Abstract
Pharmacogenomics in oncology holds the promise to personalize cancer therapy. However, its clinical application is still limited to a few genes, and, in the large majority of cancers, the correlation between genotype and clinical outcome has been disappointing. One possible explanation is that current pharmacogenomic studies do not take into account the emerging role of cancer stem cells (CSCs) in drug sensitivity and resistance. CSCs are a subpopulation of cells driven by specific signal-transduction pathways, but genetic variants affecting their activity are generally neglected in current pharmacogenomic studies. Moreover, in several malignancies, CSCs represent a rare sub-population; therefore, whole tumor profiling might mask CSC gene expression patterns. This article reviews current evidence on CSC chemoresistance and shows how common genetic variations in CSC-related genes may predict individual response to anti-cancer agents. Furthermore, we provide insights into the design of pharmacogenomic studies to address the clinical usefulness of CSC genetic profiling.
Pharmacogenomic studies and cancer
For the past two decades, cancer has been viewed as a microevolutionary process. This concept has been driven by the “stochastic model” of cancer progression, assuming that each cell within a tumor has the same possibility to develop oncogenic mutations. Successful clones override all other cancer cells. This model has been employed to explain invasiveness, metastasis and drug resistance [1].
Chemotherapy itself may act as a selective pressure, thus favoring the expansion of a resistant clone associated with acquired chemo-resistance (Figure 1). During the 1980 s, Goldie and Coldman developed a mathematical model of tumor growth after treatment, which is still considered a gold standard; this model predicts that acquired resistance can be overcome by the use of high-dose, combination chemotherapy administered at the earliest feasible time. Alternatively, some tumors may bear mutations that confer primary refractoriness (intrinsic resistance). For these cancers, which are characterized by a grim prognosis, the only option is the discovery of novel drug targets [2].
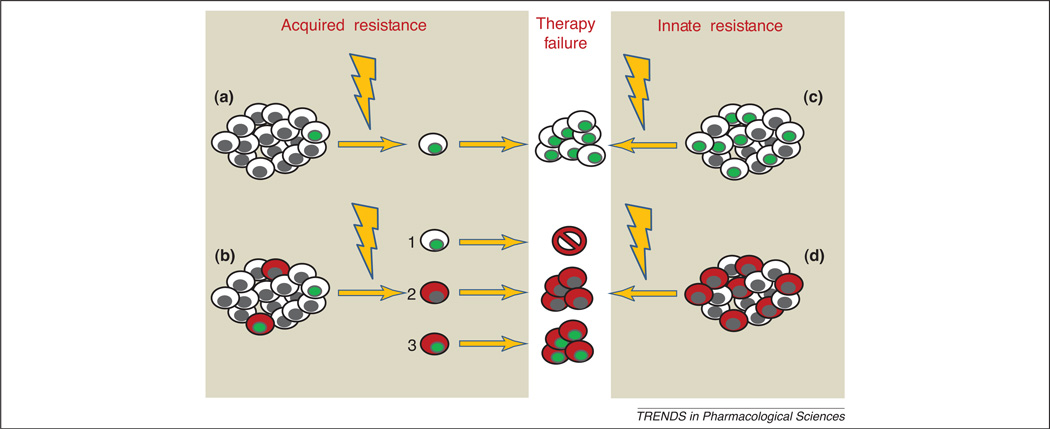
Figure 1. Models of chemoresistance.
According to the stochastic model of tumor progression, drug resistance is due to a pre-existent or acquired mutation (mutated cells are represented by green nuclei) in any tumor cell (a). According to the CSC hypothesis, acquired resistance derives from CSCs (in red) which may be intrinsically resistant or mutated after chemotherapy (b). Mutations in non-stem cell clones are ineffective, since they cannot propagate the tumor. Constitutively resistant tumors are enriched of drug-resistant CSCs. Modified from [7] with permission from Macmillan Publishers Ltd: Nature Reviews Cancer © 2005.
The stochastic paradigm has been applied to the prediction of anticancer drug response. Current pharmacogenomic studies are devoted to the identification of germinal polymorphisms, tumor-specific mutations and gene expression patterns that might predict efficacy and tolerability of cancer treatments. The main pitfall is that genomic analyses are performed on a tumor sample, with the wrong assumption that tumor is genetically homogeneous. The pharmacogenomic approach based on a “stochastic paradigm” has met some challenges, as demonstrated by the approval of tests by the USA Food and Drug Administration (FDA) [3]. These tests evaluate few common genetic variations in drug-metabolizing enzymes, including thyopurine methyltransferase (TPMT) and uridine glucoronosyl-trasferase isoform 1A1 to predict individual risk of toxicity induced by thyopurines and irinotecan, respectively. However, prospective clinical trials are required to confirm predictive power for some of them [4].
One possible pitfall of pharmacogenomic studies could be the underestimation of the contribution of cancer stem cells (CSCs) to drug response. Human cancers are organized as a hierarchy, with CSCs at the apex, because they are the only tumor-initiating cells within a malignancy [5]. It is worth noting that this model may not apply to all cancer types [6] However, from the CSC hypothesis perspective, the stochastic paradigm of chemo-resistance fails, because all mutations occurring in non-stem cancer cells would not turn into a biological advantage. Indeed, these cells could not further sustain cancer growth. In addition, CSCs are thought to be drug resistant, and to give rise to tumor progression after treatment [7].
Therefore, the CSC paradigm requires a new theoretical framework for both acquired and innate resistance [7] (Figure 1) that could change the current design of pharmacogenomic studies. The application of a new paradigm to an old problem involves the identification of new tools and new definitions, and might eventually lead to unexpected results [8]. In this article, we will explore the applicability of the pharmacogenomics to CSC paradigm, highlighting novel research findings and current limitations. We will first summarize current data on CSC drug resistance and then analyze the potential clinical applications of the CSC hypothesis.
CSC resistance: facts and legends
CSC markers
In this article, we will refer to the CSCs as a distinct population within the tumor that can be identified based on the expression of surface markers (e.g. CD133), enzymatic activity (e.g. ALDH), or drug-efflux transporters (e.g. ABCG2). CSCs are the only subpopulation capable of initiating a tumor when injected into immunologically deficient mice [9]. The xenografted tumor should phenotypically recapitulate the original one. In addition, CSCs isolated from xenografts are able to generate new tumors, a property called “long-term self-renewal.”
Based on these principles, CSCs have been identified in many solid tumors and leukemias. Several laboratories have reported the ability to isolate and propagate patient-derived CSCs from breast, colorectal, prostate and brain cancers [5]. The frequency of CSCs ranges from less than 0.1 to more than 20%, depending on cell type, selected marker and experimental conditions.
Another way to identify CSCs has been the Hoechst-33342 staining, which is not taken up by cells in the so-called side population (SP). SP cells express ABCG2, an ATP-binding cassette protein transporter, regulating the efflux of Hoechst-33342 [10]. ABCG2 expression allows these cells to transport several hydrophobic drugs, including irinotecan, imatinib and doxorubicin. Along with ABCG2, other members of the ABC family are overexpressed in the SP, thus conferring a broader spectrum of resistance [11]. More importantly, each tumor could be populated by a SP with a specific pattern of drug resistance. Additional mechanisms of SP drug resistance could be a high percentage of non-cycling cells, and high expression of DNA repair genes [10].
Despite this evidence, ABC transporter inhibitors have performed poorly in the clinical setting. However, most studies employed an ABCB1 inhibitor, whereas SP cells mainly express ABCG2 [7]. Moreover, the relationship between SP phenotype and CSCs has been questioned. Patrawala et al. [12] proposed that the higher tumorigenicity of the SP is ABCG2-independent. Besides, non-SP cells retain a percentage of CSCs., and in some tumor types, SP cells and CSCs are completely distinct populations [13]. In summary, although the SP is probably enriched with CSCs and drug-resistant cells in some cancer types, the degree of overlap between these two populations remains unknown. It is likely that SP holds a variable percentage of CSCs, depending on tumor type and experimental conditions.
An alternative approach to SP characterization is to select drug resistant clones (DRCs), and check the CSC features of these cells. For example, gemcitabine-resistant pancreatic cancer cells express CD44, CD24 and epithelial-specific antigen (ESA), three tissue-specific CSC markers, and showed a significant activation of stem cell pathways [14]. Another mechanism of chemoresistance of DRCs is linked to CSCs and is the production of angiogenic factors and cytokines, which in turn activates STAT3- and STAT5-dependent antiapoptotic signals [15].
Mouse models show that recurrence after chemotherapy is mediated by CSCs [16]. Yu et al. demonstrated that the CD44+/24− fraction is highly enriched in breast cancer tumors after neoadjuvant chemotherapy [17]. If this observation is extended to other cancers, we can conceive a model in which chemotherapy selects CSCs.
CSC-related pathways: self-renewal and beyond
CSC properties are determined by key developmental signaling pathways, each of which is composed by a distinct set of genes. The principal signaling cascades governing CSC self-renewal are Wnt, Notch and Hedgehog [18]. They comprise specific extracellular ligands, membrane receptors, and intracellular transduction pathways converging in the activation of a main transcription factor responsible of the expression of a diversity of downstream genes (Figure 2). Recent evidence indicates that each CSC-specific pathway might contribute to CSC drug resistance.
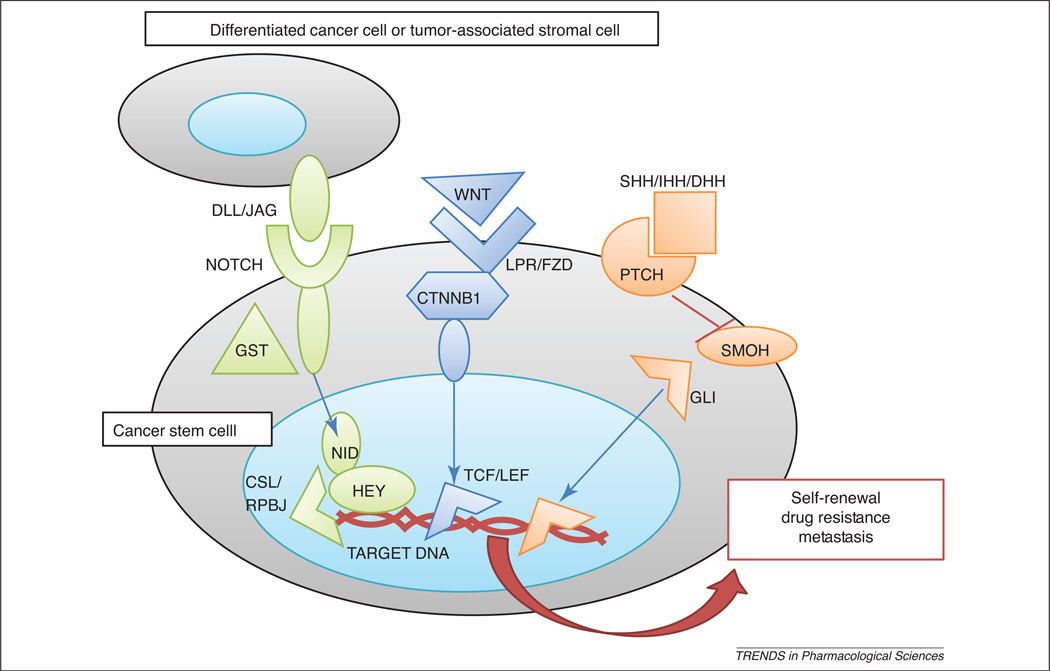
Figure 2. CSC-related pathways.
Notch, Wnt and Hedgehog pathways mediate CSC self-renewal, survival and drug resistance. Notch receptors are activated by cell-bound ligands (DLL, delta-like; JAG, jagged). This interaction activates Notch intracellular domain (NICD) cleavage by γ-secretase (GST). NID translocates to the nucleus and triggers CSL/RBPJ transcription factor (TF) and Hey repressors. Wnt molecules bind to specific receptors (LPR, lipoprotein receptor and FZD, frizzled), thereby favoring β-catenin (CTNNB) stabilization by Dishevelled (DVL) complex. CTNBB translocates to the nucleus and activate CSL/RBPJ TF. Hedgehog molecules bind to patched receptor (PTCH) and inhibit Smoothened (SMOH), which allows GLI TF stabilization and further translocation into the nucleus. Modified from [18] with permission from Bentham Science Publishers LTD.
The Notch pathway has a major role in cell fate, proliferation and survival in various organs. A critical feature of this pathway is the ability of the enzyme γ-secretase to cleave Notch Intracellular Domain (NID), which then translocates into the nucleus to trigger gene expression or gene silencing. Aberrant Notch signaling underlines a wide spectrum of human diseases, including developmental disorders and cancers [18]. The Notch network acts on normal adult stem cells, driving proliferative as well as specific cell fate signals [19], However, the opposite has been shown as well; Notch genes have been documented as oncogenes or tumor suppressors in several malignancies, and this has been hypothesized to correspond to its different functions in the normal cellular context [20]. Various reports have shown the activation of Notch pathway in CSCs from several tissues [21]. Through its antiapoptotic activity, the Notch pathway contributes to CSC chemoresistance. For example, the Notch-γ-secretase axis triggers Akt phosphorylation [22], a mechanism to escape cell death after chemotherapy (Figure 2). In keeping with this evidence, pharmacological inhibition of γ-secretase-enhanced oxaliplatin activity on colorectal cancer cells [23] and Notch3 silencing enhanced doxorubicin activity on hepatocellular carcinoma cells [24].
The Wnt pathway is an evolutionarily conserved cell paracrine signaling cascade that is extensively activated during development. In the adult organism, Wnt molecules regulate the fate of multipotent stem cells of various lineages [25]. Aberrant Wnt stimulation promotes tumorigenesis in a wide range of human malignancies. A traditional example is the inactivating mutations of the APC gene that underlies colorectal carcinogenesis [26]. Mutations in other components of the pathway (including β-catenin) have been identified in other tumour from skin, liver, prostate and ovary. The role of Wnt pathway mutations in the CSC compartment is closely related to its role in the normal somatic stem cells. In the intestine and the lung, it is crucial for the maintenance of the stem cell pool and, as a result, pathway activating mutations lead to tumorigenesis [27].
Interestingly, several Wnt pathway inhibitors, including Dicckopf-1 (DKK1), Wnt Inhibiting Factor (WIF1) and Secreted Frizzled Related Protein (SFRP) are downregulated in chemotherapy-resistant cancer cells. DKK1 sensitizes cancer cells to alkylating agent-dependent apoptosis [28]. Moreover, WIF1 enhanced chemosensitivity to paclitaxel and etoposide in prostate cancer cells [29].
The Hedgehog pathway is crucial during embryonic development. In the adult, its activity is restricted to tissue maintenance and repair, and its improper reactivation has been connected to various human cancers [30].
Several groups have linked Hedgehog pathway with the maintenance of CSCs. Liu et al. demonstrated that Hedgehog signaling plays an essential role in maintaining the CD44+CD24-/low subpopulation of breast cancer cells by regulation of a polycomb gene BMI-1 [31]. Increased Hedgehog activity was found in CD133+ cells derived from metastatic versus nonmetastatic tumors [32]. CD133+ glioma CSCs were also shown to depend on Hedgehog pathway for proliferation, survival, self-renewal and tumorigenicity [33,34].
BMI-1 activation by Hedgehog pathway is a major mechanism of resistance in some cancer cells. In nasopharyngeal carcinoma and prostate cancer, BMI-1 silencing does not affect cell survival, but strongly enhances chemotherapy- induced apoptosis [35,36].
In conclusion, the major signaling pathways governing self-renewal and differentiation in normal stem cells have been extensively documented to contribute to CSC biology. Furthermore, their roles have been demonstrated in most types of malignancies, which suggests that their targeting can be a key strategy for CSC eradication.
Pharmacogenomics of CSCs
Preclinical models suggest that tumor-initiating CSCs are more aggressive and resistant to chemotherapy than other cancer cells. However, the clinical relevance of these findings is still uncertain. Is the CSC fraction simply more tumorigenic in immunocompromised mice? [37] Is CSC chemoresistance dependent on culture conditions and drugs employed? [38
To answer these questions, clinical studies should prove that CSCs are activated in more aggressive tumors, and predict poor prognosis and response to chemotherapy.
CD133 expression predicts poor survival in colorectal cancer [39] and glioblastoma [40], whereas CD44 expression is associated to metastasis risk in gastric [41] and breast cancer [42]. Due to the prognostic role of some CSC markers and pathways, future pharmacogenomic studies should be able to discriminate between prognostic and predictive factors. This will require the comparison of populations treated with specific therapeutic regimens. Indeed, a true predictive marker must be associated to clinical outcome only in patients treated with a selected therapy [43]. Table 1 summarizes the studies suggesting a putative predictive power of CSC-related genes/pathways. These data are based on gene expression levels in cancer tissues. Some studies identified CSC markers as predictors of poor disease-free or overall survival in patients treated with specific regimens [44–47]. Interestingly, one study showed a correlation between disease-free survival and CD133 expression after neoadjuvant therapy in rectal cancer [48]. Although this study lacks a control group of untreated patients, it suggests an active role of this subpopulation in treatment resistance. Two studies fund a closer correlation between tumor regression after therapy and Wnt [49] or Hedgehog [50] pathway activation. Finally, CD133 expression in non-small cell lung cancer (NSCLC) was associated with expression of drug resistance genes [51]. Thus, it is likely that gene expression profiles of CSC-related pathways could be useful tools for future pharmacogenomic studies.
Table 1.
Clinical studies indicating an association between CSC-related pathways and treatment response
| Cancer Type | Gene/Pathway | Prediction | Ref. |
|---|---|---|---|
| Colorectal cancer | CD133 | Poor response to fluorouracil-based chemotherapy | [44] |
| Esophageal cancer | Hedgehog pathway | Poor response to chemo-radiotherapy | [50] |
| Ewing sarcoma | Wnt pathway | VIDE regimen: tumor regression | [49] |
| Glioblastoma | CD133 EGFR | Poor survival after radio-chemotherapy | [45] |
| Head/neck cancer | BMI1 | Poor survival after radio-chemotherapy | [46] |
| NSCLC | BMI1 | Poor disease-free survival after adjuvant chemotherapy | [47] |
| NSCLC | CD133 | High expression of drug-resistance genes | [51] |
| Rectal cancer | CD133 | Poor disease-free survival after neoadjuvant chemotherapy | [48] |
A more traditional strategy to perform pharmacogenomic analyses is to correlate genetic variants with response to therapy [52]. Both germinal polymorphisms and somatic mutations in cancer cells can affect drug-resistance pathways, thereby emerging as predictive markers. As shown in the previous section, Wnt, Hedgehog and Notch pathways are involved in both chemo-resistance and CSC self-renewal. Interestingly, some of these pathways are inhibited by innovative small molecules. For example, γ-secretase inhibitiors (GSIs) were shown to impair Notch-dependent signaling and CSC self-renewal in several tumors [53]. Despite their gastrointestinal toxicity, GSIs are currently being tested in Phase II clinical trials for metastatic breast and colorectal cancer, as well as recurrent glioblastoma (www.clinicaltrials.gov./), This makes GSIS among the first drugs directed against CSC-specific pathways to be studied in clinical trials. If these drugs prove to be effective in the clinical setting, it is conceivable that genetic variations in Notch pathway could predict GSI antitumor activity and/or side effects.
As a proof of principle, we searched the literature for common genetic variants in Notch pathway genes that might affect CSC self-renewal and GSI sensitivity. Table 2 shows the most significant somatic mutations and gene polymorphisms described in cancer patients. Notch 1 gene is mutated in approximately 60% T-cell acute lymphoid leukemias (T-ALL) [54]. Most alterations are missense mutations, or small deletions/insertions, resulting in ligand-independent Notch activation, or ligand hypersensitivity. Less common mutations affect the extracellular domain, or enhance γ-secretase activity. With this genetic background, there was great expectation that GSIs could be effective therapies in T-ALL. Unfortunately, the first Phase I clinical trial on a GSI inhibitor in T-ALL patients gave disappointing results, with very low response rates and dose-limiting gastrointestinal toxicity [54]. These results reveal the importance of a pharmacogenomic approach in the early phases of development of CSC-targeting drugs. Due to the highly specific nature of these drugs, it is crucial to identify molecular markers that can select patients who can actually benefit from the drug. In this case, it is conceivable that only some types of mutations (ligand-independent activation) are sensitive to GSI. Other mutations (ligand hypersensitivity) might be best targeted by different approaches (e.g. Notch1-specific antibodies). Notch signaling activation was also found in approximately 30% NSCLC cases, due to pathway inhibitor silencing, or Notch1 mutations conferring ligand-independent activity [55]. Interestingly, cells derived from Notch mutated cancers were highly sensitive to GSI. In addition, Notch pathway activation predicted poorer overall survival. Similarly, Notch3 gene amplification in high grade ovarian cancer was associated to in vitro sensitivity to GSI [56].
Table 2.
Common genetic variations in Notch Pathway genes
| Gene | Variant | Function | Rate | Ref. |
|---|---|---|---|---|
| NOTCH1 | Mutation | Activation (T-ALL) | 60% | [54] |
| NOTCH1 | Mutation | Response to GSI and prognosis (NSCLC) | 12% | [55] |
| NOTCH2 | Mutation | Unknown (Breast Cancer) | 2.1% | [57] |
| NOTCH3 | Mutation | Unknown(Colorectal Cancer) | 2% | [57] |
| NOTCH3 | Amplification | Response to GSI (Ovarian Cancer) | 19.5% | [56] |
| NOTCH2 | SNP | Increased cancer risk (Breast Cancer) | 16.8% | [58] |
| HEY1 | SNP | Response to p53 activators | 1% | [59] |
We selected both single nucleotide polymorphisms (SNP) and somatic alterations (gene amplifications, mutations) affecting cancer cell behavior. Rate refers to mutation rate in cancer samples, or minor allele frequency in Caucasians (SNP). T-ALL, T-cell acute lymphoid leukemia; NSCLC, non-small cell lung cancer; GSI, γ-secretase inhibitor.
In some solid tumors (breast and colorectal cancer), Notch or γ-secretase mutations are not common, thus questioning the efficacy of GSI [57], [58]. In these cases, Notch pathway activation may be mediated by down-stream molecules (Figure 2). For example, the activity of a Notch-dependent nuclear factor is affected by a non-synonymous SNP (Hey1 Leu94Met) [59]. Hey (hairy/enhancer- of-split related with YRPW) is a transcriptional repressor activated by NID, generally acting as a positive regulator of the p53 tumor suppressor gene. The Leu-Met substitution is able to impair p53 activation, thereby enhancing aberrant cell differentiation. In addition, the variant allele is not sensitive to the pro-apoptotic activity of p53-targeting drugs.
In conclusion, a comprehensive analysis of somatic and germinal genetic variants in CSC-related pathways could help a personalized and targeted therapeutic approach. However, because CSCs represent a small percentage of most tumors, genetic analysis must be refined through a completely new experimental system, which we will describe in the next sections.
CSCs from a new perspective
The development of new pharmacogenomic studies will require effective strategies to track CSC changes in response to anticancer therapy, as well as to isolate and characterize CSCs from patients. For example, imaging techniques could be employed to monitor CSCs in patients [60]. Multiparametric analysis and high sensitivity are required because the CSC phenotype is often defined by multiple markers, and their frequency could be as low as 1 in 1000 cells. High resolution optical imaging is ideal for superficial neoplasms (e.g. breast cancer), due to its limited penetration potential. Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET) are more suitable for internal neoplasms; both techniques will require specific contrast media or radiolabeled probes to unequivocally detect CSCs. Similar tools have been tested only in animal models [61]. Thus, future studies should test pharmacokinetic and pharmacodynamic properties of CSC-specific probes (e.g. radiolabeled anti-CD133 antibodies).
An alternative strategy would be to isolate CSCs from peripheral blood, an approach that could be readily translated to the clinic [62]. Circulating tumor cells (CTCs) are present at concentration of one cell per million in the blood of metastatic cancer patients. Disseminated tumor cells (DTCs) can be identified as micrometastasis in the bone marrow. Because CTC/DTCs and CSCs seem to share some features, recent studies investigated the presence of tumor- initiating cells in the blood and bone marrow of cancer patients. Balic et al. studied bone marrow biopsies from breast cancer patients immunostained for cytokeratin (CK), CD44 and CD24 [63]. CD44+/24-CSCs were present in all biopsies and their prevalence was 72%, i.e. much higher than in primary tumors. Similar results were obtained with CTCs from breast cancer patients [64]. These data support the idea that CSCs can escape the primary tumor site and generate micrometastasis in distant organs.
In addition, data from breast cancer patients indicate that CTCs are an interesting target for future pharmacogenomic studies. High CTC levels before and after chemotherapy predict shorter disease-free survival [65]. DTCs can survive both chemotherapy and hormonal therapy, persist in bone marrow over many years after surgery and are associated with an increased risk of late metastatic relapse [66]. If the CSC hypothesis is correct, the CSC fraction within CTC and DTC should be a more accurate predictor of patient survival after chemotherapy.
In conclusion, high resolution imaging techniques and CTC/DTC isolation could be implemented to detect CSCs during pharmacogenomic studies. The main drawback is represented by the extremely low prevalence of CSCs. This could generate false negative results if the technique is not sufficiently sensitive, or false positive results if signal amplification is excessive. In addition, data should be sufficiently reproducible to allow a time-course analysis of CSC levels before and after chemotherapy.
Design of new pharmacogenomic studies
Anticancer drug activity and efficacy measures are based on the idea that all tumor cells are equally dangerous for the patient. Antitumor activity is measured through the Response Evaluation Criteria in Solid Tumors (RECIST) [67]. RECIST criteria evaluate the MRI- or CT scan-based estimate of tumor volume after treatment. However, volumetric measurement may be misleading when measuring the activity of biological drugs [68]. In the context of the CSC paradigm, RECIST criteria are also inappropriate. Because the CSC rate can be as little as 0.1% of total tumor volume [69], an anticancer drug targeting only non-CSC could cause important tumor shrinkage with little or no benefit for disease prognosis.. Based on pre-clinical evidence and pharmacokinetic data, it is likely that current chemo-therapy regimens target both non-stem cancer cells and CSCs, being more specific for the first population. Thus, the CSC absolute number is likely reduced by chemotherapy, thereby explaining survival benefit. However, CSC rate after chemotherapy could be even increased, thus enhancing the possibility of long-term recurrence and metastasis [17].
The development of a CSC-based pharmacogenomics will require innovative technologies to estimate CSC frequency in vivo. As discussed above, high resolution CT and MRI imaging are promising technologies. The most problematic issue will be the development of CSC-specific probes. It is worth noting that for some solid tumors, the CSC phenotype is still not clearly defined. In addition, these probes should be nontoxic and able to diffuse extensively within the tumor mass.
Because none of these technologies has been tested in the clinical setting, surrogate markers of CSC activity will be needed in the near future. A surrogate marker of CSC activity could be a CSC-derived soluble factor, including cytokines. It is well known that patients suffering from many cancers show an increased blood concentration of different cytokines, particularly interleukin 6 (IL-6). Cytokines are produced by tumor associated stroma and cancer cells, and are responsible for some cancer-related symptoms, like chronic fatigue and cognitive impairment [70]. High level of IL-6 is independent predictor of short survival in some neoplasms, including breast cancer [71]. Mammospheres isolated from breast cancer patients produce high levels of IL-6. Blocking this autocrine loop impairs CSC growth and invasiveness [72]. Due to the relationship between circulating IL-6, breast cancer prognosis and CSC biology, it is likely that IL-6 levels could be used as a marker of anti-CSC activity.
As shown in Figure 3 the CSC hypothesis will have a broader impact on pharmacogenomic studies. In general, classical pharmacogenomics deals with germinal and somatic mutations [52]. In the first case, polymorphisms shared by normal and cancer cells are assessed through easily accessible samples, typically blood cells. This strategy proved to be effective in the discovery of some genetic variations affecting drug metabolism. In the second case, cancer-specific mutations or gene expression profiles are derived from tumor samples obtained by biopsy. For example, EGFR mutations have been related to non-smallcell lung cancer sensitivity to gefitinib [52].
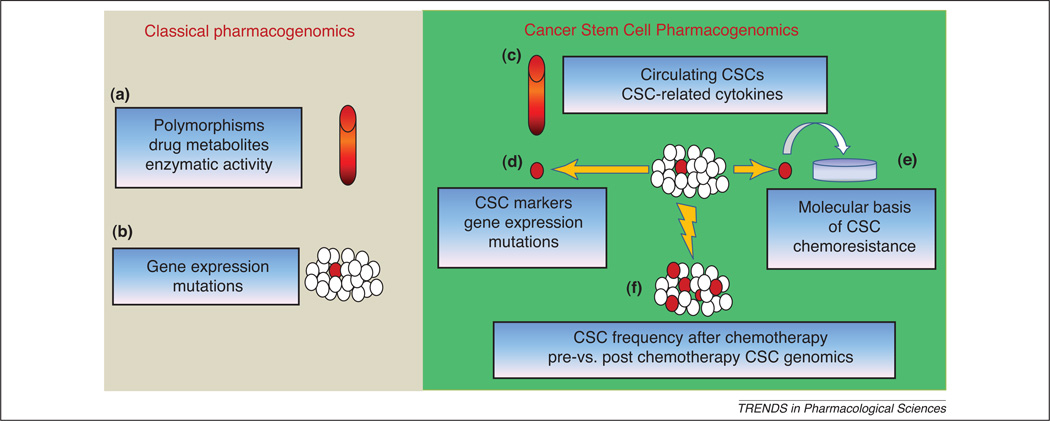
Figure 3. Design of innovative pharmacogenomic studies.
Classical pharmacogenomics deals with analysis performed on blood samples (a) or tumor specimens (b). In the first case, germinal polymorphisms or drug metabolism are studied to predict chemotherapy-related toxicity. In the second case, genomic analyses are performed on a mixed population, generally comprising a small percentage of CSCs (in red). CSC-pharmacogenomics should include detection of circulating CSCs and measurement of CSC-related cytokines from blood samples (c). In addition, CSCs isolated from primary tumors (d) are suitable for specific genomic and proteomic analysis. CSCs can also be expanded in vitro in order to perform drug sensitivity tests (e). Finally, if CSCs are more resistant, chemotherapy may increase their percentage within the tumor. Thus, analysis of CSC frequency after neoadjuvant chemotherapy, as well as comparison of pre- vs post- chemotherapy CSC genomics could predict patient response to treatment (f).
If CSCs alone drive cancer progression and chemoresistance, a more accurate tumor sampling is required. Laser microdissection (LMD) of tissue samples proved to enhance genomic profiling for pharmacogenomic studies [73]. With appropriate modifications, LMD could be implemented to specifically dissect CSC from a tumor sample. Antibodies directed against CSC markers could be employed to microdissect CSCs, thus allowing CSC-specific genomic profiling. Although it is likely that the CSC scarcity will be the main difficulty of this approach, current technologies properly enable single cell profiling [74].
An alternative way to study CSC chemoresistance would be in vitro propagation. Because LMD requires tissue fixation, it is not suitable for this purpose. CSCs typically from spheroid structures in stem cell-specific media (SCM), and express several CSC markers. Using this technique, CSC have been isolated and propagated from breast [75], brain [76] and prostate [77] cancer. SCM composition is variable from one tissue to another, and specific differences may allow normal or CSC growth [78]. It is not rare to find very different gene expression profiles from the same tissue spheroids [77], [79]. Thus, CSC expansion in a specific SCM affects gene expression profile. In addition, maintenance of CSC gene expression signatures is limited to few passages. Despite all these pitfalls, CSC spheroids could predict patient sensitivity to different drug combinations. For example, a Notch pathway inhibitor tested on glioblastoma-derived neurospheres proved to suppress brain tumorigenesis in vivo 53]. These approaches can be applied to screen CSC sensitivity to anticancer drugs.
Concluding remarks
In conclusion, pharmacogenomics will require a new definition of anticancer drug activity, more accurate sampling methods and innovative technologies to propagate CSCs. CSC-related pathways could be investigated in addition to conventional pharmacokinetics and pharmacodynamics. This approach could be applied to both chemotherapy and to novel agents targeting CSCs. In the near future, a close collaboration between CSC biologists and clinicians will hopefully drive the development of multidisciplinary studies.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Goldie JH. Drug resistance in cancer: a perspective. Cancer Metastasis Rev. 2001;20:63–68. doi: 10.1023/a:1013164609041. [DOI] [PubMed] [Google Scholar]
- 3.Maitland ML, et al. TPMT, UGT1A1 and DPYD: genotyping to ensure safer cancer therapy? Trends Pharmacol. Sci. 2006;27:432–437. doi: 10.1016/j.tips.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J. Clin. Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 5.Dick JE. Looking ahead in cancer stem cell research. Nat. Biotechnol. 2009;27:44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 6.Prestegarden L, et al. Glioma cell populations grouped by different cell type markers drive brain tumor growth. Cancer Res. 2010;70:4274–4279. doi: 10.1158/0008-5472.CAN-09-3904. [DOI] [PubMed] [Google Scholar]
- 7.Dean M, et al. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn TS. Historical structure of scientific discovery. Science. 1962;136:760–764. doi: 10.1126/science.136.3518.760. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10.Wu C, Alman BA. Side population cells in human cancers. Cancer Lett. 2008;268:1–9. doi: 10.1016/j.canlet.2008.03.048. [DOI] [PubMed] [Google Scholar]
- 11.Ho MM, et al. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 12.Patrawala L, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 13.Broadley KW, et al. Side Population is not Necessary or Sufficient for a Cancer Stem Cell Phenotype in Glioblastoma Multiforme. Stem Cells. doi: 10.1002/stem.582. [DOI] [PubMed] [Google Scholar]
- 14.Shah AN, et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 2007;14:3629–3637. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 15.Desrivieres S, et al. The biological functions of the versatile transcription factors STAT3 and STAT5 and new strategies for their targeted inhibition. J. Mammary Gland Biol. Neoplasia. 2006;11:75–87. doi: 10.1007/s10911-006-9014-4. [DOI] [PubMed] [Google Scholar]
- 16.Dylla SJ, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 18.Crea F, et al. Targeting prostate cancer stem cells. Anticancer Agents Med. Chem. 2009;9:1105–1113. doi: 10.2174/187152009789735053. [DOI] [PubMed] [Google Scholar]
- 19.Dontu G, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 21.Bolos V, et al. Notch signalling in cancer stem cells. Clin. Transl. Oncol. 2009;11:11–19. doi: 10.1007/s12094-009-0305-2. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie G, et al. Cellular Notch responsiveness is defined by phosphoinositide 3-kinase-dependent signals. BMC Cell Biol. 2006;7:10. doi: 10.1186/1471-2121-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng RD, et al. gamma-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. 2009;69:573–582. doi: 10.1158/0008-5472.CAN-08-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannini C, et al. Selective ablation of Notch3 in HCC enhances doxorubicin’s death promoting effect by a p53 dependent mechanism. J Hepatol. 2009;50:969–979. doi: 10.1016/j.jhep.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 25.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 26.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 27.Uematsu K, et al. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 28.Shou J, et al. Human Dkk-1, a gene encoding a Wnt antagonist, responds to DNA damage and its overexpression sensitizes brain tumor cells to apoptosis following alkylation damage of DNA. Oncogene. 2002;21:878–889. doi: 10.1038/sj.onc.1205138. [DOI] [PubMed] [Google Scholar]
- 29.Ohigashi T, et al. Inhibition of Wnt signaling downregulates Akt activity and induces chemosensitivity in PTEN-mutated prostate cancer cells. Prostate. 2005;62:61–68. doi: 10.1002/pros.20117. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev. Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varnat F, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clement V, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehtesham M, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 35.Alajez NM, et al. Targeted depletion of BMI1 sensitizes tumor cells to P53-mediated apoptosis in response to radiation therapy. Cell Death Differ. 2009;16:1469–1479. doi: 10.1038/cdd.2009.85. [DOI] [PubMed] [Google Scholar]
- 36.Crea F, et al. BMI1 silencing enhances docetaxel activity and impairs antioxidant response in prostate cancer. Int. J. Cancer. 2010;128:1946–1954. doi: 10.1002/ijc.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy JA, et al. Comment on “Tumor growth need not be driven by rare cancer stem cells”. Science. 2007;318:1722. doi: 10.1126/science.1149590. author reply 1722. [DOI] [PubMed] [Google Scholar]
- 38.Beier D, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 39.Artells R, et al. Tumour CD133 mRNA expression and clinical outcome in surgically resected colorectal cancer patients. Eur. J. Cancer. 2010;46:642–649. doi: 10.1016/j.ejca.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Pallini R, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin. Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 41.Okayama H, et al. CD44v6, MMP-7 and nuclear Cdx2 are significant biomarkers for prediction of lymph node metastasis in primary gastric cancer. Oncol. Rep. 2009;22:745–755. doi: 10.3892/or_00000496. [DOI] [PubMed] [Google Scholar]
- 42.Celebiler Cavusoglu A, et al. Predicting invasive phenotype with CDH1, CDH13, CD44, and TIMP3 gene expression in primary breast cancer. Cancer Sci. 2009;100:2341–2345. doi: 10.1111/j.1349-7006.2009.01333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oldenhuis CN, et al. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer. 2008;44:946–953. doi: 10.1016/j.ejca.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Ong CW, et al. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod. Pathol. 2010;23:450–457. doi: 10.1038/modpathol.2009.181. [DOI] [PubMed] [Google Scholar]
- 45.Murat A, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 46.Vormittag L, et al. Co-expression of Bmi-1 and podoplanin predicts overall survival in patients with squamous cell carcinoma of the head and neck treated with radio(chemo)therapy. Int. J. Radiat. Oncol. Biol. Phys. 2009;73:913–918. doi: 10.1016/j.ijrobp.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 47.Vrzalikova K, et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J. Cancer Res. Clin. Oncol. 2008;134:1037–1042. doi: 10.1007/s00432-008-0361-y. [DOI] [PubMed] [Google Scholar]
- 48.Yasuda H, et al. Elevated CD133, but not VEGF or EGFR, as a predictive marker of distant recurrence after preoperative chemoradiotherapy in rectal cancer. Oncol. Rep. 2009;22:709–717. doi: 10.3892/or_00000491. [DOI] [PubMed] [Google Scholar]
- 49.Schaefer KL, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur. J. Cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikawa R, et al. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br. J. Cancer. 2008;98:1670–1674. doi: 10.1038/sj.bjc.6604361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salnikov AV, et al. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. Int. J. Cancer. 2010;126:950–958. doi: 10.1002/ijc.24822. [DOI] [PubMed] [Google Scholar]
- 52.Danesi R, et al. Pharmacogenetics of anticancer drug sensitivity in non-small cell lung cancer. Pharmacol. Rev. 2003;55:57–103. doi: 10.1124/pr.55.1.4. [DOI] [PubMed] [Google Scholar]
- 53.Fan X, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma. 2009;9(Suppl 3):S205–S210. doi: 10.3816/CLM.2009.s.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westhoff B, et al. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JT, et al. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 57.Lee SH, et al. Mutational analysis of NOTCH1,2,3 and 4 genes in common solid cancers and acute leukemias. APMIS. 2007;115:1357–1363. doi: 10.1111/j.1600-0463.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 58.Fu YP, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villaronga MA, et al. HEY1 Leu94Met gene polymorphism dramatically modifies its biological functions. Oncogene. 2010;29:411–420. doi: 10.1038/onc.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart LS, El-Deiry WS. Invincible, but not invisible: imaging approaches toward in vivo detection of cancer stem cells. J. Clin. Oncol. 2008;26:2901–2910. doi: 10.1200/JCO.2008.16.9573. [DOI] [PubMed] [Google Scholar]
- 61.Heyn C, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn. Reson. Med. 2006;56:1001–1010. doi: 10.1002/mrm.21029. [DOI] [PubMed] [Google Scholar]
- 62.Riethdorf S, et al. Review: Biological relevance of disseminated tumor cells in cancer patients. Int. J. Cancer. 2008;123:1991–2006. doi: 10.1002/ijc.23825. [DOI] [PubMed] [Google Scholar]
- 63.Balic M, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin. Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 64.Theodoropoulos PA, et al. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2009;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Lang JE, et al. Significance of micrometastasis in bone marrow and blood of operable breast cancer patients: research tool or clinical application? Expert Rev. Anticancer Ther. 2007;7:1463–1472. doi: 10.1586/14737140.7.10.1463. [DOI] [PubMed] [Google Scholar]
- 66.Slade MJ, Coombes RC. The clinical significance of disseminated tumor cells in breast cancer. Nat. Clin. Pract. Oncol. 2007;4:30–41. doi: 10.1038/ncponc0685. [DOI] [PubMed] [Google Scholar]
- 67.Therasse P, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 68.Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 69.Hurt EM, et al. CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seruga B, et al. Cytokines and their relationship to the symptoms and outcome of cancer. Nat. Rev. Cancer. 2008;8:887–899. doi: 10.1038/nrc2507. [DOI] [PubMed] [Google Scholar]
- 71.Salgado R, et al. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. Int. J. Cancer. 2003;103:642–646. doi: 10.1002/ijc.10833. [DOI] [PubMed] [Google Scholar]
- 72.Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Funel N, et al. Laser microdissection and primary cell cultures improve pharmacogenetic analysis in pancreatic adenocarcinoma. Lab. Invest. 2008;88:773–784. doi: 10.1038/labinvest.2008.40. [DOI] [PubMed] [Google Scholar]
- 74.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimshaw MJ, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang X, et al. Isolation, enrichment, and maintenance of medulloblastoma stem cells. J. Vis. Exp. 2010 doi: 10.3791/2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duhagon MA, et al. Genomic profiling of tumor initiating prostatospheres. BMC Genomics. 2010;11:324. doi: 10.1186/1471-2164-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garraway IP, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70:491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dubrovska A, et al. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc. Natl. Acad. Sci. U.S.A. 2009;106:268–273. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]