHz, 1H broadband-decoupled): δ 172.6, 170.1, 168. 8, 168.6, 141.2, 130.3, 129.2, 128.1, 51.7, 49.8, 49.0, 36.8, 35.8, 20.2; IR (ATR, cm−1): 3195 (brd), 2937 (brd), 1642, 1596, 1495, 1462, 1421, 1400, 1338, 1298, 1259, 1218, 1177, 1151, 1038, 1019, 809, 781, 762, 703,; HR-MS: [M+H]+ calcd m/z = 367.1613, observed m/z = 367.1620.
X-ray Crystallography
X-ray diffraction data for peptoids 2–4 were collected on a Bruker SMART APEXII diffractometer and crystal structures were solved by direct methods using SHELXS-97.81 All non-hydrogen atoms were refined with anisotropic displacement coefficients. The crystal and diffraction parameters are provided in Table S-1 of the Supporting Information. For peptoids 2 and 3, all hydrogen atoms were located in the difference map and refined independently. For peptoid 4, all hydrogen atoms (except those of the co-crystallized water molecule) were included in the structure factor calculation at idealized positions and were allowed to ride on the neighboring atoms with relative isotropic displacement coefficients. The hydrogen atoms on the water molecule were located in the difference map and refined independently with an O-H distance restraint. The crystallographic coordinates for the structures of peptoids 2–4 are deposited at the Cambridge Crystallographic Data Centre with deposition numbers 796968–796970.
Supplementary Material
Acknowledgments
We thank the NSF (CHE-0449959), ONR (N000140710255), Greater Milwaukee Foundation, and Burroughs Welcome Fund for financial support of this work. Support for the NMR facilities at UW–Madison by the NIH (1 S10 RR13866-01 and 1 S10 RR08389-01) and the NSF (CHE-0342998 and CHE-9629688) is gratefully acknowledged. X-ray crystal structures were visualized using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). We thank Dr. Charles Fry and Dr. Monika Ivancic for assistance with NMR spectroscopy, and Lara Spencer for assistance with X-ray crystallographic work.
Footnotes
Supporting Information Available
Characterization data of intermediate compounds, 1H NMR spectra of intermediate and target peptoids, 2D NOESY spectrum of intermediate Ac-NOBn-Nph-NOBn-Dma, and detailed X-ray crystallographic data.
References and Notes
- 1.Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 2.Fowler SA, Blackwell HE. Org Biomol Chem. 2009;7:1508–1524. doi: 10.1039/b817980h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazaleyrat JP, Rage I, Mouna AM, Savrda J, Wakselman M. Bioorg Med Chem Lett. 1994;4:1281–1284. [Google Scholar]
- 4.Kordik CP, Sanfilippo PJ. Chemtracts: Org Chem. 1995;8:36–40. [Google Scholar]
- 5.Hughes J, Dockray GJ, Hill D, Garcia L, Pritchard MC, Forster E, Toescu E, Woodruff G, Horwell DC. Regul Pept. 1996;65:15–21. doi: 10.1016/0167-0115(96)00067-5. [DOI] [PubMed] [Google Scholar]
- 6.Trivedi BK, Padia JK, Holmes A, Rose S, Wright DS, Hinton JP, Pritchard MC, Eden JM, Kneen C, Webdale L, Suman-Chauhan N, Boden P, Singh L, Field MJ, Hill D. J Med Chem. 1998;41:38–45. doi: 10.1021/jm970065l. [DOI] [PubMed] [Google Scholar]
- 7.Chan C, Yin H, McKie JH, Fairlamb AH, Douglas KT. Amino Acids. 2002;22:297–308. doi: 10.1007/s007260200016. [DOI] [PubMed] [Google Scholar]
- 8.Lim HS, Archer CT, Kodadek T. J Am Chem Soc. 2007;129:7750–7751. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson LS, Burdine L, Dutta AK, Feranchak AP, Kodadek T. J Am Chem Soc. 2009;131:5760–5762. doi: 10.1021/ja900852k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn KD, Udugamasooriya GD, Roland CL, Castrillon DH, Kodadek TJ, Brekken RA. BMC Cancer. 2010;10:397–410. doi: 10.1186/1471-2407-10-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu CW, Seurynck SL, Lee KYC, Barron AE. Chem Biol. 2003;10:1057–1063. doi: 10.1016/j.chembiol.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Seurynck SL, Patch JA, Barron AE. Chem Biol. 2005;12:77–88. doi: 10.1016/j.chembiol.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Dohm MT, Seurynck-Servoss SL, Seo J, Zuckermann RN, Barron AE. Biopolymers. 2009;92:538–553. doi: 10.1002/bip.21309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peretto I, Sanchez-Martin RM, Wang X-h, Ellard J, Mittoo S, Bradley M. Chem Commun (Cambridge, U K) 2003:2312–2313. doi: 10.1039/b306438g. [DOI] [PubMed] [Google Scholar]
- 16.Foged C, Franzyk H, Bahrami S, Frokjaer S, Jaroszewski JW, Nielsen HM, Olsen CA. Biochim Biophys Acta, Biomembr. 2008;1778:2487–2495. doi: 10.1016/j.bbamem.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Eggenberger K, Birtalan E, Schroeder T, Braese S, Nick P. ChemBioChem. 2009;10:2504–2512. doi: 10.1002/cbic.200900331. [DOI] [PubMed] [Google Scholar]
- 18.Unciti-Broceta A, Diezmann F, Ou-Yang CY, Fara MA, Bradley M. Bioorg Med Chem. 2009;17:959–966. doi: 10.1016/j.bmc.2008.02.068. [DOI] [PubMed] [Google Scholar]
- 19.Goodson B, Ehrhardt A, Ng S, Nuss J, Johnson K, Giedlin M, Yamamoto R, Moos WH, Krebber A, Ladner M, Giacona MB, Vitt C, Winter J. Antimicrob Agents Chemother. 1999;43:1429–1434. doi: 10.1128/aac.43.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Au VS, Bremner JB, Coates J, Keller PA, Pyne SG. Tetrahedron. 2006;62:9373–9382. [Google Scholar]
- 21.Chongsiriwatana N, Patch JA, Zuckermann RN, Marcano Y, Czyzewski A, Barron AE. Pept Sci. 2006;43:220–221. [Google Scholar]
- 22.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Proc Natl Acad Sci U S A. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryge TS, Frimodt-Moller N, Hansen PR. Chemotherapy (Basel, Switz) 2008;54:152–156. doi: 10.1159/000119707. [DOI] [PubMed] [Google Scholar]
- 24.Meinike K, Hansen PR. Protein Pept Lett. 2009;16:1006–1011. doi: 10.2174/092986609789055421. [DOI] [PubMed] [Google Scholar]
- 25.Bremner JB, Keller PA, Pyne SG, Boyle TP, Brkic Z, David DM, Robertson M, Somphol K, Baylis D, Coates JA, Deadman J, Jeevarajah D, Rhodes DI. Bioorg Med Chem. 2010;18:2611–2620. doi: 10.1016/j.bmc.2010.02.033. [DOI] [PubMed] [Google Scholar]
- 26.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Bioorg Med Chem Lett. 1994;4:2657–2662. [Google Scholar]
- 27.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Drug Dev Res. 1995;35:20–32. [Google Scholar]
- 28.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 29.Culf AS, Ouellette RJ. Molecules. 2010;15:5282–5335. doi: 10.3390/molecules15085282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuckermann RN, Kodadek T. Current Opinion in Molecular Therapeutics. 2009;11:299–307. [PubMed] [Google Scholar]
- 31.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev (Washington, D C) 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 32.Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo B, Kirshenbaum K. Curr Opin Chem Biol. 2008;12:714–721. doi: 10.1016/j.cbpa.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Butterfoss GL, Renfrew PD, Kuhlman B, Kirshenbaum K, Bonneau R. J Am Chem Soc. 2009;131:16798–16807. doi: 10.1021/ja905267k. [DOI] [PubMed] [Google Scholar]
- 35.Pauling LC, RB, Branson HR. Proc Natl Acad Sci U S A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson JS. Nature. 1977;268:495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- 37.Voet DVJG. Biochemistry. 3. Wiley; Hoboken, NJ: 2004. [Google Scholar]
- 38.Gorske BC, Bastian BL, Geske GD, Blackwell HE. J Am Chem Soc. 2007;129:8928–8929. doi: 10.1021/ja071310l. [DOI] [PubMed] [Google Scholar]
- 39.Gorske BC, Stringer JR, Bastian BL, Fowler SA, Blackwell HE. J Am Chem Soc. 2009;131:16555–16567. doi: 10.1021/ja907184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moehle K, Hofmann HJ. Biopolymers. 1996;38:781–790. doi: 10.1002/(SICI)1097-0282(199606)38:6%3C781::AID-BIP9%3E3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 41.Sui Q, Borchardt D, Rabenstein DL. J Am Chem Soc. 2007;129:12042–12048. doi: 10.1021/ja0740925. [DOI] [PubMed] [Google Scholar]
- 42.Armand P, Kirshenbaum K, Goldsmith RA, Farr-Jones S, Barron AE, Truong KTV, Dill KA, Mierke DF, Cohen FE, Zuckermann RN, Bradley EK. Proc Natl Acad Sci U S A. 1998;95:4309–4314. doi: 10.1073/pnas.95.8.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradley E, Kerr J, Richter L, Figliozzi G, Goff D, Zuckermann R, Spellmeyer D, Blaney J. Molecular Diversity. 1997;3:1–15. doi: 10.1023/a:1009698309407. [DOI] [PubMed] [Google Scholar]
- 44.Huang K, Wu CW, Sanborn TJ, Patch JA, Kirshenbaum K, Zuckermann RN, Barron AE, Radhakrishnan I. J Am Chem Soc. 2006;128:1733–1738. doi: 10.1021/ja0574318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirshenbaum K, Barron AE, Goldsmith RA, Armand P, Bradley EK, Truong KTV, Dill KA, Cohen FE, Zuckermann RN. Proc Natl Acad Sci U S A. 1998;95:4303–4308. doi: 10.1073/pnas.95.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah NH, Butterfoss GL, Nguyen K, Yoo B, Bonneau R, Rabenstein DL, Kirshenbaum K. J Am Chem Soc. 2008;130:16622–16632. doi: 10.1021/ja804580n. [DOI] [PubMed] [Google Scholar]
- 47.Stringer JR, Crapster JA, Guzei IA, Blackwell HE. J Org Chem. 2010;75:6068–6078. doi: 10.1021/jo101075a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CW, Kirshenbaum K, Sanborn TJ, Patch JA, Huang K, Dill KA, Zuckermann RN, Barron AE. J Am Chem Soc. 2003;125:13525–13530. doi: 10.1021/ja037540r. [DOI] [PubMed] [Google Scholar]
- 49.Bradley EK. J Magn Reson, Ser B. 1996;110:195–197. [Google Scholar]
- 50.Wu CW, Sanborn TJ, Huang K, Zuckermann RN, Barron AE. J Am Chem Soc. 2001;123:6778–6784. doi: 10.1021/ja003154n. [DOI] [PubMed] [Google Scholar]
- 51.Gorske BC, Jewell SA, Guerard EJ, Blackwell HE. Org Lett. 2005;7:1521–1524. doi: 10.1021/ol0502984. [DOI] [PubMed] [Google Scholar]
- 52.Gorske BC, Blackwell HE. J Am Chem Soc. 2006;128:14378–14387. doi: 10.1021/ja065248o. [DOI] [PubMed] [Google Scholar]
- 53.Fowler SA, Luechapanichkul R, Blackwell HE. J Org Chem. 2009;74:1440–1449. doi: 10.1021/jo8023363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lossen H. Liebigs Ann Chem. 1869:150. [Google Scholar]
- 55.Muri EMF, Nieto MJ, Sindelar RD, Williamson JS. Current Medicinal Chemistry. 2002;9:1631–1653. doi: 10.2174/0929867023369402. [DOI] [PubMed] [Google Scholar]
- 56.Domagal-Goldman SD, Paul KW, Sparks DL, Kubicki JD. Geochimica et Cosmochimica Acta. 2009;73:1–12. [Google Scholar]
- 57.Farkas E, Batka D, Csapo E, Buglyo P, Haase W, Sanna D. Polyhedron. 2007;26:543–554. [Google Scholar]
- 58.Marmion C, Griffith D, Nolan K. European Journal of Inorganic Chemistry. 2004;2004:3003–3016. [Google Scholar]
- 59.Kolasa T. Tetrahedron. 1983;39:1753–1754. [Google Scholar]
- 60.Bracher BH, Small RWH. Acta Crystallographica Section B-Structural Crystallography and Crystal Chemistry. 1970;B 26:1705. [Google Scholar]
- 61.Kaczor A, Proniewicz LM. Journal of Molecular Structure. 2004;704:189–196. [Google Scholar]
- 62.Kakkar R, Grover R, Chadha P. Organic & Biomolecular Chemistry. 2003;1:2200–2206. doi: 10.1039/b300466j. [DOI] [PubMed] [Google Scholar]
- 63.Larsen IK. Acta Crystallographica Section B-Structural Science. 1988;44:527–533. [Google Scholar]
- 64.Smith WL, Raymond KN. J Am Chem Soc. 1980;102:1252–1255. [Google Scholar]
- 65.Aubry A, Dupont V, Marraud M. Acta Crystallographica Section C. 1995;51:1577–1579. [Google Scholar]
- 66.Dupont V, Lecoq A, Mangeot JP, Aubry A, Boussard G, Marraud M. J Am Chem Soc. 1993;115:8898–8906. [Google Scholar]
- 67.Takeuchi Y, Marshall GR. J Am Chem Soc. 1998;120:5363–5372. [Google Scholar]
- 68.Aleman C. The Journal of Physical Chemistry A. 2001;105:6717–6723. [Google Scholar]
- 69.Jencks WP. J Am Chem Soc. 1958;80:4585–4588. [Google Scholar]
- 70.Kolasa T, Chimiak A. Tetrahedron. 1977;33:3279–3284. [Google Scholar]
- 71.Maire P, Blandin V, Lopez M, Vallee Y. Synlett. 2003;2003:0671, 0674. [Google Scholar]
- 72.Moeller BLM, IJ, Conn EE. Acta Chem Scand B. 1977;31:343–344. [Google Scholar]
- 73.Ottenheijm HCJ, Herscheid JDM. Chemical Reviews. 1986;86:697–707. [Google Scholar]
- 74.Kolasa T, Chimiak A. Tetrahedron. 1974;30:3591–3595. [Google Scholar]
- 75.Bax A, Davis DG. Journal of Magnetic Resonance (1969) 1985;63:207–213. [Google Scholar]
- 76.Bothner-By AA, Stephens RL, Lee J, Warren CD, Jeanloz RW. J Am Chem Soc. 1984;106:811–813. [Google Scholar]
- 77.Neuhaus DWMP, editor. The Nuclear Overhauser Effect in Structural and Conformational Analysis. Wiley-VCH; New York: 2000. [Google Scholar]
- 78.Nam KT, Shelby SA, Choi PH, Marciel AB, Chen R, Tan L, Chu TK, Mesch RA, Lee BC, Connolly MD, Kisielowski C, Zuckermann RN. Nature Materials. 2010;9:454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]
- 79.Still WC, Kahn M, Mitra A. J Org Chem. 1978;43:2923–2925. [Google Scholar]
- 80.Goddard TD, Kneller DG. SPARKY. 3.110. University of California; San Francisco: [Google Scholar]
- 81.Sheldrick GM. Acta Cryst. 2008;A64:112–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.