Abstract
Background
The single spanning transmembrane amyloid precursor protein (APP) and its proteolytic product, amyloid-beta (Aβ) peptide, have been intensely studied due to their role in the pathogenesis of Alzheimer’s disease. However, the biological role of the secreted ectodomain of APP, which is also generated by proteolytic cleavage, is less well understood. Here, we report Tol2 red fluorescent protein (RFP) transposon gene trap integrations in the zebrafish amyloid precursor protein a (appa) and amyloid precursor-like protein 2 (aplp2) genes. The transposon integrations are predicted to disrupt the appa and aplp2 genes to primarily produce secreted ectodomains of the corresponding proteins that are fused to RFP.
Results
Our results indicate the Appa-RFP and Aplp2 fusion proteins are likely secreted from the central nervous system and accumulate in the embryonic veins independent of blood flow.
Conclusions
The zebrafish appa and aplp2 transposon insertion alleles will be useful for investigating the biological role of the secreted form of APP.
Keywords: Tol2 gene trap, endothelial cells, vein, vasculature, central nervous system
INTRODUCTION
Alzheimer’s disease (AD) is the most prevalent form of human dementia, accounting for 60–70% cases worldwide. The neural pathology of AD includes senile plaques, neurofibrillary tangles, and loss of neurons. In addition, there is a significant vascular pathology in AD characterized by amyloid deposits in cerebral vessel walls (cerebral amyloid angiopathy), as well as structural abnormalities in the microvasculature (Revesz et al., 2003; Bailey et al., 2004; Kumar-Singh, 2008; Storkebaum et al., 2011). The amyloid precursor protein (APP) is known to be the source of the hydrophobic peptide amyloid β (Aβ) that is a major component of amyloid deposits in the brains of AD patients (Kang et al., 1987; Newman et al., 2007; Philipson et al., 2010). Membrane bound APP is composed of a large extracellular amino-terminal domain, a single transmembrane domain, and a short cytoplasmic domain (reviewed in Gralle and Ferreira, 2007; Jacobsen and Iverfeldt, 2009). The processing of APP involves regulated intramembrane proteolysis, which can be divided into two major pathways. Approximately 90% of APP proteolytic processing is through the nonamyloidogenic pathway, in which cleavage of the extracellular domain by α-secretase releases a soluble form of APP (sAPPα) into the extracellular space. Subsequent cleavage by γ-secretase releases the sAPPα into the cytoplasm. The remaining 10% of APP processing occurs by means of the amyloidogenic pathway in which the extracellular domain is cleaved at a different residue by β-secretase. This releases an alternative extracellular soluble form, sAPPβ. Cleavage of the remaining membrane bound protein by γ-secretase releases the hydrophobic Aβ peptide into the extracellular space. Although there are extensive studies on APP and the Aβ peptide, the in vivo biological function and localization of secreted sAPP is not completely known, nor is the contribution of sAPP to the neural and vascular pathogenesis of AD.
Studies in Drosophila and mammalian cell culture systems have implicated sAPP in the regulation of neurite outgrowth (Small et al., 1994), neuronal survival (Araki et al., 1991) and neuroprotection (Goodman and Mattson, 1994). In addition, the sAPP peptide is sufficient to rescue molting and morphogenesis defects from loss of APL-1 in Caenorhabditis elegans and is suggested to function in a cell-nonautonomous manner (Hornsten et al., 2007). In mice, a knock-in allele that produces sAPPα exclusively rescues the postnatal lethality in APP/APLP2 double mutants (Weyer et al., 2011), suggesting that much of the normal biological function of the APP gene family can be mediated through the soluble extracellular domains. It is also intriguing that patients with AD display reduced levels of the sAPPα cleavage peptide (Lannfelt et al., 1995), raising the possibility that the sAPPα could contribute to the pathogenesis of AD. In humans and mice, the APP genes are predominantly expressed in neural tissues, and there is little evidence for expression in cell types other than neurons or ganglia (Goldgaber et al., 1987; Tanzi et al., 1987; Arai et al., 1991).
The application of Tol2 DNA transposons for gene trap, enhancer trap, and germline mutagenesis screens is well established in zebrafish (Balciunas and Ekker, 2005; Balciunas et al., 2006; Kawakami, 2007; Largaespada, 2009; Suster et al., 2009; Ivics and Izsvak, 2010). A major advantage of using gene trap transposons as a mutagen is that the integrated transposon acts as a molecular tag that facilitates gene cloning. Zebrafish are particularly well suited to gene trap insertional mutagenesis due to the optical clarity of the embryo and the amenability of the organism to large-scale screens. Gene trap transposons are engineered to intercept splicing of the endogenous gene transcript and produce fluorescent proteins that act as reporters of the normal expression pattern of the mutated gene. This approach, using red fluorescent protein (RFP) or green fluorescent protein (GFP) trap Tol2 transposons, has identified many genes with tissue specific patterns of interest in the developing zebrafish embryo (Balciunas et al., 2004; Kawakami et al., 2004, 2010; Parinov et al., 2004; Choo et al., 2006; Kawakami, 2007; Sivasubbu et al., 2007; Asakawa and Kawakami, 2009). Recent modification of the Tol2 gene trap system has yielded a powerful tool for expression as well as functional annotation of the zebrafish genome (Clark et al., 2011). Live cell imaging of trapped fluorescent proteins in the zebrafish embryo can yield novel insights into the biological role of the gene product in a particular cellular or developmental process.
In the present study, we carried out a germline mutagenesis screen using the Tol2 gene trap GBT-R15 (Petzold et al., 2009) to identify novel genes involved in vascular development in the zebrafish embryo. We present an analysis of two of the integration lines that show accumulation of RFP in the vasculature. Cloning of the integration sites revealed one line had an insertion in the gene amyloid beta (A4) precursor protein a (appa; Musa et al., 2001) and the other in the amyloid beta (A4) precursor-like protein 2 (aplp2; Jelen et al., 2007). Both appa and aplp2 are members of the APP family that encode type I transmembrane proteins expressed in cells of the central nervous system. In situ hybridization in the zebrafish embryo demonstrates that appa and aplp2 are expressed in the neurons and ganglia of the central and peripheral nervous system. The locations of the gene trap insertion sites are predicted to create fusion proteins that are similar to the normal processing of APP by the α- or β-secretase pathway to produce the soluble secreted form of APP. Our analysis of the Appa-RFP and Aplp2-RFP proteins in living zebrafish embryos shows the secreted soluble proteins accumulate in the venous compartment of the vasculature. The appa and aplp2 gene trap lines isolated in this study will be useful for investigating the biological roles of the secreted soluble APP proteins.
RESULTS AND DISCUSSION
Isolation of appa and aplp2 Tol2 Gene Trap Alleles
To identify genes involved in early vascular development, we performed a germline mutagenesis screen using the Tol2 RFP gene trap GBT-R15 (Petzold et al., 2009). The GBT-R15 transposon contains a splice acceptor, an RFP lacking the first initiating AUG codon, and a transcriptional terminator. Out of 227 founder adults, 37 produced progeny with tissue and cell-type specific patterns of RFP gene expression in the embryo (frequency of 16%). This frequency is comparable to previously published enhancer trap and gene trap reports in zebrafish (Balciunas and Ekker, 2005). Four of the 37 lines showed RFP expression in the embryonic vasculature, named V1–V4. To identify the Tol2 integration site in the vascular gene trap lines, we performed 5′-rapid amplification of cDNA ends (RACE) on total RNA isolated from 2 days post fertilization (dpf) RFP expressing embryos. The 5′-RACE product from line V2 contained the first 4 exons of the zebrafish amyloid beta (A4) precursor protein a (appa) gene fused to the 5′ end of RFP. The V3 line 5′-RACE product contained the first 12 exons of the amyloid beta (A4) precursor-like protein 2 (aplp2) gene fused to the 5′ end of RFP. These results indicate that in gene trap line V2 the Tol2 transposon had integrated into intron 4 of the appa gene (Fig. 1A), while in line V3 the transposon had integrated into intron 12 of the aplp2 gene (Fig. 1A). Zebrafish appa (Ensembl(Zv9):ENSDARG00000059036) and aplp2 (Ensembl(Zv9):ENSDARG00000054864) are orthologues of human APP and APLP2, respectively, and are both members of the amyloid beta (A4) precursor protein family. The gene trap insertion alleles for the two lines isolated in this study are designated as appais22Gt and aplp2is23Gt.
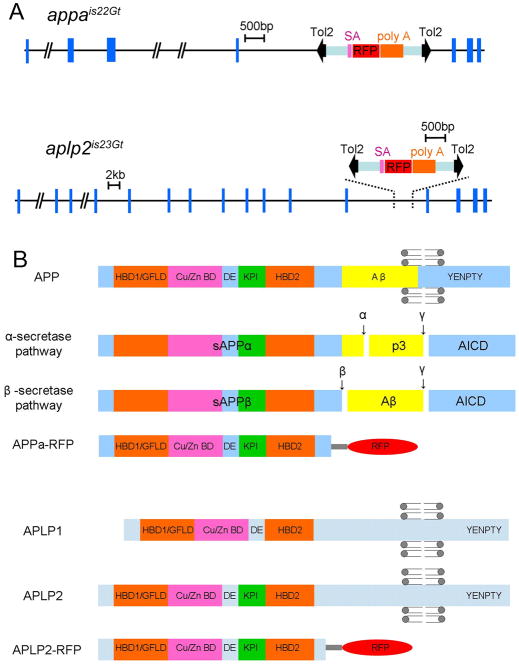
Fig. 1.
Genomic organization of zebrafish appa and aplp2 genes showing the location of Tol2 gene trap integration sites and predicted polypeptides. A: Genomic structure of appa and aplp2. Blue boxes indicate exons. Location of Tol2 gene trap insertion site is shown. Black arrowheads indicate transposon arms. Purple, red and orange boxes represent the splice acceptor (SA), red fluorescent protein cDNA (RFP), and transcriptional termination/poly adenylation sequences (poly A), respectively. B: Schematic of domain organization of the App proteins and their proteolytic processing. The predicted Appa-RFP and Aplp2-RFP proteins contain nearly the entire ectodomain. Grey circles with attached lines represent the lipid bilayer positioned at the transmembrane domain. HBD1/GFLD: heparin-binding domain/growth factor like domain; Cu/Zn BD: copper- ad zinc- binding domain; DE: acidic-rich domain; KPI: Kunitz protease inhibitor domain; α, β and γ: α-, β- and γ-secretase cleavage sites; p3, α-secretase proteolytic product; Aβ: β-secretase proteolytic product, amyloid β peptide; AICD, APP intracellular domain; YENPTY, conserved C-terminal interaction motif.
To confirm the location of the Tol2 integration sites in lines appais22Gt and aplp2is23Gt, genomic Southern blot analyses were performed with probes specific to the appa and aplp2 genes, and the Tol2 GBT-R15 transgene. Using a gene-specific probe a larger restriction fragment length polymorphism (RFLP) is detected in line appais22Gt and aplp2is23Gt (Supp. Fig. S1A,B, which is available online), as expected based on the predicted location of the transposon integration site in each gene (Supp. Fig. S1). Genomic Southern analysis on lines appais22Gt and aplp2is23Gt using a probe complementary to the RFP cDNAdetected a single RFLP of the expected size in each line (Supp. Fig. S2A,B). Together these results demonstrate that the RFP expression patterns observed in lines appais22Gt and aplp2is23Gt are the result of a single Tol2 gene trap integration in the appa and aplp2 genes, respectively.
Zebrafish App family
The human APP gene family encodes single-pass transmembrane proteins that are cleaved to produce extracellular and intracellular soluble peptides. The zebrafish appa and aplp2 genes encode proteins that have the same predicted structure, with a large extracellular domain containing multiple subdomains, a single transmembrane domain, and a short cytoplasmic tail (Musa et al., 2001; Fig. 1B). The conserved extracellular domain contains two heparin-binding domains (HBD), a growth factor like domain (GFLD), a metal-binding domain (Cu/Zn BD), an acidic domain (DE) and a Kunitz protease inhibitor domain (KPI; Fig. 1B). In addition, the locations of predicted cleavage sites recognized by α-, β-, and γ-secretases are conserved (Fig. 1B, Supp. Fig. S3). This indicates that the same pathways likely process the zebrafish proteins as human APP to create the soluble and secreted forms of APP. The location of the Tol2 gene trap integration in each gene suggests that the Appa-RFP and Aplp2-RFP gene trap proteins are secreted. The transposon had integrated into an intron upstream of the exons that code for the single-pass transmembrane domain (Fig. 1A,B; Supp. Fig. S3). The accumulation of the Appa-RFP and Aplp2-RFP trap proteins in the vasculature in the embryo also supports this (see below). The resulting fusion proteins are predicted to contain an N-terminal signal sequence that targets them to the secretory pathway.
In zebrafish, the APP family is composed of four genes (Fig. 2A). There are two orthologues of APP, named appa and appb, probably originating as a result of the ancient genome duplication in the teleost lineage (Musa et al., 2001; Joshi et al., 2009). There is a single orthologue of the APLP1 and APLP2 genes, named aplp and aplp2 (Jelen et al., 2007). Zebrafish appa is located on chromosome 1 and encodes a 738 amino acid protein with 80% similarity to human APP (Supp. Fig. S3). Zebrafish aplp2 is located on chromosome 18 and encodes a 764 amino acid protein with 78% similarity to human APLP-2 (Supp. Fig. S3). Phylogenetic analysis indicates that APP and APLP2 orthologues are more closely related to each other than to APLP or the single APP orthologues in C. elegans (apl-1) and Drosophila (appl; Fig. 2A).
Fig. 2.
Phylogenetic and expression analysis of zebrafish appa and aplp2. A: Phylogenetic tree of amyloid precursor proteins. Zebrafish Appa and Aplp2 are shown in bold. The horizontal bars represent the percentage of amino acid substitutions required to generate the corresponding tree. Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis. B: Relative expression levels of appa and aplp2 throughout zebrafish development and in adult tissues as evaluated by reverse transcriptase-polymerase chain reaction. Expression of the ribosomal protein S6 kinase b, polypeptide 1b (rps6kb1) gene was used as a control.
Expression patterns of Zebrafish appa and aplp2
The APP genes are widely expressed in human adult and fetal tissues (Goldgaber et al., 1987; Tanzi et al., 1987); however, tissues are composed of multiple cell types, and there is little evidence that the genes are expressed in cell types other than neurons. Although there is one report of human APP protein localization in endothelial cells of the gut (Cabal et al., 1995), there are no reports in the literature of APP mRNA expression in vascular cells. A previous study showed that appa gene expression is first detectable at the mid-gastrula stage in the zebrafish embryo (Musa et al., 2001). We used reverse transcriptase-polymerase chain reaction (RT-PCR) to examine the relative expression level of appa and aplp2 in the developing embryo and adult tissues (Fig. 2B). A low level of appa expression is detected in 1 to 12 hours post fertilization (hpf) embryos. This level increases substantially at 24 hpf, and remains high through 5 dpf. In contrast, the level of aplp2 is relatively low from day 1 through day 5. In adult tissues, both appa and aplp2 are widely expressed except in the muscle and liver. The level of expression of appa is higher in the adult brain compared with aplp2, while aplp2 is higher than appa in the ovary and intestine. These results suggest that zebrafish appa and aplp2 are widely expressed throughout development, similar to the human APP gene family.
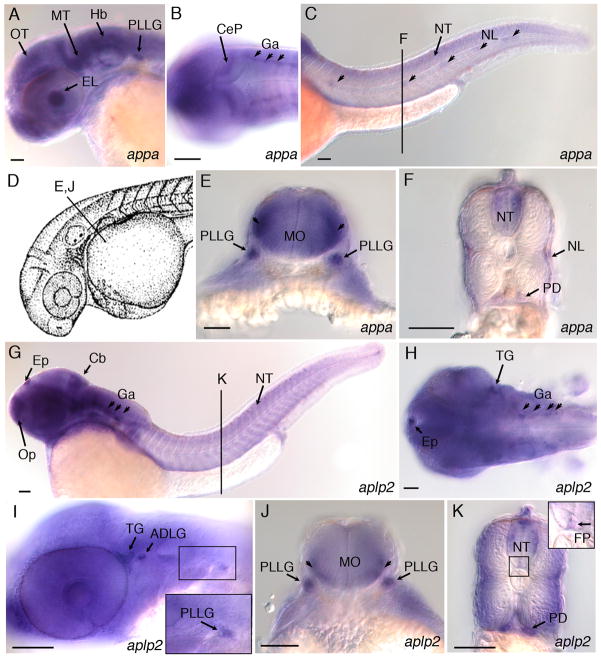
To identify the cell types that express zebrafish appa and aplp2 during early development, we performed whole-mount in situ hybridization on wild-type embryos. Zebrafish appa has previously been shown to be expressed in the developing nervous system at 24 hpf (Musa et al., 2001). We also observed expression of appa, as well as aplp2, throughout the central nervous system at 36 hpf (Fig. 3). Control experiments using appa and aplp2 sense probes showed no detectable pattern above background (Supp. Fig. S4). Both appa and aplp2 show very similar patterns in the embryo and are expressed throughout the forebrain, midbrain and hindbrain (Fig. 3A,G) and along the length of the neural tube (Fig. 3C,F,K). Expression was also detected in cranial ganglia (Fig. 3B,H), the trigeminal ganglia (Fig. 3H,I) and anterodorsal and posterior lateral line ganglia (Fig. 3E,I,J). A cross-section through the trunk shows expression of both genes in the pronephric ducts and the neural tube (Fig. 3F,K). appa is expressed in the neuromasts of the lateral line (Fig. 3C,F), while a low level of aplp2 is detected in the floor plate (Fig. 3K, inset). Neither gene is expressed at detectable levels in the dorsal aorta, caudal vein, vessels in the head or intersegmental vessels of the trunk.
Fig. 3.
Expression of appa and aplp2 in developing central and peripheral nervous system of 36 hpf zebrafish embryos. A: Lateral view, appa expression in the eye lens, optic tectum, midbrain tegmentum, hindbrain, and posterior lateral line ganglion. B: Dorsal view, appa expression in the cerebellar plate and ganglia. C: Lateral view, appa expression in the neural tube and lateral line neuromasts (arrowheads). Line labeled “F” refers to position of cross-section shown in panel F. D: Camera lucida drawing of 35 hours post fertilization (hpf) zebrafish embryo (Kimmel et al., 1995). Line labeled “E, J” refers to position of cross-sections in panels E and J. E: appa expression in the posterior lateral line ganglia and lateral regions of medulla oblongata (arrowheads). F: appa expression in the neural tube, neuromasts of the lateral line, and pronephric ducts. G: Lateral view of aplp2 expression in the epiphysis, olfactory placode, cerebellum, ganglia, and neural tube. Line labeled “K” refers to position of cross-section shown in panel K. H: Dorsal view, aplp2 expression in the epiphysis, trigeminal ganglia, and ganglia in the hindbrain. I: Lateral view, aplp2 expression in the trigeminal ganglia, anterodorsal and posterior lateral line ganglia. J: aplp2 expression in posterior lateral line ganglion and lateral regions of medulla oblongata (arrowheads). K: aplp2 expression in lateral regions of the neural tube, floor plate, and pronephric duct. Inset shows higher magnification view of boxed region of neural tube. Arrow points to expression of aplp2 in the floor plate region. ADLG, anterodorsal lateral line ganglia; Cb, cerebellum; CeP, cerebellar plate; EL, eye lens; Ep, epiphysis; FP, floor plate; Ga, ganglion; Hb, Hindbrain; MO, medulla oblongata; MT, midbrain tegmentum; NL, neuromasts of lateral line; NT, neural tube; Op, olfactory placode; OT, optic tectum; PD, pronephric duct; PLLG, posterior lateral line ganglion; TG, trigeminal ganglia. Scale bars = 50 μm.
Secreted Appa- and Aplp2-RFP Fusion Proteins Accumulate at the Zebrafish Embryonic Vasculature
We examined the localization of the Appa-RFP and Aplp2-RFP fusion proteins by fluorescence and confocal microscopy in living 36 hpf embryos. Appa-RFP localization is primarily in the caudal vein but can be detected in some of the intersegmental vessels (Fig. 4A,B). A low level of expression can be detected throughout the central nervous system in the head and trunk (Fig. 4A). A similar pattern of RFP fluorescence was detected for the Aplp2-RFP protein in aplp2is23Gt embryos (Fig. 5A,B). Several veins are in close association with each region of the central nervous system and pronephric ducts where appa and aplp2 were detected by in situ hybridization. These vessels include the cerebral and cardinal veins, intersegmental vessels, and the axial vein. It is possible that App-RFP fusion proteins are secreted by neurons and accumulate in the vessels located nearby. In situ hybridization with an RFP-specific probe on appais22Gt and aplp2is23Gt 36 hpf embryos reveals a pattern similar to the endogenous appa and aplp2 genes. A high level of expression was present in the central nervous system and pronephric ducts, however, no signal was detected in the endothelial cells of the blood vessels (Supp. Fig. S5). These results are consistent with the prediction that the Appa-RFP and Aplp2-RFP fusion proteins are secreted from neuronal cells and cells of the pronephric ducts and subsequently accumulate at the embryonic vasculature.
Fig. 4.
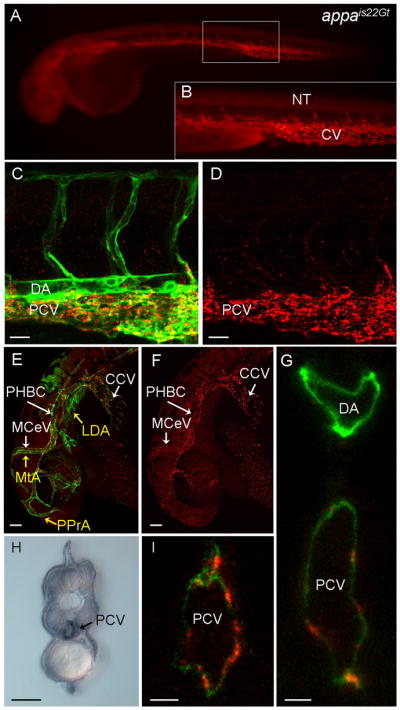
Red fluorescent protein (RFP) expression in the Appais22Gt gene trap line. A–G,I: Fluorescence images of live Appais22Gt; Tg(flk1:moesin-gfp)is1 embryos. A: Appa-RFP was detected throughout the veins and weakly in CNS and neural tube in 34 hours post fertilization (hpf) embryos. B: Enlargement of boxed area in (A) shows RFP fluorescence in the caudal vein and intersegmental vessels. C–F: Appa-RFP localization at 32 hpf embryos in the trunk (C,D) and the head (E,F). Appa-RFP fluorescence overlaps with GFP expression in endothelial cells in the veins (C,E), but not the arterial vessels (MTA, PPrA, LDA marked with yellow arrows in E). H: Immunolocalization with anti-RFP antibody shows Appa-RFP is localized to the caudal vein but absent from the dorsal aorta. G,I: Confocal images of cross-section through the trunk of a 36 hpf embryo shows Appa-RFP accumulation in the posterior caudal vein. CCV, common cardinal vein; CV, caudal vein; LDA, lateral dorsal aorta; MCeV, midcerebral vein; MtA, metencephalic artery; NT, neural tube; PHBC, primordial hindbrain channel; PPrA, primitive prosencephalic artery. Scale bars = 20 μm in C,D, 50 μm in E–I.
Fig. 5.
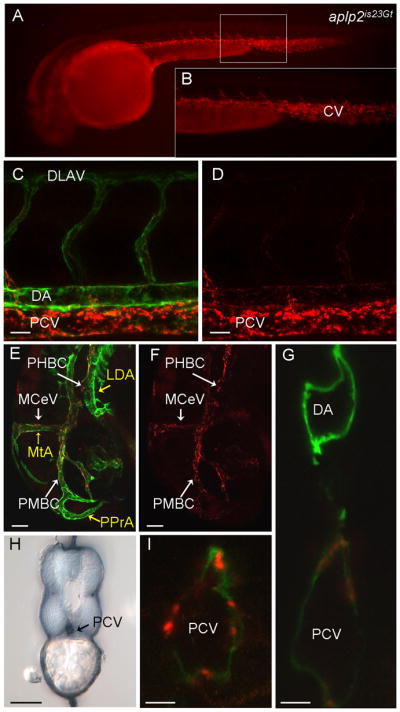
Red fluorescent protein (RFP) expression in the aplp2is23Gt gene trap line. Fluorescence images of live aplp2is23Gt; Tg(flk1:moesin-gfp)is1 embryos. A: Aplp2-RFP fluorescence in the caudal vein and intersegmental vessels in a 34 hours post fertilization (hpf) embryo. B: Enlargement of boxed area in (A). C–F: RFP fluorescence at 32 hpf was observed in the trunk (C,D) and the head (E,F). C,E: Aplp2-RFP fluorescence overlaps with green fluorescent protein (GFP) expression in endothelial cells in the veins, but not the arterial vessels (MTA, PPrA, LDA marked with yellow arrows in E). Weak expression is detected in the dorsal aorta. H: Immunolocalization with anti-RFP antibody shows Appa-RFP was in the caudal vein but absent from the dorsal aorta. G,I: Confocal images of cross-sections through the trunk of 36 hpf embryo shows Aplp2-RFP accumulation at the posterior caudal vein. CV, caudal vein; LDA, lateral dorsal aorta; MCeV, midcerebral vein; MtA, metencephalic artery; PHBC, primordial hindbrain channel; PMBC, primordial midbrain channel; PPrA, primitive prosencephalic artery. Scale bars = 20 μm in C,D, 50 μm in E–I.
To confirm that Appa-RFP and Aplp2-RFP localization is restricted to the venous vessels, we created double transgenic embryos using a line that expresses a GFP fusion protein in endothelial cells of both arteries and veins. The transgenic line Tg(flk1:moesin-egfp)is1 (Wang et al., 2010) expresses a Moesin1-GFP fusion protein under the control of the flk1 promoter, which results in expression in the endothelial cells of the vasculature (Jin et al., 2005). The appais22Gt and aplp2is23Gt lines were crossed to Tg(flk1:moesin-egfp)is1 and living embryos imaged by confocal microscopy. Similar results were observed in appais22Gt/+; Tg(flk1:moesin-egfp)is1/+ (Fig. 4) and aplp2is23Gt/+; Tg(flk1:moesin-egfp)is1/+ embryos (Fig. 5). Comparison of GFP and RFP localization in the trunk showed overlap only in the posterior caudal vein and the ventral region of the intersegmental vessels (Figs. 4C,D, 5C,D). In the head, GFP was detected throughout the veins and arteries, whereas RFP was primarily in the veins, including the common cardinal vein, midcerebral vein (MCeV), and the primordial hindbrain channel (PHBC; Figs. 4E,F, 5E,F). We confirmed the localization of RFP in the veins by immunolocalization of RFP in fixed 36 hpf transgenic embryos. A cross-section through the trunk shows RFP enriched at the posterior caudal vein but absent from the neural tube and posterior lateral line ganglia (Figs. 4H, 5H). Confocal microscopy of double transgenic living embryos suggested that Appa-RFP and Aplp2-RFP fusion proteins were localized to both the luminal and abluminal surface of the endothelial cells of the posterior caudal vein, as well as in intracellular regions possibly corresponding to endocytic vesicles (Figs. 4G,I, 5G,I).
Examination of Appa-RFP and Aplp2-RFP protein localization in embryos at earlier stages of development is consistent with the in situ hybridization data indicating expression is primarily restricted to neural tissues. Appa-RFP and Aplp2-RFP showed accumulation in locations in which blood vessels had not yet formed. At 24 hpf, Appa-RFP is detected at a low level throughout the neural tube and central nervous system (Supp. Fig. S6A–D). In the head Appa-RFP is enriched in an area around the lens in the eye (Supp. Fig. S6A,B, yellow arrows). Appa-RFP is also enriched at the somite boundaries in the trunk (Supp. Fig. S6C,D, yellow arrows), before the intersegmental vessels have completely formed. Later, at 28 hpf, Appa-RFP is detected in the region of the posterior caudal vein (Supp. Fig. 6G,H, white arrows). Similar results were observed with Aplp2-RFP protein localization (data not shown). Taken together, these results suggest that the Appa-RFP and Aplp2-RFP fusion proteins are not expressed in vascular cells, but instead are expressed in neurons, ganglia, and cells of the pronephros and are secreted. These results are consistent with studies of human β-APP that detected the presence of the proteins in neural tissues and only a few nonneural tissues, namely the pituitary and adrenal glands and cardiac muscle (Arai et al., 1991). The expression patterns in zebrafish indicate that following production in the neurons, the secreted Appa-RFP and Aplp2-RFP fusion proteins become enriched at the venous vessels in the embryo after 28 hpf.
Appa-RFP and Aplp2-RFP Are Localized to Blood Vessels in the Absence of Blood Flow
We next tested whether blood flow was required for the accumulation of the fusion proteins in the veins by crossing the gene trap alleles into a silent heartmn0031Gt (sihmn0031Gt) mutant background (Clark et al., 2011), which lacks a heartbeat and blood flow due to a mutation in the cardiac troponin T2a gene, tnnt2a (Sehnert et al., 2002). The appais22Gt/+; sihmn0031Gt/+ or aplp2is23Gt/+; sihmn0031Gt/+ double heterozygous fish were further crossed with Tg(flk1:moesin1-egfp)is1/+; sihmn0031Gt/+ to generate homozygous appais22Gt/+; sihmn0031Gt/mn0031Gt or aplp2is23Gt/+; sihmn0031Gt/mn0031Gt mutants in a Tg(flk1:moesin1-egfp)is1/+ background. Despite the absence of a heartbeat or normal blood flow in sihmn0031Gt/mn0031Gt homozygous mutant embryos, Appa-RFP and Aplp2-RFP accumulated at the venous vessels, similar to control heterozygous sihmn0031Gt/+ siblings with a normal heart beat (Supp. Fig. S7). These results demonstrate that the accumulation of Appa-RFP and Aplp2-RFP at the veins occurs by a mechanism independent of blood flow.
We followed the expression of the Appa-RFP and Aplp2-RFP fusion proteins in older animals and found that RFP accumulated in the kidneys beginning at 5 dpf; however, RFP was still detected at the venous vessels even at 10 dpf (data not shown). Adult fish were dissected and the organs examined for RFP fluorescence on a standard epi-illumination microscope. RFP expression was detected in cells embedded in the membrane covering the brain (Supp. Fig. S8A,B,E–H). These cells are likely to be microglia, which are the resident macrophages of the central nervous system. RFP expressing cells were also present in the connective tissue in the ovary and the membrane covering the spleen (data not shown). RFP was not detected in muscle, liver, skin, intestine, or spleen. Consistent with the expression of Appa-RFP and Aplp2-RFP in older larvae, RFP was highly expressed throughout the kidney tubules (Supp. Fig. S8C–E,J).
The localization of Appa-RFP and Aplp2-RFP is distinct from the mRNA expression patterns of appa and aplp2, suggesting that the fusion proteins are secreted by nonvascular cells and then accumulate at the veins. Because both fusion proteins contain nearly the entire N-terminal extracellular domain, similar to the soluble secreted form of sAPP, it is possible that the conserved subdomains (i.e., heparin binding/growth factor like, metal binding, acidic, or KPI domains) are responsible for the binding of the fusion proteins to the extracellularmatrix at the veins. Studies of the extracellular domain of APP indicate APP functions as an adhesion molecule/contact receptor, has the ability to self-dimerize, and binds to specific components of the extracellular matrix including collagen, heparin-sulfate proteoglycans, and F-spondin (Gralle and Ferreira, 2007; Jacobsen and Iverfeldt, 2009). Release of the intracellular domain of APP by the γ-secretase pathway is stimulated by binding of the extracellular domain to the GPI-linked ligand TAG1 (Ma et al., 2008), similar to the processing of other type-1 transmembrane proteins such as Notch. Fibulin-1, a member of the fibulin family of secreted glycoproteins that are components of basement membrane, has been shown to bind APP’s amino-terminal growth factor-like domain; this interaction contributes to the neurotrophic activities of APP (Ohsawa et al., 2001). In addition, Fibulin-1 and fibulin-5 have been shown to suppress angiogenesis in tumors derived from mice injected with HT1080 cancer cells (Xie et al., 2008). Fibulin-1 knock-out mice presented with a severe perinatal lethal phenotype with structural defects in endothelial cells of the microvasculature (Kostka et al., 2001). It is possible that sAPP might bind fibulin-1 in the vascular basement membrane in zebrafish, and this interaction could account for the localization of the Appa-RFP and Aplp2-RFP fusion proteins at the embryonic veins. Binding to other components of the extracellular matrix could also contribute to the localization of Appa-RFP and Aplp2-RFP in the zebrafish embryo.
Previous analyses of the zebrafish appb gene demonstrated that endogenous appb is expressed in the central nervous system (Musa et al., 2001), while an appb promoter-GFP transgene revealed GFP present throughout the nervous system and the embryonic vasculature (Lee and Cole, 2007). Our present study demonstrates that endogenous appa and aplp2 are expressed primarily in the central and peripheral nervous system, and in the pronephric ducts of the developing kidney, but not in the vasculature. Our results for appa are consistent with previous analyses of zebrafish appa and appb (Musa et al., 2001), and of human and mouse APP genes (Goldgaber et al., 1987; Tanzi et al., 1987; Arai et al., 1991). Studies on Presenilin-1 (PS1), one of three genes (APP, PS1, and Presenilin-2) known to be mutated in familial AD, suggest that signaling between neuronal and vascular tissues plays a role in the pathology of AD. Gama-Sosa et al. have shown that transgenic mice overexpressing a mutant form of human PS1 develop vascular pathologies (Gama Sosa et al., 2010). The PS1 transgene is specifically expressed in neural cells and is absent from vascular endothelial cells (Gama Sosa et al., 2010). Our similar findings on zebrafish appa and aplp2 expression suggest the appais22Gt and aplp2is23Gt alleles could provide new insights into the role of secreted App proteins in vascular biology and the mechanism linking the neural and vascular pathologies of Alzheimer’s disease.
In a recent study, the knockdown of appb was shown to cause defective convergent extension movements in zebrafish embryos, while knockdown of appa had no apparent effect on development (Joshi et al., 2009). The zebrafish appais22Gt and aplp2is23Gt insertion alleles isolated in the present study are homozygous viable, consistent with the previously published morpholino knockdown experiments. The lack of an obvious phenotype could be due to functional redundancy among APP family members in zebrafish. This could also be a result of low amounts of wild-type appa and aplp2 transcripts present in homozygous appais22Gt/is22Gt and aplp2is23Gt/is23Gt embryos, as detected by RT-PCR (Supp. Figs. S9C, S10C). This indicates that the gene trap alleles do not completely disrupt normal splicing of the gene and are not null alleles. Moreover, the production of soluble Appa-RFP and Aplp2-RFP fusion proteins may compensate for the loss of the full-length Appa and Aplp2 proteins, similar to studies in mice (Weyer et al., 2011). The ligand binding and adhesive activity of the extracellular domain of APP is important for its role in synaptogenesis, cell adhesion, and neurite outgrowth, but the exact biological function remains unclear (Jacobsen and Iverfeldt, 2009). The secreted App-RFP proteins expressed by the zebrafish appais22Gt and aplp2is23Gt gene trap alleles isolated in our study will be useful for investigating further the biological function of APP.
EXPERIMENTAL PROCEDURES
Zebrafish Husbandry and Strains
Wild-type and WIK strains of zebrafish were housed in an AHAB system (Aquatic Ecosystems, Inc., Apopka, FL) and maintained under a 14-hr light /10-hr dark cycle at 27°C. The WIK zebrafish strain was obtained from the Zebrafish International Research Center (http://zebrafish.org/zirc/home/guide.php). The silent heartmn0031Gt allele contains a Tol2 gene trap integration in the cardiac tropinin T gene (Clark et al., 2011) and was obtained from Darius Balciunas, Temple University. The Tg(flk1:moesin-egfp)is1 line was described previously (Wang et al., 2010). Embryos were collected following fertilization and allowed to develop in a 28.5°C incubator under standard laboratory conditions. For in situ hybridization experiments, the embryos were placed in fish water (60.5 mg salts/l) containing 0.003% 1-phenyl-2-thiourea (PTU) to inhibit pigment formation in melanocytes. Staging of embryos was as published (Kimmel et al., 1995).
Embryo Injections for the Germline Insertional Mutagenesis Screen
Capped Tol2 transposase mRNA was synthesized in vitro from 1 μg of linearized pT3TS-Tol2 plasmid template (Balciunas et al., 2006) using the mMESSAGE mMACHINE High Yield Capped RNA transcription kit (Ambion, Cat. No. AM1348) and purified using the RNeasy MinElute Cleanup Kit (Qiagen). The GBT-R15 Tol2 RFP gene trap plasmid (Petzold et al., 2009) was propagated in bacteria and purified using the QIAPrep spin column kit (Qiagen). The mRNA and plasmid DNA were diluted in water, and 50 pg of GBT-R15 plasmid DNA and 300 pg of Tol2 mRNA were coinjected into 1-cell embryos. 227 injected embryos were raised to adulthood. Adults were intercrossed or outcrossed with WIK, and the F1 progeny were screened for RFP expression from the Tol2 gene trap at 24 and 48 hpf. F1 embryos for each Tol2 integration line were raised to adulthood, and subsequent generations maintained by outcrossing with WIK.
Computational Analysis
Protein sequence alignment was performed using Clustal W (Swiss Institute of Bioinformatics). Phylogenetic tree construction was carried out using MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software (Tamura et al., 2007).
In Situ Hybridization and Confocal Imaging
Whole-mount in situ hybridization on zebrafish embryos and larvae was performed using digoxigenin antisense RNA probes as described previously (Larson et al., 2004). appa and aplp2 cDNAs were amplified by PCR using the following primers: appa, forward 5′-TGTGGAGTTCGTGTGCTGTC-3′; appa, reverse 5′-CGTGATGACGATGATGGTTG-3′; aplp2, forward 5′-CCCTGTGGCATCGATAAGTT-3′; and aplp2, reverse 5′-CCGTACTGCCTCTTCCTCAG-3′. The PCR products were cloned into the pCRII TOPO vector. A 541-bp fragment of the RFP was amplified from the GBT-R15 plasmid using primers rfp-L: 5′-CGAGGACGTCATCAAGGAGT-3′ and rfp-R: 5′-CTTGGCCATGTAGGTGGTCT-3′, and cloned into the pCR4 TOPO vector. The cloned appa, aplp2, and RFP cDNAS were used as template for synthesizing digoxigenin-labeled antisense and sense RNA probes. Digoxigenin-labeled RNA probes were synthesized by in vitro transcription in the presence of dig-labeled UTP (Roche) and purified as described (McGrail et al., 2010). In situ labeled embryos were analyzed on a Zeiss Axioscope using DIC optics and photographed with a Nikon Coolpix camera. Living embryos were mounted in 1% low-melting agarose and imaged on a Zeiss LSM 700 confocal microscope. Confocal z-series were analyzed with Zeiss AxioVision software. The identification of blood vessels in the embryonic vasculature was determined according to zebrafish anatomy described at http://zfish.nichd.nih.gov/zfatlas/FinalDesign1/DiagPage.html and http://zfin.org/action/anatomy/term-detail?anatomyItem.zdbID=ZDB-ANAT-010921-585.
Immunohistochemistry
Zebrafish embryos were selected for RFP expression and were fixed overnight in 4% paraformaldehyde in phosphate buffered saline (PBS) at 4°C. Embryos were dechorionated, dehydrated through an ethanol series and stored at −20°C. Embryos were transferred to acetone at −20°C for 15 min and washed in H2O with 0.1% Triton X-100 at room temperature for 5 min. Following two rinses in PBS/0.1% Triton X-100 (PBSTx), embryos were blocked for one hour at room temperature in 2% sheep serum, 1% bovine serum albumin (BSA), 1% dimethyl sulfoxide (DMSO), 0.1% Triton X-100 in PBS (blocking solution). DsRed rabbit polyclonal antibody (Clontech) was diluted 1:400 in blocking solution and absorbed to embryos overnight at 4°C. Embryos were washed at room temperature eight times over two hours in 1% BSA, 1% DMSO, 0.1% TX-100 in PBS and placed in blocking solution for 30 min. An anti-rabbit horseradish peroxidase (HRP) antibody (Zymed) was diluted 1:500 in blocking solution and absorbed to the embryos overnight at 4°C. Following incubation with the secondary antibody, the embryos were washed as above and equilibrated for 15 min in diaminobenzidine (DAB) buffer (Roche). Embryos were then stained with DAB staining solution (Roche). Embryos were imaged as above for in situ hybridization.
5′-RACE Analysis
Total RNA was isolated from 5 to 10 RFP-positive embryos at 2 to 3 dpf using Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA amplification was performed using a 5′ RACE System (Invitrogen) according to the manufacturer’s instructions. First strand cDNA was synthesized using a primer complementary to RFP: 5′-GTAGATGAACTCGCCGTCCT-3′. After cDNA purification, a string of cytosines was added to the 3′ end of the cDNA by the terminal deoxy transferase reaction. The dC-tailed cDNA was amplified by nested PCR using an RFP complementary primer: 5′-AATGGATCCGGAGCCGTACTGGAACTGAG-3′ and an abridged anchor primer that recognizes the dC tail at the 3′ end of the cDNA. Nested PCR was performed with an annealing temperature of 55°C for 30–35 cycles using Taq DNA polymerase (Promega). PCR products were Topo-TA (Invitrogen) cloned and sequenced at the DNA Facility at Iowa State University.
Genotyping and RT-PCR Analysis
Genotyping was based on PCR amplification of the junction fragments between the flanking regions of the Tol2 transposon integration site and the Tol2 transposon. The locations of primers are shown in Figure S2A and Figure S3A. Genomic DNA was extracted from 3 to 4 dpf embryos using the DNeasy Blood and Tissue DNA Extraction kit (Qiagen). The primers used to amplify from appa gene were V2 jf-L: 5′-CCCATCATTCCTTCCTCAAGCA-3′ and V2 jf-R: 5′-TCCAGGCGCAATGCTGTGTTAC-3′. For aplp2 gene, primers were V3 jf-L: 5′-CAACACTTCCAGTTTCCCGCCTA-3′ and V3 jf-R: 5′-CAGAAAGTTCAACGGTGGGGCAA-3′. The PCR primer used to amplify out from the Tol2 transposon was Tol2-L: 5′-CGGCTGCCTGTGAGAGGCTT-3′. The PCR products were gel purified and sequenced to confirm the amplified products.
RT-PCR analysis of total RNA was carried out from staged embryos, larvae, and adult organs as described (McGrail et al., 2010). The sequences of primers used for PCR were as follows: appa, forward 5′-GTGGAGGCCATGCTGAACGA-3′ in exon 4; appa, reverse 5′-CGATGATGCCGTGGTGGATG-3′ in exon 9; aplp2, forward 5′-CGCCATGTTCTGCGGGAAAC-3 in exon 2′; aplp2, reverse 5′-CAACCTCCACGATGCCGTGA-3′ in exon 16. Control primer sequences for ribosomal protein S6 kinase b (rps6kb1) were forward 5′-CATGGCGACGGTGCGTTCAT-3′ and reverse 5′-AGCTTGCCGCCCGTCTGAAA-3′.
Genomic Southern Blot Analyses
Genomic DNA was extracted from adult fish and nonradioactive Southern blot analysis performed as previously described (McGrail et al., 2011). The sequences of primers used to amplify digoxigenin-labeled PCR probes were as follows. A 297-bp probe for the appa gene was amplified from genomic DNA using primers appa-L: 5′-TCTGGACTGAAGCCTGATGA-3′ and appa-R: 5′-GGGGTTTTCATAGCCGTTCT-3′. A 292-bp probe complementary to the aplp2 gene was amplified from genomic DNA using primers aplp2-L: 5′-GCTCTGGCATGAACAGTCTG-3′ and aplp2-R: 5′-CCGTACTGCCTCTTCCTCAG-3′. A 541-bp probe complementary to the RFP cDNA was amplified from the GBT-R15 plasmid using primers rfp-L: 5′-CGAGGACGTCATCAAGGAGT-3′ and rfp-R: 5′-CTTGGCCATGTAGGTGGTCT-3′. Images of blots were captured on a ChemiDoc XRS imaging system.
Supplementary Material
Acknowledgments
Grant sponsor: Center for Integrated Animal Genomics; Grant sponsor: Roy J. Carver Charitable Trust; Grant number: 07-2991; Grant sponsor: NIH-NCRR; Grant number: P40 RR012546.
The authors thank Dr. Darius Balciunas for zebrafish harboring the sihmn0031Gt allele. The WIK strain of zebrafish used in this study was obtained from the Zebrafish International Resource Center. This work was supported by the Center for Integrated Animal Genomics, the Roy J. Carver Charitable Trust, and Iowa State University startup funds awarded to J.J.E., H.K.L., and M.M. wrote the manuscript; H.K.L., Y.W., J.J.E., M.M., K.J.C., S.C.E. designed the screen and experiments; K.J.C. and S.C.E. provided plasmids; Y.W. and M.M. performed injections; Y.W. and Q.W. performed screening; M.M. designed RACE-PCR cloning strategy; Y.W., J.B., and C.K.K. cloned RACE products; H.K.L. and K.E.N.W. cloned genomic-transposon junction fragments; K.E.N.W. performed phylogenetic and RT-PCR analyses; H.K.L. and Y.W. performed confocal analysis; Y.W. performed Southern analyses; H.K.L. performed genotyping, immunohistochemistry, and in situ expression analyses.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Arai H, Lee VM, Messinger ML, Greenberg BD, Lowery DE, Trojanowski JQ. Expression patterns of beta-amyloid precursor protein (beta-APP) in neural and nonneural human tissues from Alzheimer’s disease and control subjects. Ann Neurol. 1991;30:686–693. doi: 10.1002/ana.410300509. [DOI] [PubMed] [Google Scholar]
- Araki W, Kitaguchi N, Tokushima Y, Ishii K, Aratake H, Shimohama S, Nakamura S, Kimura J. Trophic effect of beta-amyloid precursor protein on cerebral cortical neurons in culture. Biochem Biophys Res Commun. 1991;181:265–271. doi: 10.1016/s0006-291x(05)81412-3. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K. The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish. Methods. 2009;49:275–281. doi: 10.1016/j.ymeth.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Rivara CB, Rocher AB, Hof PR. The nature and effects of cortical microvascular pathology in aging and Alzheimer’s disease. Neurol Res. 2004;26:573–578. doi: 10.1179/016164104225016272. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Ekker SC. Trapping fish genes with transposons. Zebrafish. 2005;1:335–341. doi: 10.1089/zeb.2005.1.335. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal A, Alonso-Cortina V, Gonzalez-Vazquez LO, Naves FJ, Del Valle ME, Vega JA. beta-Amyloid precursor protein (beta APP) in human gut with special reference to the enteric nervous system. Brain Res Bull. 1995;38:417–423. doi: 10.1016/0361-9230(95)02006-d. [DOI] [PubMed] [Google Scholar]
- Choo BG, Kondrichin I, Parinov S, Emelyanov A, Go W, Toh WC, Korzh V. Zebrafish transgenic Enhancer TRAP line database (ZETRAP) BMC Dev Biol. 2006;6:5. doi: 10.1186/1471-213X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, Balciunas D, Pogoda HM, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, Ni J, Nielsen AL, Patowary A, Scaria V, Sivasubbu S, Xu X, Hammerschmidt M, Ekker SC. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama Sosa MA, Gasperi RD, Rocher AB, Wang AC, Janssen WG, Flores T, Perez GM, Schmeidler J, Dickstein DL, Hof PR, Elder GA. Age-related vascular pathology in transgenic mice expressing presenilin 1-associated familial Alzheimer’s disease mutations. Am J Pathol. 2010;176:353–368. doi: 10.2353/ajpath.2010.090482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science. 1987;235:877–880. doi: 10.1126/science.3810169. [DOI] [PubMed] [Google Scholar]
- Goodman Y, Mattson MP. Secreted forms of beta-amyloid precursor protein protect hippocampal neurons against amyloid beta-peptide-induced oxidative injury. Exp Neurol. 1994;128:1–12. doi: 10.1006/exnr.1994.1107. [DOI] [PubMed] [Google Scholar]
- Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O’Connor G, Plasterk R, Li C. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci U S A. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z. The expanding universe of transposon technologies for gene and cell engineering. Mob DNA. 2010;1:25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KT, Iverfeldt K. Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors. Cell Mol Life Sci. 2009;66:2299–2318. doi: 10.1007/s00018-009-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelen N, Ule J, Zivin M, Darnell RB. Evolution of Nova-dependent splicing regulation in the brain. PLoS Genet. 2007;3:1838–1847. doi: 10.1371/journal.pgen.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Joshi P, Liang JO, DiMonte K, Sullivan J, Pimplikar SW. Amyloid precursor protein is required for convergent-extension movements during Zebrafish development. Dev Biol. 2009;335:1–11. doi: 10.1016/j.ydbio.2009.07.041. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Abe G, Asada T, Asakawa K, Fukuda R, Ito A, Lal P, Mouri N, Muto A, Suster ML, Takakubo H, Urasaki A, Wada H, Yoshida M. zTrap: zebrafish gene trap and enhancer trap database. BMC Dev Biol. 2010;10:105. doi: 10.1186/1471-213X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kostka G, Giltay R, Bloch W, Addicks K, Timpl R, Fassler R, Chu ML. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–7034. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Singh S. Cerebral amyloid angiopathy: pathogenetic mechanisms and link to dense amyloid plaques. Genes Brain Behav. 2008;7(suppl 1):67–82. doi: 10.1111/j.1601-183X.2007.00380.x. [DOI] [PubMed] [Google Scholar]
- Lannfelt L, Basun H, Wahlund LO, Rowe BA, Wagner SL. Decreased alpha-secretase-cleaved amyloid precursor protein as a diagnostic marker for Alzheimer’s disease. Nat Med. 1995;1:829–832. doi: 10.1038/nm0895-829. [DOI] [PubMed] [Google Scholar]
- Largaespada DA. Transposon mutagenesis in mice. Methods Mol Biol. 2009;530:379–390. doi: 10.1007/978-1-59745-471-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Wadman SA, Chen E, Kerley L, Clark KJ, Eide M, Lippert S, Nasevicius A, Ekker SC, Hackett PB, Essner JJ. Expression of VE-cadherin in zebrafish embryos: a new tool to evaluate vascular development. Dev Dyn. 2004;231:204–213. doi: 10.1002/dvdy.20102. [DOI] [PubMed] [Google Scholar]
- Lee JA, Cole GJ. Generation of transgenic zebrafish expressing green fluorescent protein under control of zebrafish amyloid precursor protein gene regulatory elements. Zebrafish. 2007;4:277–286. doi: 10.1089/zeb.2007.0516. [DOI] [PubMed] [Google Scholar]
- Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, Karagogeos D, Watanabe K, Dawe GS, Xiao ZC. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- McGrail M, Batz L, Noack K, Pandey S, Huang Y, Gu X, Essner JJ. Expression of the zebrafish CD133/prominin1 genes in cellular proliferation zones in the embryonic central nervous system and sensory organs. Dev Dyn. 2010;239:1849–1857. doi: 10.1002/dvdy.22307. [DOI] [PubMed] [Google Scholar]
- McGrail M, Hatler JM, Kuang X, Liao HK, Nannapaneni K, Watt KE, Uhl JD, Largaespada DA, Vollbrecht E, Scheetz TE, Dupuy AJ, Hostetter JM, Essner JJ. Somatic mutagenesis with a Sleeping Beauty transposon system leads to solid tumor formation in zebrafish. PLoS One. 2011;6:e18826. doi: 10.1371/journal.pone.0018826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa A, Lehrach H, Russo VA. Distinct expression patterns of two zebrafish homologues of the human APP gene during embryonic development. Dev Genes Evol. 2001;211:563–567. doi: 10.1007/s00427-001-0189-9. [DOI] [PubMed] [Google Scholar]
- Newman M, Musgrave IF, Lardelli M. Alzheimer disease: amyloidogenesis, the presenilins and animal models. Biochim Biophys Acta. 2007;1772:285–297. doi: 10.1016/j.bbadis.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Ohsawa I, Takamura C, Kohsaka S. Fibulin-1 binds the amino-terminal head of beta-amyloid precursor protein and modulates its physiological function. J Neurochem. 2001;76:1411–1420. doi: 10.1046/j.1471-4159.2001.00144.x. [DOI] [PubMed] [Google Scholar]
- Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- Petzold AM, Balciunas D, Sivasubbu S, Clark KJ, Bedell VM, Westcot SE, Myers SR, Moulder GL, Thomas MJ, Ekker SC. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci U S A. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson O, Lord A, Gumucio A, O’Callaghan P, Lannfelt L, Nilsson LN. Animal models of amyloid-beta-related pathologies in Alzheimer’s disease. FEBS J. 2010;277:1389–1409. doi: 10.1111/j.1742-4658.2010.07564.x. [DOI] [PubMed] [Google Scholar]
- Revesz T, Ghiso J, Lashley T, Plant G, Rostagno A, Frangione B, Holton JL. Cerebral amyloid angiopathies: a pathologic, biochemical, and genetic view. J Neuropathol Exp Neurol. 2003;62:885–898. doi: 10.1093/jnen/62.9.885. [DOI] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Sivasubbu S, Balciunas D, Amsterdam A, Ekker SC. Insertional mutagenesis strategies in zebrafish. Genome Biol. 2007;8(suppl 1):S9. doi: 10.1186/gb-2007-8-s1-s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DH, Nurcombe V, Reed G, Clarris H, Moir R, Beyreuther K, Masters CL. A heparin-binding domain in the amyloid protein precursor of Alzheimer’s disease is involved in the regulation of neurite outgrowth. J Neurosci. 1994;14:2117–2127. doi: 10.1523/JNEUROSCI.14-04-02117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storkebaum E, Quaegebeur A, Vikkula M, Carmeliet P. Cerebrovascular disorders: molecular insights and therapeutic opportunities. Nat Neurosci. 2011;14:1390–1397. doi: 10.1038/nn.2947. [DOI] [PubMed] [Google Scholar]
- Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Gusella JF, Watkins PC, Bruns GA, St George-Hyslop P, Van Keuren ML, Patterson D, Pagan S, Kurnit DM, Neve RL. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235:880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kaiser MS, Larson JD, Nasevicius A, Clark KJ, Wadman SA, Roberg-Perez SE, Ekker SC, Hackett PB, McGrail M, Essner JJ. Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development. 2010;137:3119–3128. doi: 10.1242/dev.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer SW, Klevanski M, Delekate A, Voikar V, Aydin D, Hick M, Filippov M, Drost N, Schaller KL, Saar M, Vogt MA, Gass P, Samanta A, Jaschke A, Korte M, Wolfer DP, Caldwell JH, Muller UC. APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 2011;30:2266–2280. doi: 10.1038/emboj.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Palmsten K, MacDonald B, Kieran MW, Potenta S, Vong S, Kalluri R. Basement membrane derived fibulin-1 and fibulin-5 function as angiogenesis inhibitors and suppress tumor growth. Exp Biol Med (Maywood) 2008;233:155–162. doi: 10.3181/0706-RM-167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.