Abstract
The SMN protein is essential and participates in the assembly of macromolecular complexes of RNA and protein in all cells. The best-characterized function of SMN is as an assembler of spliceosomal small nuclear ribonucleoproteins (snRNPs). SMN performs this function as part of a complex with several other proteins called Gemins. snRNPs are assembled in the cytoplasm in a stepwise manner and then are imported to the nucleus where they participate globally in the splicing of pre-mRNA. Mutations in the SMN1 gene result in the motor neuron disease, spinal muscular atrophy (SMA). Most of these mutations result in a reduction in the expression levels of the SMN protein, which, in turn, results in a reduction in snRNP assembly capacity. This review highlights current studies that have investigated the mechanism of SMN-dependent snRNP assembly, as well as the downstream effects on pre-mRNA splicing that result from a decrease in SMN.
Keywords: spinal muscular atrophy, survival motor neuron, SMN, SMA, snRNP, Gemin
1. Introduction
Spinal muscular atrophy (SMA) is a severe motor neuron disease characterized by skeletal muscle weakness, respiratory insufficiency and death (Crawford and Pardo, 1996). SMA is caused by mutations in the Survival Motor Neuron 1 (SMN1) gene (Lefebvre et al., 1995). Most of these SMN1 mutations are deletions; however, ~2% are missense mutations (Alias et al., 2009). Humans also possess a second copy of the gene, SMN2, containing a single nucleotide change in exon 7. While this change is translationally silent, it does affect splicing of the SMN pre-mRNA, resulting in aberrant skipping of exon 7 in approximately 85% of the SMN2 transcripts (Cartegni and Krainer, 2002; Gennarelli et al., 1995; Kashima and Manley, 2003; Lorson et al., 1999; Monani et al., 1999). Thus, those afflicted with SMA have reduced levels of functional full-length SMN protein expressed in cells. The precise biochemical pathways that result in motor neuron dysfunction and death when SMN protein expression is reduced are obscure. On the other hand, much progress has been made in understanding the major cellular function of the SMN protein. This work was initiated by the Dreyfuss laboratory, which first recognized the role of SMN in the biogenesis of spliceosomal snRNPs (Liu and Dreyfuss, 1996).
The SMN protein is a multi-functional protein, broadly acting in the assembly of protein-RNA complexes (RNPs) (Eggert et al., 2006; Fischer et al., 2011; Liu et al., 1997; Meister et al., 2002; Paushkin et al., 2002; Simic, 2008; Wan et al., 2005). SMN accomplishes this in a modular way, bringing together several RNA binding proteins with several “substrate” RNAs, facilitating the assembly of specific proteins on the target RNAs. In this review, we describe the role of the SMN protein in the stepwise assembly of spliceosomal small nuclear RNPs (snRNPs), highlight the relationship between defective snRNP assembly and SMA phenotype, and consolidate our current understanding of the downstream RNA processing consequences of defective snRNP biogenesis.
2. snRNP assembly
Splicing of pre-mRNA transcripts is carried out in eukaryotes by 9 snRNPs (Nilsen, 2003; Will and Luhrmann, 2001). These large RNA-protein complexes recognize the 5’ and 3’ splice sites, as well as the branch points, on most eukaryotic introns, and directly catalyze the splicing reaction, yielding mature, spliced mRNAs. Each snRNP contains a single copy of one of 9 uridine-rich small-nuclear RNAs (U1, U2, U4, U5, U6, U11, U12, U4atac, and U6atac snRNAs), bound to a set of accompanying proteins. In addition to proteins specific to each snRNP, all snRNPs except U6 also contain 7 common core proteins, known as Sm proteins. The seven Sm proteins (Sm B/B’, D1, D2, D3, E, F and G) form a heptameric ring around the uridine-rich Sm site found on all snRNAs (Achsel et al., 2001; Kambach et al., 1999; Stark et al., 2001). Formation of this Sm core is required for the in vivo stability, nuclear import, and splicing activity of the snRNPs.
At one time, it was thought that snRNAs and Sm proteins spontaneously assembled to form spliceosomal snRNPs in cells. However, in 1996, Dreyfuss and colleagues demonstrated that the SMN protein existed in a protein complex with spliceosomal snRNP proteins and subsequently demonstrated that SMN was required for the assembly of spliceosomal snRNPs in cells (Liu and Dreyfuss, 1996; Pellizzoni et al., 2002b). They also found that SMN does not act in this function alone, but within a complex with seven other proteins called Gemins. SMN itself lies at the heart of the SMN complex, contacting most of the Gemins. Gemin2, a 32-kDa protein most often found with SMN, binds to Sm proteins (Fischer et al., 1997; Liu et al., 1997). Gemin3 is a DEAD-box RNA-dependent RNA helicase and ATPase (Charroux et al., 1999). Gemin5 is a large protein containing WD repeat domains (Gubitz et al., 2002). Gemins 6 and 7 adopt Sm folds (Baccon et al., 2002; Ma et al., 2005; Pellizzoni et al., 2002a), and little is known about the structures of the remaining Gemins 4 and 8.
It is now clear that the SMN complex directly binds to snRNAs and to Sm proteins, and then assembles the Sm core onto the snRNA in an ATP-dependent manner (Fischer et al., 1997; Meister et al., 2001a; Pellizzoni et al., 2002b). Thus, SMN plays a profound cellular function that has the potential to influence pre-mRNA splicing in all cells. Recent research has focused on understanding how the SMN complex binds to snRNAs and to Sm proteins.
3. snRNA binding activity
The SMN complex binds snRNA through one of its associated proteins, Gemin5. Gemin5 binds directly and with high affinity and specificity to snRNAs (Battle et al., 2006; Lau et al., 2009). The 170-kDa Gemin5 protein comprises at least two domains, an N-terminal WD-repeat domain and a C-terminal half containing no identifiable sequence homology to any other proteins (Gubitz et al., 2002). Gemin5 primarily uses its WD-repeat domain to bind directly to the snRNA (Lau et al., 2009). While the WD-repeat domain accounts for the majority of the affinity for snRNAs, the novel C-terminal domain is required to achieve the full snRNA-binding specificity of the intact SMN complex (Battle et al., 2006; Golembe et al., 2005; Lau et al., 2009). Gemin5 makes specific contacts to several positions of the Sm site on snRNAs, including high-affinity contacts with the first adenosine as well as the first and third uridines, although the other uridines also contribute to the binding (Battle et al., 2006). While, in this respect, Gemin5 seems to mimic the binding specificity of Sm proteins, Gemin5 further requires the presence of a stem-loop immediately 3’ of the Sm site, the so-called “snRNP code” (Battle et al., 2006; Golembe et al., 2005).
The majority of Gemin5 is in the cytoplasm, where, depending on conditions, roughly 25–75% of Gemin5 resides outside of the SMN complex (Battle et al., 2007a; Pellizzoni, 2007). Inhibition of snRNP assembly, either in cell extracts with ATPase inhibitors or in vivo with cell stressors or protein synthesis inhibitors, induces Gemin5 to dissociate from SMN and accumulate in these cytoplasmic SMN-free complexes (Battle et al., 2007a; Yong et al., 2010). It is this SMN-free Gemin5 that binds to newly exported snRNAs prior to association with SMN (Figure 1). Further, Gemin5 binds precursor snRNAs with extraneous sequence still on their 3’ ends, suggesting that 3’ end maturation occurs on the SMN complex (Yong et al., 2010).
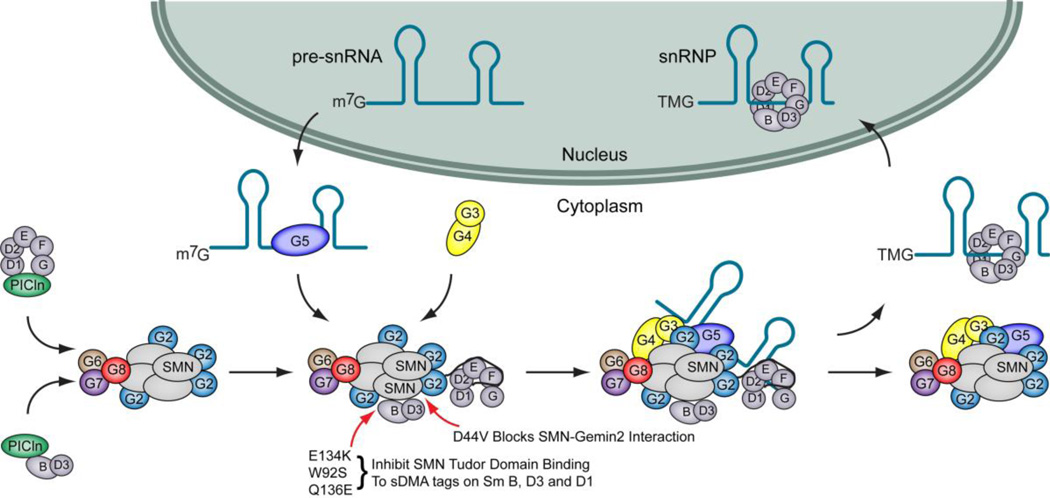
Fig. 1.
The role of SMN in snRNP biogenesis. Following transcription, snRNAs are exported to the cytoplasm where they are bound by Gemin5 and delivered to SMN for Sm core assembly. Sm proteins are methylated in the cytoplasm by PRMT5 and then transferred to pICln, which delivers them to the SMN complex. The SMN complex binds Sm proteins through at least 2 binding modes: 1) SMN binds to sDMA tails on Sm B, D1 and D3, and 2) Gemin2 recognizes the Sm-fold of the Sm D1/D2/E/F/G pentamer. Several SMA patient mutations disrupt these binding activities. SMN forms oligomers and is depicted here as a tetramer for simplicity. The SMN complex then assembles the Sm proteins into a heptameric ring on the snRNA. Assembled snRNPs are then imported into the nucleus for final maturation and function in pre-mRNA splicing.
4. Interactions of SMN with Sm proteins
Sm proteins, like snRNAs, are never free in the cytoplasm. After synthesis, Sm proteins are processed by several complexes based on the protein methyltransferase, PRMT5, and the small protein, pICln (Brahms et al., 2000; Brahms et al., 2001; Friesen et al., 2001b; Friesen et al., 2002; Meister et al., 2001b) (Figure 1). Sm proteins are first targeted to the 20S methylosome, where arginine residues on the C-terminal tails of Sm D1, D3 and B/B’ are converted by PRMT5 to symmetrical dimethylarginine (Meister et al., 2001b). Following methylation, a pentamer of Sm proteins, D1/D2/E/F/G, are sequestered by pICln (Meister et al., 2001b). Recent EM structures reveal that the pentamer-pICln complex is a closed ring similar to the heptameric Sm ring, except that pICln takes the place of the B/D3 subunits (Chari et al., 2008; Chari et al., 2009). SMN and Gemin2 (and possibly Gemin8) catalyze release of the Sm pentamer from pICln (Chari et al., 2008; Chari et al., 2009).
The Sm pentamer is then transferred to the SMN complex, where it is bound by Gemin2. Recently, the structure of the Sm pentamer bound to Gemin2 and the Gemin2-binding domain of SMN was solved by X-ray crystallography (Zhang et al., 2011). When bound to Gemin2, the partial Sm ring is in an open configuration, in contrast to the pICln-bound Sm pentamer (Zhang et al., 2011). Gemin2 wraps around the open ring, contacting all five Sm proteins. Interestingly, the N-terminal tail of Gemin2 extends into the snRNA-binding pocket on the Sm pentamer, blocking the RNA-binding surface. Further, although the pentamer ring is open at one end, the intersubunit angles are reduced relative to those observed in the heptameric Sm structures, leaving no room for either the B/D3 dimer or snRNA (Figure 1). The Gemin2 binding domain of SMN binds to Gemin2 distal to the Sm binding surface, and does not contact the Sm proteins. A notable SMA-causing missense mutation occurs at position D44, which is observed in the crystal structure at the SMN-Gemin2 interface (Sun et al., 2005; Zhang et al., 2011). The D44V patient mutation disrupts snRNP assembly by disrupting the interaction between SMN and the Sm-fold binding Gemin2 protein (Ogawa et al., 2007; Zhang et al., 2011).
In addition to binding the Sm-fold of the pentamer, the SMN complex also binds to the symmetrical-dimethylarginine (sDMA) tails found on Sm B/B’, D1 and D3 (Friesen et al., 2001a). This binding is directly mediated by the Tudor domain of SMN (Friesen et al., 2001a). NMR structures have detailed the interaction of sDMA peptides with the SMN Tudor domain, explaining the specificity for sDMA, as well as elucidating the mechanism by which several SMA patient missense mutations interfere with snRNP assembly (Selenko et al., 2001; Sprangers et al., 2003; Tripsianes et al., 2011). The SMN Tudor domain forms a cage of aromatic residues that specifically bind sDMA (Tripsianes et al., 2011). Glu134 stabilizes the conformation of the cage through a hydrogen bond network (Selenko et al., 2001; Tripsianes et al., 2011). The SMN E134K mutation disrupts this network, causing significant reduction in the ability of the Tudor domain to bind sDMA (Tripsianes et al., 2011).
Still other SMA-associated SMN mutations occurring in the Tudor domain disrupt Sm binding in different ways. For instance, W92S and Q136E cause the Tudor domain to misfold, disrupting the ability of SMN to bind sDMA (Kotani et al., 2007; Shpargel and Matera, 2005; Tripsianes et al., 2011). Three more SMN missense mutations, G95R, I116F and A111G, are oriented towards the interior of the hydrophobic core formed by the beta barrel structure of the SMN Tudor domain (Selenko et al., 2001; Tripsianes et al., 2011). In the context of the Tudor domain alone, these mutations cause smaller changes to the structure and do not greatly affect sDMA binding, although there may be larger defects in the context of full-length SMN.
Although these mutations suggest that the ability to bind sDMA is critical to the SMA-relevant SMN function, the precise role for sDMA in Sm core assembly remains unclear. The simplest explanation may be that while Gemin2 holds the D1/D2/E/F/G pentamer, the sDMA tails either allow the recruitment of the B/D3 dimer to SMN, or they allow proper spatial orientation of the Sm proteins for transfer to snRNA, although these activities do not seem to be strictly required in in vitro systems (Chari et al., 2008; Zhang et al., 2011). Further, it must be noted that while these mutations disrupt productive SMN interactions with Sm proteins necessary for Sm core assembly, these mutations could also potentially disrupt the interaction of the SMN Tudor domain with other methylated arginine-bearing proteins and thereby affect non-snRNP-related functions, as well as cause loss of misfolded SMN proteins in their entirety (Côté and Richard, 2005; Jones et al., 2001; Pellizzoni et al., 2001; Pillai et al., 2003; Schumperli and Pillai, 2004) Also, SMN does not act alone, but rather as an obligate oligomer, and the effects of these mutations may be ameliorated by oligomerization of mutant SMN with the wildtype SMN expressed from SMN2. The SMN A111G mutation, for example, seems to be relatively inactive in snRNP assembly on its own, but is able to complement activity in the presence of small amounts of wildtype SMN expressed from the SMN2 gene (Burghes and Beattie, 2009; Workman et al., 2009).
Sm core assembly proceeds readily in vitro with purified Sm proteins and snRNAs, and without SMN (Kleinschmidt et al., 1989; Meister et al., 2001a; Pellizzoni et al., 2002b; Raker et al., 1996; Raker et al., 1999; Sumpter et al., 1992). This is not the case when the reaction occurs in cell extracts (Fischer et al., 1997; Meister et al., 2001a; Pellizzoni et al., 2002b). This suggests that, in vivo, the SMN complex functions as a specificity factor, sequestering Sm proteins and snRNAs until they are finally assembled together (Pellizzoni et al., 2002b). Gemin5 binds to snRNAs in a complex absent of either SMN or Sm proteins (Battle et al., 2007b; Yong et al., 2010). It is likely that this complex serves to sequester snRNAs until they are delivered to SMN for assembly. pICln, in turn, binds to Sm proteins in the absence of SMN, blocking their snRNA-binding activity (Pu et al., 1999). Once pICln delivers the Sm proteins to the SMN-Gemin2 subunit, Gemin2 blocks the snRNA binding surface (Zhang et al., 2011). Presumably, SMN acts as an adaptor in vivo to bring both the snRNA and Sm proteins together where, finally, the Sm proteins and snRNA are released for assembly.
5. Downstream consequences
SMN itself does not appear to function directly in pre-mRNA splicing. Rather, reduced SMN lowers the capacity of cells to assemble snRNPs, the general pre-mRNA splicing machinery, which leads to altered levels of spliceosomal components and defects in splicing. It remains unclear how a defect of splicing results in a motor neuron-specific phenotype. Two broad theories have emerged to explain this cell type-specificity (Burghes and Beattie, 2009; Pellizzoni, 2007). 1) Altered splicing in motor neurons could result in the absence of critical pre-mRNA splicing events that are unique to motor neurons. 2) SMN may be critical in other RNA-protein complexes in motor neurons, and a loss in this function may explain motor neuron selectivity. It is also likely that these two possibilities are not mutually exclusive and may be tied to the unique developmental profile of motor neurons.
It is possible to quantify the capacity of cell extracts to assemble spliceosomal snRNPs. Overall, snRNP assembly activity correlates well with both SMN levels and disease phenotype (Gabanella et al., 2007; Wan et al., 2005; Workman et al., 2009). In vitro snRNP assembly assays are conducted in cell extracts containing excess amounts of exogenous snRNA and ATP (Pellizzoni et al., 2002b; Wan et al., 2005). In vivo, excess ATP and RNA are not necessarily present, and it is known that snRNP levels are further dependent on the rates of snRNA transcription, snRNP turnover, nuclear import, and final snRNP maturation in the nucleus. As such, the in vitro snRNP assembly assays measure the capacity of the SMN complex in an extract to assemble Sm cores, and do not necessarily directly reflect in vivo snRNP levels. Nevertheless, much has been learned by measuring snRNP assembly capacity in cell and animal models of SMA.
Studies of SMN expression and snRNP assembly in mice have revealed that SMN levels and snRNP assembly activity changes during CNS development (Gabanella et al., 2005). snRNP assembly capacity is high in the spinal cord until approximately 2 weeks postnatally in mice, when it is reduced approximately 10-fold compared to the E18 level. These data suggest that SMN function in snRNP assembly is developmentally regulated, and that a reduction of SMN in SMA could preferentially affect neuronal cells during a critical developmental window. In cellular and animal models of SMA, reduction of SMN protein or expression of SMA patient missense mutations predicts low snRNP assembly activity (Gabanella et al., 2005; Wan et al., 2005; Winkler et al., 2005). Further, in SMA animal models, reintroduction of SMN to rescue the phenotype rescues snRNP assembly activity (Gabanella et al., 2005; Winkler et al., 2005).
One of the most exciting new ideas to emerge in addressing the cell-specificity of SMA is that, when SMN is reduced in an organism, the various snRNPs are not reduced to the same extent, but rather, changes in the relative amounts of individual snRNPs are specific to different cell types (Gabanella et al., 2007; Zhang et al., 2008). These cell type-specific changes in the relative amounts of the various snRNPs should, therefore, give rise to cell type-specific changes in pre-mRNA splicing. This was determined to be the case in several recent studies that have looked directly at the effect of reduced SMN on snRNP levels in SMA mice (Gabanella et al., 2007; Zhang et al., 2008). The U11 and U12 minor snRNPs are preferentially lowered in spinal cord and brain of some SMA animals, indicating that perhaps a reduction of the minor splicing pathway may be a neuron-specific defect. These data suggest that neurons may reduce cell type-specific transcripts that contain introns processed by the minor splicing pathway, which could make neurons hypersensitive to a reduction in minor snRNP levels. Interestingly, voltage-gated ion channels have a large number of U12-type introns and thus, may be affected by a change in minor snRNP levels (Wu and Krainer, 1999). To date, mRNAs spliced by the minor snRNPs have not been found to be preferentially defective (Baumer et al., 2009; Zhang et al., 2008).
Presumably, the proximal cause of motor neuron degeneration is not from defects in snRNPs per se, but rather from a downstream defect in pre-mRNA splicing. In one of the most complete studies thus far, the transcriptome was analyzed for changes in splicing as a result of low SMN in SMA mouse models (Zhang et al., 2008). Exon microarray data were obtained for brain, spinal cord and kidney of normal and SMA mouse littermates. As expected from the variation in snRNP levels among the different tissues, the defective transcripts vary dramatically among the three tissues. The exon array detected 259 splicing defects in the spinal cord of the SMA animals, with somewhat more defects observed in kidney and fewer in the brain. Unfortunately, none of these defective genes are as yet obvious candidates to explain the neurodegenerative phenotype. Various other studies of mRNA transcripts and snRNA levels have also been inconclusive as to the disease-relevant consequence of SMN reduction (Baumer et al., 2009; Boulisfane et al., 2011; Campion et al., 2010). A pre-symptomatic, early symptomatic and late symptomatic analysis of SMA mice found that splicing changes are not as dramatic in pre-symptomatic or early symptomatic mice as in late stage mice (Baumer et al., 2009), indicating that some of the splicing defects observed in later mice could be a secondary effect of the disease state and not a primary cause.
6. Conclusions and future directions
The mystery of motor neuron selectivity in SMA may be discovered within the framework of its function as the assembler of spliceosomal snRNPs. The resolution and the timing of experiments to define the motor neuron transcriptome in animal and cell culture models of SMA are important considerations that may provide invaluable information about essential splicing events. While it appears from mRNA measurements that SMN expression levels in motor neurons of wildtype mice at early developmental stages are lower than in other neuronal populations (Ruggiu et al., 2012), there have been no longitudinal studies of motor neuron gene expression in animals. The use of laser-capture microdissection paired with quantitative mRNA analysis will elucidate the exact changes in downstream expression patterns that may help in the discovery of the relevant changes leading to motor neuron death, thus providing a model of splicing pattern changes (Corti et al., 2008). Further, the functional consequences of any expression changes presumed to be causal will have to be studied.
An important concept that has recently emerged from the many, dramatically successful preclinical experiments involving gene and anti-sense oligonucleotide-based therapies is that rescue of an SMA phenotype in mice is more effective when therapy is initiated as early as possible after birth (Dominguez et al., 2011; Foust et al., 2010; Hua et al., 2011; Le et al., 2011; Lutz et al., 2011; Passini et al., 2010; Porensky et al., 2011; Valori et al., 2010). Longitudinal transcriptome analysis of this crucial immediate postnatal period may need to be performed as early as possible because it is not known exactly when changes in gene expression that lead to motor neuron death occur and, as animals become weakened from the disease, it becomes challenging to distinguish between secondary expression changes that result from weakness and the sustained disease state and primary changes that are causal.
Motor neurons are particularly vulnerable to alterations in RNA-protein interactions (Baumer et al., 2010; Kolb et al., 2010; Lemmens et al., 2010). The extreme specialization of the lower motor neuron, with a transcriptome that is, most likely, largely devoted to the growth and maintenance of an axon that can be 20,000 times as long as the cell body is wide suggests that an understanding of critical RNA-protein interactions in the axoplasm may be required to uncover the basis of this vulnerability. Nevertheless, snRNP capacity insufficiency resulting from inadequate expression of SMN protein may be the primary cause of motor neuron dysfunction and death in SMA. Experiments to reduce the abundance of spliceosomal snRNPs in animal models would provide powerful proof of principle in support of this contention.
Highlights.
SMN functions in the assembly of spliceosomal snRNPs.
Reduction of SMN protein causes defects in snRNP biogenesis.
Mutation of SMN causes the neuromuscular disease, spinal muscular atrophy.
It is not known how defects of snRNP biogenesis cause motor neuron degeneration.
Acknowledgements
Dr. Kolb receives funding from the NINDS (K08NS067282), the Families of Spinal Muscular Atrophy and the Fred F. and Herman M. Dreier Fund at The Columbus Foundation. Dr. Battle receives funding from the NINDS (NS077010).
Abbreviations
- SMA
spinal muscular atrophy
- SMN
survival motor neuron protein
- SMN1
Survival Motor Neuron 1 gene
- SMN2
Survival Motor Neuron 2 gene
- RNPs
ribonucleoprotein complexes
- snRNPs
small nuclear ribonucleoproteins
- snRNA
small nuclear RNA
- sDMA
symmetrically dimethylated arginine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achsel T, Stark H, Luhrmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alias L, Bernal S, Fuentes-Prior P, Barcelo MJ, Also E, Martinez-Hernandez R, Rodriguez-Alvarez FJ, Martin Y, Aller E, Grau E, Pecina A, Antinolo G, Galan E, Rosa AL, Fernandez-Burriel M, Borrego S, Millan JM, Hernandez-Chico C, Baiget M, Tizzano EF. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. JBiol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Wang J, Dreyfuss G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 2007a;282:27953–27959. doi: 10.1074/jbc.M702317200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2007b;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Ansorge O, Almeida M, Talbot K. The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert Rev. Mol. Med. 2010;12:e21. doi: 10.1017/S1462399410001523. [DOI] [PubMed] [Google Scholar]
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, Brabandere VD, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B9 and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion Y, Neel H, Gostan T, Soret J, Bordonne R. Specific splicing defects in Spombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 2010;29:1817–1829. doi: 10.1038/emboj.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Chari A, Paknia E, Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 2009;21:387–393. doi: 10.1016/j.ceb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A Novel DEAD Box Protein that Interacts with SMN, the Spinal Muscular Atrophy Gene Product, and Is a Component of Gems. JCell Biol. 1999;147:1181–1193. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Saladino F, Bordoni A, Fortunato F, Del Bo R, Papadimitriou D, Locatelli F, Menozzi G, Strazzer S, Bresolin N, Comi GP. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J. Clin. Invest. 2008;118:3316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Dominguez E, Marais T, Chatauret N, Benkhelifa-Ziyyat S, Duque S, Ravassard P, Carcenac R, Astord S, Pereira de Moura A, Voit T, Barkats M. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- Eggert C, Chari A, Laggerbauer B, Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN–SIP1 Complex Has an Essential Role in Spliceosomal snRNP Biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 2001a;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 2001b;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 2002;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein Assembly Defects Correlate with Spinal Muscular Atrophy Severity and Preferentially Affect a Subset of Spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarelli M, Lucarelli M, Capon F, Pizzuti A, Merlini L, Angelini C, Novelli G, Dallapiccola B. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem. Biophys. Res. Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. Specific Sequence Features, Recognized by the SMN Complex, Identify snRNAs and Determine Their Fate as snRNPs. Mol. Cell. Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt AM, Patton JR, Pederson T. U2 small nuclear RNP assembly in vitro. Nucleic Acids Res. 1989;17:4817–4828. doi: 10.1093/nar/17.12.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb SJ, Sutton S, Schoenberg DR. RNA processing defects associated with diseases of the motor neuron. Muscle Nerve. 2010;41:5–17. doi: 10.1002/mus.21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Sutomo R, Sasongko TH, Sadewa AH, Gunadi, Minato T, Fujii E, Endo S, Lee MJ, Ayaki H, Harada Y, Matsuo M, Nishio H. A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J. Neurol. 2007;254:624–630. doi: 10.1007/s00415-006-0410-x. [DOI] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- Le TT, McGovern VL, Alwine IE, Wang X, Massoni-Laporte A, Rich MM, Burghes AH. Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet. 2011;20:3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lemmens R, Moore MJ, Al-Chalabi A, Brown RH, Jr, Robberecht W. RNA metabolism and the pathogenesis of motor neuron diseases. Trends Neurosci. 2010;33:249–258. doi: 10.1016/j.tins.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The Spinal Muscular Atrophy Disease Gene Product, SMN, and Its Associated Protein SIP1 Are in a Complex with Spliceosomal snRNP Proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, Murphy AJ, Winberg ML, Monani UR. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest. 2011;121:3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dostie J, Dreyfuss G, Van Duyne GD. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure (Camb) 2005;13:883–892. doi: 10.1016/j.str.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001a;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 2001b;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Ogawa C, Usui K, Aoki M, Ito F, Itoh M, Kai C, Kanamori-Katayama M, Hayashizaki Y, Suzuki H. Gemin2 plays an important role for stabilization of the SMN complex. J. Biol. Chem. 2007;282:11122–11134. doi: 10.1074/jbc.M609297200. [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O'Riordan CR, Klinger KW, Shihabuddin LS, Cheng SH. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002a;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002b;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AH. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol. Cell. Biol. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, McGovern VL, Lotti F, Saieva L, Li DK, Kariya S, Monani UR, Burghes AH, Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell. Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: farreaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Bühler D, Fischer U, Sattler M. SMN Tudor domain structure and its interaction with the Sm proteins. Nature. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic G. Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 2008;116:223–234. doi: 10.1007/s00401-008-0411-1. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR Structures of the SMN Tudor Domain: Conformational Variation in the Binding Site for Symmetrically Dimethylated Arginine Residues. J. Mol. Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Sumpter V, Kahrs A, Fischer U, Kornstadt U, Luhrmann R. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol. Biol. Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- Sun Y, Grimmler M, Schwarzer V, Schoenen F, Fischer U, Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum. Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ, Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The Survival of Motor Neurons Protein Determines the Capacity for snRNP Assembly: Biochemical Deficiency in Spinal Muscular Atrophy. Mol. Cell. Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AH. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Krainer AR. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol. 1999;19:3225–3236. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 Delivers snRNA Precursors to the SMN Complex for snRNP Biogenesis. Mol. Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, Dreyfuss G. Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell. 2011;146:384–395. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]