Abstract
This study investigates differences in expression of clock and clock-controlled genes (CCGs) between human breast epithelial and breast cancer cells and breast tumor xenografts in circadian intact rats and examines if the pineal hormone melatonin influences clock gene and CCG expression. Oscillation of clock gene expression was not observed under standard growth conditions in vitro, however, serum shock (50% horse serum for 2 h) induced oscillation of clock gene and CCG expression in MCF-10A cells, which was repressed or disrupted in MCF-7 cells. Melatonin administration following serum shock differentially suppressed or induced clock gene (Bmal1 and Per2) and CCG expression in MCF10A and MCF-7 cells. These studies demonstrate the lack of rhythmic expression of clock genes and CCGs of cells in vitro and that transplantation of breast cancer cells as xenografts into circadian competent hosts re-establishes a circadian rhythm in the peripheral clock genes of tumor cells.
Keywords: melatonin, clock genes, circadian, serum shock, breast cancer
Introduction
Circadian rhythms play an important role in mammalian physiology and behavior and are driven by a central clock that resides within the suprachiasmatic nuclei (SCN) of the hypothalamus.1 Through multiple output pathways the SCN synchronizes peripheral clocks (oscillators) throughout the body.2 The molecular aspect of this biological clock is based on the transcriptional/translational feedback loop of clock proteins Period (PER), Cryptochrome (CRY), CLOCK, and brain and muscle arnt-like protein-1 (BMAL1).3 Two basic helix-loop-helix (bHLH)-PAS-containing transcription factors, CLOCK and BMAL1, form a heterodimer that binds E-box enhancer elements in promoters of target genes driving the transcription of Per1, Per2, Per3, Cry1, Cry2, Dec1, and Dec2 genes.4 After the PER and CRY proteins have been translated in the cytoplasm, they form heterodimeric complexes that translocate to the nucleus and inhibit their own transcription by binding to the Bmal1 promoter to inhibit Bmal1 gene expression.
Nuclear orphan receptors have emerged as functional links between the regulatory loops of the molecular clock. The retinoic acid related orphan receptor alpha (RORα) is implicated in clock control, as it induces Bmal1 transcription through binding to ROR response elements (RORE) in the Bmal1 promoter.5 Conversely, REV-ERBα inhibits Bmal1 transcription by competing with RORα for binding to RORE sequences in the Bmal1 promoter.6 Analysis of Bmal1 defective mice has revealed the indispensable role of BMAL1 as the mainspring of the molecular clockwork. Targeted disruption of Bmal1 results in complete loss of both circadian behavior and expression of the core clock regulators, PER1 and PER2, in the SCN, suggesting that expression of PER1 and PER2 is tightly coupled to the transcriptional activity of BMAL1.7,8
In mammals, a network of feedback loops function not only in the master circadian clock in the SCN of the hypothalamus, but also in almost all peripheral tissues as peripheral oscillators.9 These rhythms in peripheral tissue oscillators were thought to depend exclusively on neuroendocrine/neuronal output from the master clock in the SCN, however, core body temperature rhythms appear to be a primary synchronizer of the peripheral oscillator rhythm.10 The pineal gland is controlled by the SCN via a multisynaptic pathway making its hormone melatonin a neuroendocrine output of the circadian clock.11 A number of studies have demonstrated that the circadian clock regulates cell cycle and cell growth,12,13 providing circadian synchronization for cell proliferation and even apoptosis. Thus, disruption of circadian rhythms and the clock can alter cell cycle and cell growth to potentiate tumorigenesis. Recent studies demonstrate that circadian disruption in women by late night shift work or in rodents by exposure to light-at-night (LAN) increases breast cancer incidence and mammary tumorigenesis.14–17 We also reported that breast tumor cells fail to express the clock protein PER2, and re-expression of PER2 induces p53 up-regulation, cell cycle arrest, and, subsequently, apoptosis in MCF-7 breast cancer cells.18
The present study evaluated the differences in clock and clock controlled gene (CCG) expression in human mammary epithelial cells (MCF-10A) and human breast cancer cells (MCF-7), to determine whether melatonin, an output of the central clock, can regulate the expression of peripheral oscillator genes in both normal and tumor cells. This study tests our hypothesis that differences exist in the expression of peripheral oscillators in breast epithelial vs. breast cancer cells and that the peripheral oscillators in cell lines are non-oscillatory under long-term standard cell culture conditions, but can be induced to oscillate in vitro in response to serum-shock, and will show circadian rhythmicity as xenografts in a circadian intact host. Finally, these studies tested our hypothesis that the clock gene, Per2, is central to the regulation of cell cycle, apoptosis, and oncogenesis in breast epithelial cells.
Materials and Methods
Cell culture
Immortalized MCF-10A human breast epithelial cells were purchased from American Tissue Type Culture Collection (Rockville, MD) and MCF-7 breast tumor cells were obtained from the laboratory of the late William L. McGuire (San Antonio, TX). SR- MCF-7 breast cancer cells were isolated from serially passaged MCF-7 tissue-isolated tumor xenografts.19 MCF-10A cells were grown in MEGM medium (Lonza, Basel Switzerland) and MCF-7 and SR- MCF-7 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) [Gibco BRL, Grand Island, NY], 100 U/mL penicillin, 100 mg/mL streptomycin, and 2 mM glutamine (PSG). All cells were incubated in a humidified atmosphere of 5% CO2 and 95% air at a constant temperature of 37 °C.
Antibodies, plasmids and recombinant DNAs
Monoclonal antibodies against human CLOCK, BMAL1, PER1, PER2, CRY1 and CRY2 proteins were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and a mouse antibody against GAPDH was purchased from Sigma Chemical Co. (St. Louis, MO). The Bmal1-luciferase reporter construct (pGL3-Bmal1) was provided by Dr. John Hogenesch (U. Pennsylvania School of Medicine, Philadelphia, PA), the pCDNA3.1- Rorα1 expression vector was provided by Dr. Vincent Giguere (McGill U., Montreal, Canada), and the pcDNA 3.1 vector was purchased from Invitrogen (Carlsbad, CA). Per2 shRNA and lentiviral and control particles were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Protein isolation and Western blot analysis
MCF-10A and MCF-7 cells were cultured in 10 cm culture dishes and, upon reaching confluence, were harvested and total cellular protein extracted in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 0.1% Triton X-100, 1 mM PMSF, 0.02% sodium azide, 1 mg/mL aprotinin, and 1 mg/mL leupeptin) for 30 min at 4 °C and centrifuged at 10,000 × g for 10 min. Total cellular protein was isolated and quantitated using the Bio-Rad protein assay system (BioRad, Hercules, CA). One hundred micrograms of protein per sample was electrophoretically separated on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred onto a nylon membrane (Amersham Life Science) by electroblotting. Membranes were blocked with 5% (w/v) nonfat dry milk in TBST (150 mM NaCl, 10 mM Tris-HCl, 0.1% TWEEN), incubated with antibodies directed against the CLOCK, BMAL1, PER1, PER2, CRY1, CRY2, or GAPDH (loading control) overnight at 4 °C, washed with TBST, and incubated for 1 h at room temperature with horseradish peroxidase-conjugated rabbit-anti-mouse IgG or goat-anti-rabbit IgG (Amersham Life Science) secondary antibody. Immunoreactive proteins were visualized using the enhanced chemiluminescence system (ECL; Amersham Life Science) and Kodak X-OMAT AR film.
Transient transfection and luciferase assays
MCF-7 or MCF-10A cells were seeded onto 35 mm 6-well plates at a density of 2 × 105 cells/well. After 24 h in culture, cells were transfected with 0.2 μg of the pCMVβ construct and 0.5 μg of the pcDNA3.1-Rorα1 expression plasmid for studies of Bmal1 mRNA expression or the Rorα1 expression plasmid and 0.5 μg of the Bmal1-LUC construct for Bmal1 reporter analyses using 3.6 μL of FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) per well in serum-free medium for 6 h. Following transfection, cells were incubated with DMEM supplemented with 10% FBS and treated with either melatonin (1 nM) or vehicle (0.001% ethanol) for 16 h. The cells were harvested and lysed in 200 μL of cell lysis buffer (100 nM potassium phosphate, pH 7.8, 0.2% triton X-100, and 1 mM dithiothreitol) and the lysate measured for protein content and β-galactosidase activity. Luciferase activity, which reflects Bmal1 transcriptional activity, was determined by measuring luminescence activity on the Model 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA).
Timed analysis of breast tumor and breast epithelial cell clock genes and CCG expression under normal and serum shocked culture conditions
Approximately 5 × 105 MCF-10A, MCF-7, or SR-MCF- 7 cells were seeded on 10 cm cell culture dishes. At confluence (after 3 days for MCF-7/SR-MCF-7 and 4 days for MCF-10A cells) the cells were refed with RPMI-1640supplemented with 50% horse serum (serum shock) or 10% FBS (regular medium). After 2 h in serum-rich or regular media, cells were rinsed and re-fed with serum-free RPMI-1640 + PSG and treated with either diluent (0.001% ethanol) or melatonin (1 nM). At the indicated times (every 4 h over a 24–48 h period) dishes were washed twice with ice-cold PBS, lysed, and harvested in 1 mL TRIzol reagent (Invitrogen, Carlsbad, CA) by scraping with a rubber policeman and RNA extracted from cell lysates.
Tissue-isolated SR- MCF-7 cells/xenografts and circadian/clock evaluation
Adult female nude rats (170 g; 36/group) bearing “tissue-isolated” human MCF-7 steroid receptor negative (SR-) cancer xenografts were maintained on a control 12 L (345 lux; 141.5 μW/cm2):12D photoperiod as previously described.10,19 When tumors reached an estimated weight of 5–6 g, animals were sacrificed at 6 circadian time-points over a 24-hr period (beginning at 0400 hrs; n = 6/timepoint) and tumors freeze-clamped and stored at −80 °C for analysis of mRNA levels of the Clock genes Bmal1, Clock, Cry1 and the CCGs Sirt1 and c-Myc.
RNA extraction and cDNA synthesis
Total cellular RNA was extracted with TRIzol reagent according to the manufacturer’s protocol. First strand synthesis was achieved using 3 to 5 μg of total RNA, 5 U/μL reverse transcriptase (Superscript™, Invitrogen, Carlsbad, CA), 1 × RT buffer (50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2), 100 μM dNTP mix (equimolar, dGTP, dCTP, dATP and dTTP), 10 μM DTT, and 25 ng/μL random decamers (Geneworks): 42 °C, 1 h. Each representative cDNA pool was then aliquoted and stored at −20 °C until used.
Quantitative real time polymerase chain reaction (qPCR)
Performance and optimal annealing temperatures of the PCR primers (Table 1) were tested with gradient PCR. An initial DNA denaturation step at 95 °C for 5 min was followed by 35 cycles of denaturation at 95 °C for 30 s, primer annealing and extension at 50 °C–70 °C for 20 s. The primer specificity was confirmed by sequence searches against human DNA databases and analyzed electrophoretically on agarose gels. qPCR was performed using an iCycler iQ apparatus (Bio-Rad) associated with the iCycler Optical System Interface software (version 2.3; Bio-Rad). All PCRs were performed in triplicate in a volume of 20 μL, using 96-well optical-grade PCR plates and an optical sealing tape (Bio-Rad). The differences in the expression of all the clock and CCG transcripts were normalized with respect to GAPDH expression. The thermal cycling conditions used an initial DNA denaturation step at 95 °C for 8 min followed by 35 cycles of denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 1 min. The relative level of expression was calculated with the formula 2−Δct.
Table 1.
Primers of clock and clock controlled genes for qPCR.
| Forward | Reverse | |
|---|---|---|
| Clock | 5′-AAGTTAGGGCTGAAAGACGACG-3′ | 5′-GAACTCCGAGAAGAGGCAGAAG-3 |
| Bmal1 | 5′-GGCTCATAGATGCAAAAACTGG-3′ | 5′-CTCCAGAACATAATCGAGATGG-3′ |
| Per1 | 5′-TGGCTATCCACAAGAAGATTC-3′ | 5′-GGTCAAAGGGCTGGCCCG-3′ |
| Per2 | 5′-GGCCATCCACAAAAAGATCCTGC-3′ | 5′-GAAACCGAATGGGAGAATAGTCG-3′ |
| Cry1 | 5′-CTCCATGGGCACTGGTCTCAGTG-3′ | 5′-TCCCCACCAATTTCAGCTGCAAC-3′ |
| Cry2 | 5′-CCAAGAGGGAAGGGCAGGGTAGAG-3′ | 5′-AGGATTTGAGGCACTGTTCCGAGG-3′ |
| RORα1 | 5′-GAGGTATCTCAGTCACGAAG-3′ | 5′-AACAGTTCTTCTGACGAGGACAGG-3′ |
| Rev-erbα | 5′-ACAGCTGACACCACCCAGATC-3′ | 5′-CATGGGCATAGGTGAAGATTTCT-3′ |
| Sirt1 | 5′-GAGATAACCTTCTGTTCGGTGATGAA-3′ | 5′-CGGCAATAAATCTTTAAGAATTGTTCG-3′ |
| MT1 | 5′-GCCACAGTCTCAAGT ACGACA | 5′-CTGGAGAACCAGGATCCATAT |
| Dec2 | 5′-GCTGGAGCCCCTCGCCTACT-3′ | 5′-CCGCTGTCGGTGTCCGTGTC-3′ |
| ID2 | 5′-TCAGCCTGCATCACCAGAGA-3′ | 5′-CTGCAAGGACAGGATGCTGAT-3′ |
| C-Myc | 5′-GCCACGTCTCCACACATCA-3′ | 5′-TCTTGGCAGCAGGATAGTC-3′ |
| GAPDH | 5′-CAACTACATGGTCTACATG-3′ | 5′-CTCGCTCCTGGAAGATG-3′ |
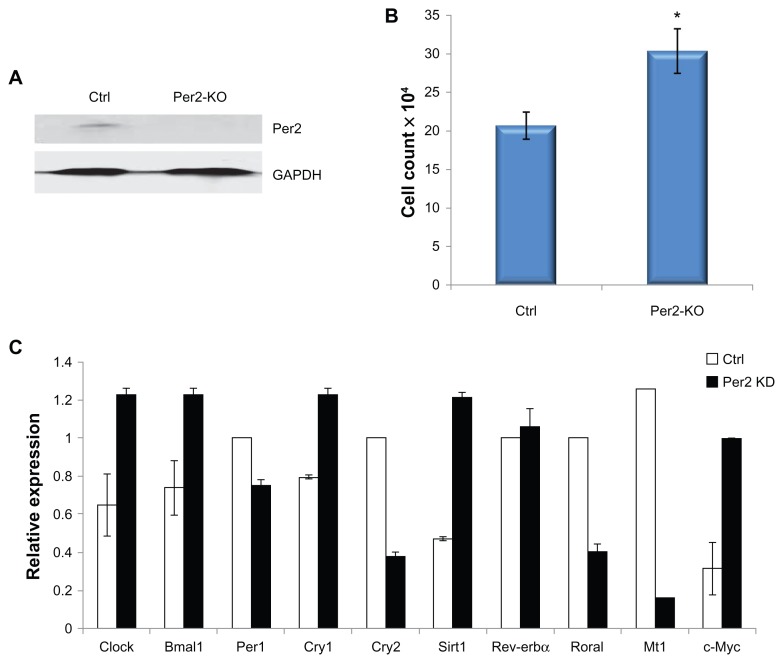
Knock-down of PER2 expression and cell proliferation
Expression of PER2 was knocked down using a Per2 shRNA lentiviral approach. Stably transformed MCF-10A PER2-knock down (KD) and control cell lines were generated according to the manufacturers protocol. For cell proliferation studies control MCF-10A or MCF-10A PER2-KD cells were plated in 6-well plates at a density of 2 × 104 cells/well in MEGM medium and culture for 4 days. Viable cells were counted using the trypan blue dye exclusion method in a heamocytometer.
Statistical analysis
Statistical differences in Bmal1 mRNA expression and transcriptional activity and PER2-KD cell proliferation were determined by two-way analysis of variance (ANOVA) followed by a Newman–Kuel’s post hoc test. P-values < 0.05 were considered statistically significant.
Results
Expression of circadian clock proteins and mRNA in human breast epithelial and breast cancer cell lines
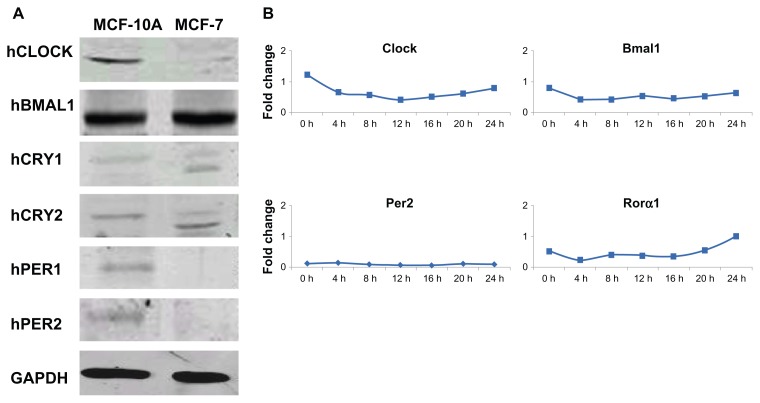
We have reported that human breast cancer cells fail to express PER2 and that re-expression of PER2 in breast cancer cells inhibits cell proliferation, up-regulates p53 expression, and induces apoptosis,18 providing evidence that perturbations of the circadian clock promote the development and progression of breast cancer.20,21 The current study investigates the protein expression levels of 6 canonical clock proteins CLOCK, BMAL1, PER1, PER2, CRY1, and CRY2 by Western blot analysis in both human breast epithelial (MCF-10A) and breast cancer (MCF-7) cell lines. In non-synchronized breast epithelial cells all 6 cononical clock proteins were expressed (Fig. 1A). However, in MCF-7 breast cancer cells, PER1 and PER2 were not expressed or expressed at levels below the limit of detection. CLOCK, BMAL1, CRY1, and CRY2 proteins were expressed in both cell lines, but the amplitude of the expression of clock genes was reduced compared to MCF-10A cells.
Figure 1.
Expression of clock proteins and mRNA in human breast epithelial and breast cancer cell lines under standard culture conditions. (A) Total cellular protein was isolated from MCF-10A immortalized human breast epithelial cell line and MCF-7 human breast tumor cell line. 100 μg total cellular protein was electrophoretically separated and subjected to Western blot analysis using antibodies directed against the various human clock proteins. (B) Lack of clock gene oscillation in MCF-7 cells under standard culture conditions.
Notes: MCF-7 cells were grown 4 days to almost complete confluence, the cells were refed with complete media (RPMI-1640 supplemented with 10% FBS) and after 2 h this medium was replaced with serum-free medium. At the indicated times, cells were washed and collected. RNA was extracted and qPCR was performed to measure gene expression. GAPDH served as an internal control. Clock gene expression was standardized on the basis of GAPDH expression, and the relative level of each clock gene is plotted in the graph.
Clock, Bmal1, Per2, and Rorα mRNA expression in MCF-10A, MCF-7, and SR- MCF-7 cells was examined over a 24 h period to determine possible rhythmic expression under standard culture conditions (media supplemented with 10% FBS). As shown in Figure 1B, no rhythmic expression of Clock, Bmal1, Per2 and Rorα mRNA was seen over a 24 h period in MCF-7 cells.
Rhythmic expression of circadian clock genes and CCGs following serum shock in breast epithelial and breast cancer cells
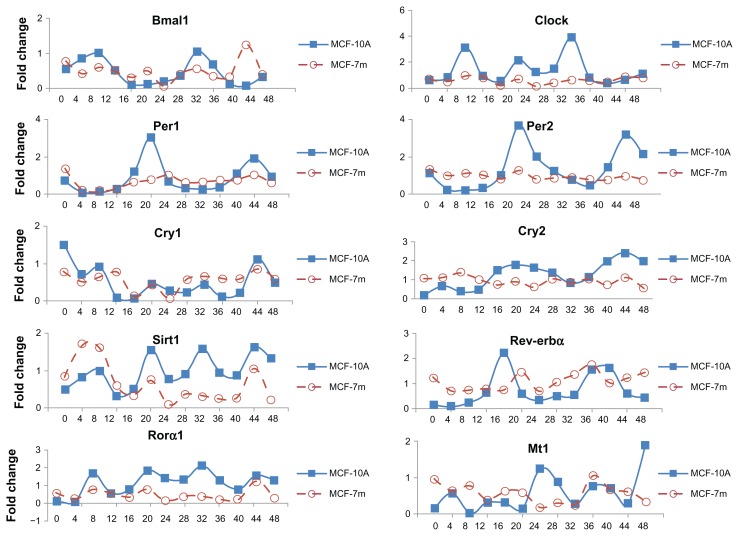
To determine possible differences in oscillation of clock and CCGs in MCF-10A and MCF-7 cells, synchronization by serum shock, an approach known induce the oscillation of clock gene expression in mammalian cells grown in culture, was employed.22 MCF-10A and MCF-7 cells were transferred to medium containing 50% horse serum for 2 h and returned to serum-free medium. Cells were harvested every 4 h (through 24 or 48 h) and the expression of mRNAs from clock and CCGs examined by qPCR. Three independent runs were conducted, however, we have presented data from a single run, as the runs showed very similar results, but the modest differences in timing and fold change between the runs generate very large standard diviations. In MCF-10A cells following serum shock, mRNA expression of the clock genes Clock, Bmal1, and Rorα, the positive regulatory arm of the circadian transcriptional complex, displayed peak expression levels at 8 h and 32 h following synchronization. Per1, Per2, and Rev-erbα, the negative regulatory arm of the circadian transcriptional complex, showed peak mRNA expression at 20 h and 44 h following synchronization (Fig. 2). Thus, the peak expression of Clock and Bmal1 was 12 to 14 h in advance of Per1, Per2, and Rev-erbα expression. Cry2 expression followed the general pattern of Per2, peaking at approximately 20 h and 44 h. The CCG Sirt1 and Rorα1 showed multiple peaks of expression at 8, 20, 32 and 44 h, while the MT1 melatonin receptor mRNA peak times were 4, 28, 36 and 48 h and were almost anti-phase of Cry1 expression.
Figure 2.
Oscillation profiles of clock genes and CCGs in MCF-10A and MCF-7 cells following serum shock.
Notes: MCF-10A and MCF-7 cells were grown 3 to 4 days to confluence, the medium was exchanged with serum-rich medium (RPMI-1640 supplemented with 50% horse serum) and after 2 h this medium was replaced with serum-free medium. At the indicated times, cells were washed and collected. RNA was extracted and qPCR was performed to measure gene expression. GAPDH served as an internal control. Clock gene expression was standardized on the basis of GAPDH expression, and the relative level of each clock gene is plotted in the graph.
Overall, serum shock generated an oscillating profile of clock and CCG mRNA as a function of time in breast epithelial cells. MCF-7 breast cancer cells showed oscillation of many clock and CCG genes, however a number of genes showed a disrupted or altered pattern of expression compared to MCF-10A cells. For example, Clock and Rorα1 have similar rhythmic pattern of expression in MCF-10A and MCF-7 cells, but with greatly reduced amplitude in MCF-7 cells, while Per1 and Per2 mRNAs are minimally expressed and show no rhythm in MCF-7 cells.
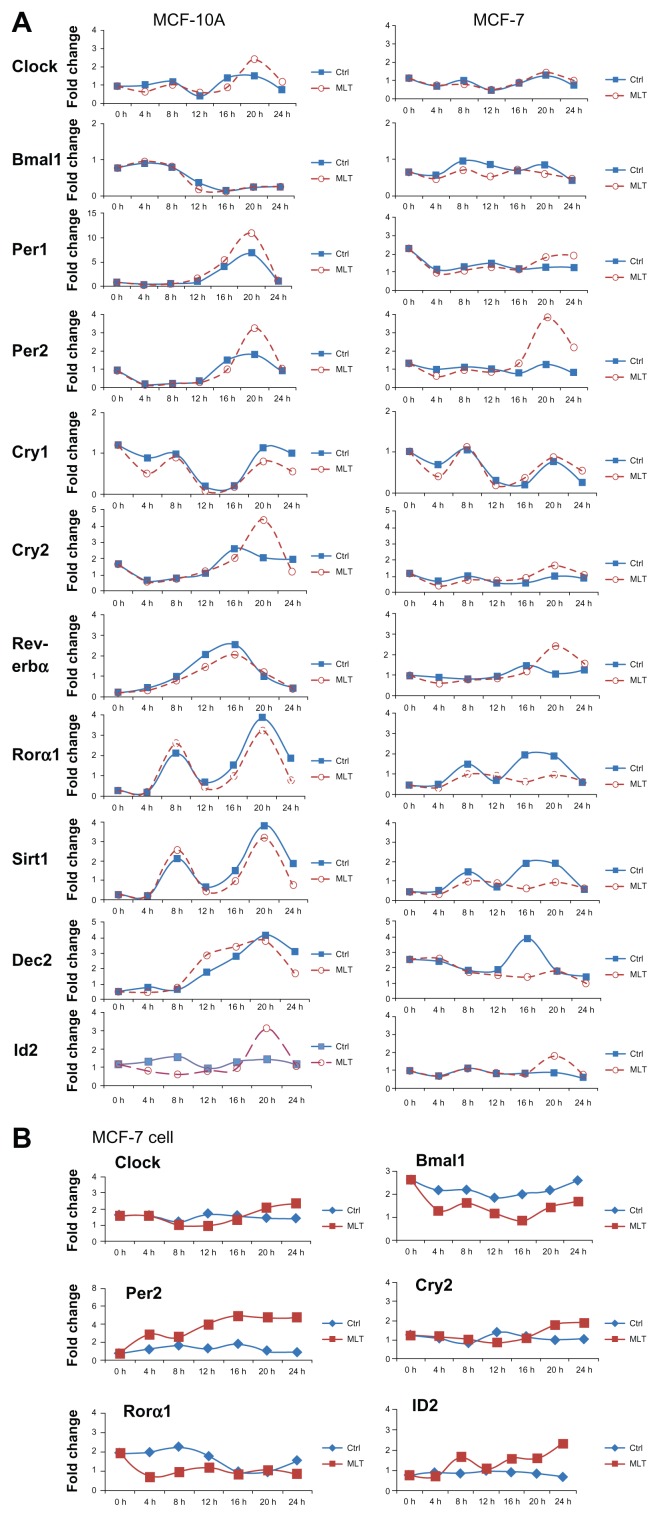
Effects of melatonin on clock genes and CCGs in normal and serum shocked MCF-10A and MCF-7 cells
To determine whether melatonin, an output of the circadian clock,11 influences the expression of peripheral oscillator (clock) genes in both breast epithelial and breast cancer cells, serum-shocked MCF-10A and MCF-7 cells were challenged over time with melatonin (1 nM). Total RNA was extracted every 4 h up to 24 hours and mRNA expression analyzed by qPCR. As shown in Figure 3A, within the first 12 h following melatonin administration most core clock genes in MCF-10A cells did not show a significant alteration in the amplitude or timing of their rhythm. However, at 20 h following melatonin administration, large increases in the amplitude of mRNA expression was evident in the Per2 (3-fold increase) and Cry2 (2-fold increase), as well as the CCG basic helix-loop-helix (bHLH) protein inhibitor of DNA binding 2 (Id2) [2-fold increase]. No oscillation was observed in clock genes or CCGs in MCF-7 breast cancer cells under standard growth conditions (Fig. 3B). Treatment of MCF-7 cells under standard culture conditions with melatonin modestly repressed Bmal1 and Rorα1 mRNA levels but induced the mRNA expression of Per2 and Id2. Unlike serum shocked MCF-7 cells, the effects of melatonin on gene expression happened very rapidly rather that 18 to 20 h later, and the mRNA levels remained generally suppressed or elevated over the entire 24 h period.
Figure 3.
Effects of melatonin on the expression and oscillation of clock genes and CCGs in MCF-10A and MCF-7 cells grown following serum shock or in standard culture conditions. MCF-10A and MCF-7 cells were grown 3 to 4 days to confluence, then the medium was exchanged with (A) serum-rich medium (RPMI-1640 with 50% horse serum) or (B) standard cell culture media supplemented with 10% FBS, and after 2 hr this medium was replaced with serum-free medium and treated with diluent (0.001%) ethanol or melatonin (1 nM).
Notes: Total mRNA was extracted every 4 h thereafter up to 24 h and analyzed by qPCR, with GAPDH expression serving as an internal control. Clock and clock controlled gene expression was standardized on the basis of GAPDH expression, and the relative level of each clock gene is plotted in the graph.
As shown in Figure 3A and B, the response of MCF-7 breast cancer cells to melatonin following serum shock was similar to that of the normal breast epithelial cells with an increase in the amplitude in expression of Per2 (2.5-fold increase) and Id2 (1.2-fold increase) at 20 h. However, unlike MCF-10A cells, melatonin induced a 1.5-fold increase in the amplitude of the mRNA of Rev-erbα (at 20 h) and repressed the amplitude of other clock genes and CCGs including Bmal1 (1.5-fold decrease after 8 h), Rorα1 (1.4-fold decrease at 16 h), and the bHLH transcription factor and core clock gene Dec2 (2.5-fold by 16 h) in MCF-7 cells.
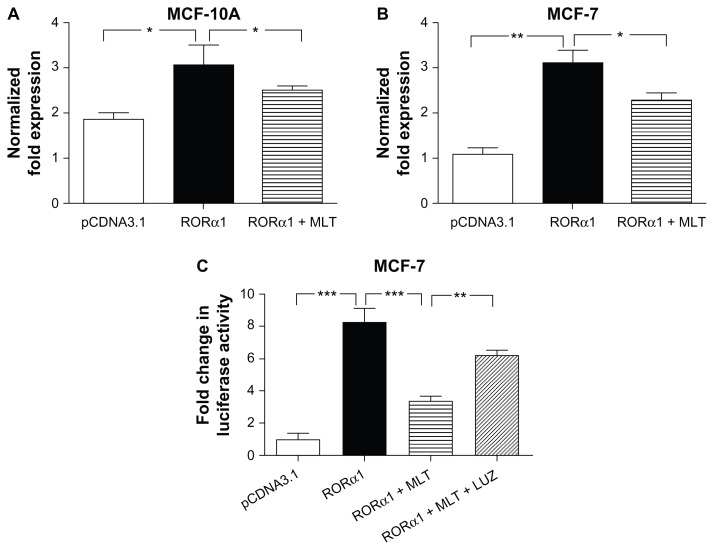
Melatonin repression of RORα1 transactivation and Bmal1 transcription
RORα has been shown to compete with Rev-erbα for binding to ROREs promoter of Bmal1 promoter to drive Bmal1 transcription.23 Although melatonin is not a ligand for the RORα, we have reported that it represses the transcriptional activity of a number of nuclear receptors including the RORα1 in MCF-7 cells.24 To investigate whether melatonin affects the expression of Bmal1 gene expression through regulation of RORα1 and Rev-erbα expression or transactivation, we employed qPCR and Bmal1 promoter luciferase reporter analyses. As seen in Figure 4, elevated expression of Rorα1 by transient transfection enhances endogenous Bmal1 mRNA expression in both MCF-10A (Fig. 4A) and MCF-7 cells (Fig. 4B). Administration of melatonin significantly suppresses Rorα1 stimulated Bmal1 promoter activity and endogenous Bmal1 mRNA expression in breast cancer cells (Fig. 4C). Finally, repression of Bmal1 expression by melatonin is mediated via its MT1 G protein coupled receptor, as treatment with the MT1/MT2 melatonin receptor antagonist luzindole (1uM) prior to melatonin (1nM) administration blocked melatonin’s inhibition of Rorα1 induced Bmal1 promoter activity and endogenous Bmal1 mRNA expression (Fig. 4C).
Figure 4.
Melatonin represses the stimulatory effect of Rorα1 on Bmal1 transcription. Real-time PCR analysis of Bmal1 mRNA expression was examined in MCF-10A (A) and MCF-7 (B) cells 24 h following transient transfection with an RORα1 expression construct and administration of 1 nM melatonin (MLT). MCF-7 (C) cells were transfected with the Bmal1-Luc reporter and RORα1 expression constructs ± the melatonin receptor antagonist luzindole (LUZ) (1 uM) for 30 min prior to melatonin (1nM) administration.
Notes: Cell lysates were collected after 18 h and Luciferase assays were performed. Data were analyzed for statistical significance using a one-way ANOVA followed by Tukey’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.001, n = 3 independent experiments.
Effect of per2 knockdown in MCF-10A cells
Our studies (Fig. 3A) show that melatonin enhances Per2 mRNA expression in serum-shocked breast epithelial and breast cancer cells. To investigate the influence of Per2 on MCF-10A cells, PER2 protein expression was knocked down using a Per2 shRNA lentiviral approach. As shown in Figure 5A, PER2 protein expression is significantly suppressed following PER2 knockdown in MCF-10A cells. Heamocytometer cell counts (Fig. 5B) demonstrate that knockdown of PER2 significantly enhances MCF-10A cell growth. Further study of gene expression by qPCR analysis shows that PER2 knockdown markedly increases the expression of Sirt1 and c-Myc mRNA, but represses the expression of the MT1 melatonin receptor (Fig. 5D).
Figure 5.
Per2 knockdown in MCF-10A cells alters clock gene and CCG expression. MCF-10A Per2 knockdown or control cell lines were generated by infecting MCF-10A cells with Per2 shRNA or control lentiviral particles (Santa Cruz Biotechnology) according to the vendor’s protocol. (A) Stably infected cell lines were tested by Western blotting to determine the level of Per2 knockdown, with GAPDH expression used as a measure of protein loading. (B) Control and Per2 knockdown MCF-10A cells were plated as described in Materials and Methods and cell proliferation was determined by counting the number number of cells on a heamoctyometer after 4 days. (C) qPCR expression of clock and CCGs was determined as control and Per2 knockdown MCF-10A cells were harvested, mRNA extracted, and the clock gene expression for Clock, Bmal1, Per1, Cry1, Cry2, Rev-erbα and clock controlled genes MT1, c-Myc, and Sirt1 were analyzed by qPCR.
Notes: *P < 0.05, n = 3.
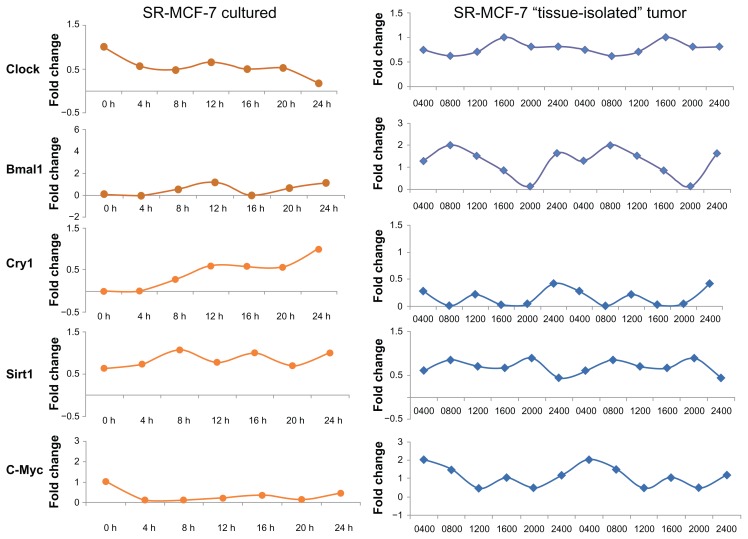
Differential expression of circadian clock genes and CCGs in SR- MCF-7 cell line and SR- breast tumor xenografts from circadian competent nude female rats
SR- MCF-7 breast cancer cells grown in culture, like parental MCF-7 breast cancer cells, do not demonstrate an oscillatory pattern of expression of clock genes (Bmal1, Clock, and Cry1) or CCGs (Sirt1 or C-Myc) [data not shown]. Given that serum shock is able to induce peripheral oscillator rhytmicity in MCF-7 cells in culture, we asked if transplantation of MCF-7 breast tumor cells as a “tissue-isolated” xenograft into circadian intact athymic nude female rats might restart the rhythmicity of peripheral oscillator genes and CCGs in a circadian fashion in SR- MCF-7 tumor cells. Figure 6 demonstrates that establishment of breast tumor cell line xenografts in circadian intact nude rats is able to re-establish a circadian rhythm of Clock (Bmal1, Clock, and Cry1) and CCG (Sirt1 and C-Myc) mRNA expression.
Figure 6.
Oscillation profiles of clock genes and CCGs in SR- MCF-7 cells grown in normal culture media or as “tissue-isolated” xenografts in circadian intact athymic nude female rats. (A) SR- MCF-7 cells were grown 4 days to confluence, the medium was exchanged with fresh medium (RPMI-1640 supplemented with 10% FBS) and after 2 h this medium was replaced with serum-free medium. At the indicated times, cells were washed and collected. RNA was extracted and qPCR was performed to measure gene expression. GAPDH served as an internal control. Clock gene expression was standardized on the basis of GAPDH expression, and the relative level of each clock gene is plotted in the graph. (B) Adult female nude rats bearing “tissue-isolated” human SR- MCF-7 cancer xenografts were maintained on either a control 12L (345 lux; 141.5 μW/cm2):12D photoperiod as previously described.17
Notes: When tumors reached an estimated weight of 5–6 g, animals were sacrificed at 6 circadian time-points over a 24-hr period (beginning at 0400 hrs; n = 6/timepoint) and tumors were freeze-clamped and stored at −80 −C for analysis of mRNA levels of the Clock genes Bmal1, Clock, Cry1 and the clock-associated genes Sirt1 and cMyc.
Discussion
Peripheral oscillatory machinery is present in cells throughout the body and drive many physiologic and disease processes, and are independent, cell-autonomous oscillators that can be synchronized by the master clock in the SCN via outputs of the SCN including core body temperature and melatonin.9,10,25 We and others18,20,21 have reported that the expression of the circadian clock gene Per2 is lost or diminished in human breast tumor cells, and that re-expression of Per2 in these cells suppresses cyclin D1 expression and cell proliferation and induces p53 expression and apoptosis.
The circadian system regulates many cell signaling pathways in both normal and malignant tissues to impact cell cycle, cell proliferation, apoptosis, cell signaling, and metabolism.2 As cells in long-term culture have been removed from the entraining influence of the master clock in the SCN, it is probable that their peripheral oscillators have become non- rhythmic and, thus, these models provide an incomplete picture of the timing and circadian changes associated with various signaling pathways influencing cell cycle, cell proliferation, apoptosis, and metabolism under true physiologic conditions. Previous reports have found that the addition of serum-rich media to cultured mammalian cells triggers the rhythmic expression of clock and CCGs.22
To determine if normal breast epithelial cells and breast cancer cells have a different expression pattern of clock and CCG expression, we first examined the protein expression of core clock genes in the MCF- 10A human breast epithelial and MCF-7 human breast cancer cell lines. As shown in Figure 1, MCF- 10A breast epithelial cells express all the core clock proteins (CLOCK, BMAL1, CRY1, CRY2, PER1, and PER2), while MCF-7 human breast cancer cells express the core clock proteins (CLOCK, BMAL1, CRY1, and CRY1), but fail to express PER1 and PER2. Furthermore, under standard culture conditions, Clock, Bmal1, Per2 and Rorα genes do not demonstrate an oscillatory pattern of mRNA expression in MCF-7 breast cancer cells and Per2 mRNA levels are extremely low. Thus, cells in long-term culture have the components of the molecular clock, but the rhythm of the peripheral oscillator appears to be compromised without input from the master clock in the SCN.
Following serum-shock of MCF-10A cells, qPCR analysis showed a rhythmic pattern of mRNA expression (Fig. 2), with large increases in the expression amplitude of a number of clock genes (Clock, Per1, Per2, Cry2, Rev-erbα, and Rorα1), and the CCGs (Sirt1 and MT1). Serum shock of MCF-7 breast cancer cells also induced oscillation of some genes including Bmal1, Clock, Cry1, Cry2, Rev-erbα, Sirt1, and MT1, however, the amplitude of the expression of a number of these genes was greatly diminished compared to normal breast epithelial cells. Interestingly, both Clock and Rorα1 display an oscillatory pattern with three peaks, compared to most other genes that show only two peaks in a 24 h time frame. At present, the importance of the different oscillation patterns is not clear. As well, the levels of Clock protein and mRNA is considerably lower in MCF-7 as compared to MCF- 10A cells and there is very little oscillation following serum shock in MCF-7 breast cancer cells suggesting that the expression of Clock may be dysregulated in cancer. Furthermore, the rhythmic expression patterns of Bmal1, Cry1, and Cry2 were shifted in cancer cells, suggesting inherent and significant differences in the peripheral oscillators between normal and malignant breast cells.
Melatonin is an output of the central clock and has been reported to regulate the central circadian clock,11,25 thus, we asked if melatonin administration alters the rhythm of clock and CCGs in cells following serum shock. As shown in Figure 3, melatonin treatment of serum-shocked MCF-10A cells enhances the amplitude of the mRNA expression of Per1, and induced an even greater increase in the amplitude of Per2 and Cry2, while repressing the amplitude of Cry1 and the CCG Sirt1. As previously noted, Per1 and Per2 mRNA and protein are not expressed in MCF-7 breast tumors.16 Thus, the ability of melatonin to induce Per1 and Per2 mRNA expression in serum shocked MCF-7 breast cancer cells is very intriguing given the importance of the PER2 protein as a tumor suppressor in breast cancer.18 In addition, melatonin also increased the expression of Cry2 and Rev-erbα, and the CCG Id2, but suppressed the amplitude of the clock genes Bmal1 and Rorα1, and the CCG Dec2 expression in serum-shocked MCF-7 cells. Melatonin treatment of MCF-7 cells grown under standard culture conditions, without oscillating Clock genes, showed a similar pattern of induction (Per2 and Id2) or suppression (Bmal1 and Rorα1), however the onset of induction or repression of these genes was almost immediate, as early as 4 h, and persisted throughout the 24 h sampling period. In serum shocked MCF-7 cells, induction or suppression of gene expression did not typically occur until 18 or 20 h later and had a rapid offset, suggesting a an influence of the clock on mRNA transcription and stability.
We have reported that melatonin represses the transcriptional activity of RORα, a known inducer of Bmal1 gene expression.6 Our studies show that melatonin, via activation of the MT1 receptor, inhibits RORα1 transactivation to suppress Bmal1 promoter activity and endogenous Bmal1 mRNA expression (Fig. 4). It is possible that melatonin, via regulation of RORα and BMAL1, can alter BMAL1/CLOCK dimer formation to modulate the expression of E-box genes including Pers, Crys, Dec2, and Id2. Furthermore, melatonin, via regulation of various transcription factors including nuclear receptors (RORα, REV-ERBα ERα, GR) or bHLH transcription factors (DEC2 or ID2), may differentially regulate the expression of other clock genes and/or CCGs.26–28 Thus, it is likely that multiple mechanisms are involved in melatonin’s regulation of these different clock genes.
Numerous studies have demonstrated that breast tumor cell lines and primary breast tumors fail to express or express very low levels of PER1 and PER2.29,30 The functional significance of PER2 in our earlier report18 noted that re-introduction of PER2 into MCF-7 cells induced p53 expression and decreased the expression of cyclin D1, leading to significant decreases in tumor cell proliferation and ultimately apoptosis. In the current study, knockdown of PER2 in MCF-10A breast epithelial cells clearly induces cell proliferation, probably through enhancement of cyclin D1 expression, and repression of p53 expression. Currently, the mechanism(s) for the loss of Per1 and Per2 mRNA expression in breast tumors is not well understood; however hypermethylation may play a key role in the transcriptional repression of these genes.30,31 The ability of melatonin to induce the expression of Per1 and Per2 mRNA in MCF-7 breast tumor cells suggests it may impact gene methylation or possibly mediate DNA unwinding by histone regulation. We have not attempted to correlate mRNA expression of each gene with the protein levels since they may be quite differently regulated as it is well known that the protein levels can be regulated by translation or ubiquitination.
Previous studies have shown that PER1 and PER2 can reduce cyclin D1 expression and enhance the expression of the ATM pathway, including CHK1 and CHK2, and the p53 DNA repair pathways.32–34 The induction of Per1 and Per2 mRNA expression by melatonin, under serum-shocked conditions, suggests there may be an associated enhancement of DNA-repair and/or apoptotic pathways. In addition, knockdown of PER2 protein levels increased c-Myc expression in MCF-10A cells (Fig. 5C), while forced PER2 expression in cancer cells has been reported to decrease c-Myc expression,35 suggesting another pathway by which melatonin regulation of PER2 may impact both cell proliferation, DNA-repair, and protect the normal breast epithelium from genomic instability.
DEC2 is a bHLH transcription factor that binds E-box elements36 and has also been reported to function as a co-repressor of the retinoid X receptor (RXRs) in chondrocytes.37 Given that RXR dimerizes with retinoic acid receptors (RAR), vitamin D3 receptor, and peroxisome proliferation activating receptor gamma to inhibit the proliferation and induce the differentiation and apoptosis of human breast cancer cells,38,39 it is possible that reduced expression of DEC2 in response to melatonin could promote the anti-tumor activity of several nuclear receptor signaling pathways that inhibit breast cancer progression. Although we have not demonstrated that melatonin can inhibit DEC2’s repression of RXR transcriptional activity, we have reported that melatonin potentiates RAR and RXR transcriptional activity in breast cancer cells.40,41
The role of the inhibitor of DNA binding 2 (ID2) in cancer is still rather nebulous. ID2 is a member of the Id family of HLH transcription factors that lacks the basic DNA-binding region and acts as a dominant-negative factor, dimerizing with other bHLH factors to prevent their binding to DNA target sequences.42 Several reports suggest that Id2 is a Myc regulated gene and can regulate cell proliferation.43 However, others suggest that ID2 plays a role in mammary gland differentiation44 and suppresses breast cancer cell proliferation. Recently, ID2 has been reported to regulate c-Myc and cyclin D1 in breast cancer.45,46 The ability of melatonin to induce Id2 mRNA expression in serum-shocked MCF-10A and MCF-7 cells may be causally related to its ability to suppress breast cancer cell proliferation through regulation of cyclin D1. Furthermore, the induction of Id2 gene expression by melatonin may also play an important role in our recent observations that melatonin represses mouse mammary gland development by suppressing ductal branching and terminal end bud formation in pubertal mammary glands.47
In conclusion, clock and CCGs are regulators of the cell cycle, cell proliferation, apoptosis, and even metabolic events, and thus are involved in the balance of survival in human breast epithelial and cancer cells. These studies demonstrate for the first time that the breast epithelial and breast cancer cell in longterm culture do express core clock genes, but that the rhythmic expression of these genes at the mRNA level is suppressed or absent. Given that the clock genes and CCGs impact the expression of many signaling pathways involved in cell proliferation and cell survival (extracellular receptor kinases Erk1/2, PI3K/AKT, and Myc), DNA-repair/apoptosis (p53, ATM, Chk1, Chk2), and even cell metabolic regulators (SIRT1 and PGC1α), the lack of a functional oscillator in these cells in culture provides a very different view of their regulation (particularly timing) compared to the in vivo situation. We have demonstrated that short-term culture of these cells in the presence of high serum levels can transiently restart the rhythmic expression of clock and CCG mRNAs. More importantly, we have demonstrated that development of human breast cancer cell xenografts in circadian intact hosts show activated peripheral oscillators in the tumor cells allowing the re-establishment of a circadian rhythm of expression of clock genes and CCGs. Thus, melatonin, through its regulation of key clock genes and CCGs plays an important role in controlling the cell cycle, proliferation, and apoptosis in both normal and malignant breast epithelium, and is able to inhibit the expression of Bmal1 and induce the expression of Per2 mRNA in serum shocked cells. As Per2 is a tumor suppressor gene that can induce p53 but repress cyclin D1, Myc, and Sirt1 gene expression, its induction by melatonin, an output of the central clock in the SCN, adds further support to the critical nature of the circadian system in the etiology of breast cancer.
Footnotes
Author Contributions
Conceived and designed the experiments: SMH. Analysed the data: LM. Wrote the first draft of the manuscript: LM. Contributed to the writing of the manuscript: all authors. Agree with manuscript results and conclusions: all authors. Jointly developed the structure and arguments for the paper: SMH, LM, DEB. Made critical revisions and approved final version: NM, SMH. All authors reviewed and approved of the final manuscript.
Competing Interests
Authors disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
This study was supported by CA-054152-14 and Army DOD grant to SMH, and an American Association of Laboratory Animal Science Grant for Laboratory Animal Science (GLAS) award to RTD.
References
- 1.Moore R, Eichler V. Loss of circadian adrenal corticosterone rhythm following superchiasmatic lesions in the rat. Brain Res. 1972;42:201–6. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 2.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Teri H. The human mouse and Period 1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–33. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Ann Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17:3867–77. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by Rev-erb and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 7.Kondratov RV, Kondratova AA, Gorbacheva VY, Mykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Lee J, Kwon I, et al. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J Cell Sci. 2010;123:3547–57. doi: 10.1242/jcs.070300. [DOI] [PubMed] [Google Scholar]
- 9.Whitmore D, Foulkes NS, Stahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent circadian oscillators. Nat Neurosci. 1998;1:701–7. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- 10.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–85. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalsbeek A, Palm IF, La Fleur SE, et al. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–69. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 12.Bjarnason GA, Jordan RC, Wood PA, et al. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oikonomou K, Cross FR. Frequency control of cell cycle oscillators. Curr Opin Genet Dev. 2010;20:605–12. doi: 10.1016/j.gde.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–62. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 15.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–7. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Schernhammer ES, Rosner B, Willett WC. Epidemiology of urinary melatonin in women and its relation to other hormones and night work. Cancer Epidemiol Biomark Prev. 2004;13:936–43. [PubMed] [Google Scholar]
- 17.Blask DE, Brainard GC, Dauchy RT, et al. Melatonin-depleted blood from premenopausal women exposed to light at night stimulates growth of breast cancer xenografts in nude rats. Cancer Res. 2005;65:11174–84. doi: 10.1158/0008-5472.CAN-05-1945. [DOI] [PubMed] [Google Scholar]
- 18.Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4–12. doi: 10.1186/1740-3391-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauchy RT, Dupepe LM, Ooms TG, et al. Eliminating Animal Facility Light-at-Night (LAN) contamination and Its Impact on Circadian Regulation of Rodent Physiology, Tumor Growth and Metabolism: A Challenge in the Relocation of a Modern Cancer Research Laboratory. J Am Assoc Lab Anim Sci. 2011;50:326–36. [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman AE, Yi CH, Zheng T, et al. Clock in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analysis. Cancer Res. 2010;70:1459–68. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423–31. doi: 10.1007/s10549-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 22.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 23.Guillamond F, Dardente H, Giguere V, Cermakian N. Differential control of Baml1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 24.Dai J, Ram PT, Yuan L, Spriggs LL, Hill SM. Transcriptional repression of RORalpha activity in human breast cancer cells by melatonin. Mol Cell Endocrinol. 2001;176:111–20. doi: 10.1016/s0303-7207(01)00449-x. [DOI] [PubMed] [Google Scholar]
- 25.Hunt AE, Al-Ghoul WM, Gillette MU, Dubocovich ML. Activation of MT(2) melatonin receptors in rat suprachiasmatic nucleus phase advances in the circadian clock. Am J Physiol Cell Physiol. 2001;260:C110–8. doi: 10.1152/ajpcell.2001.280.1.C110. [DOI] [PubMed] [Google Scholar]
- 26.Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 27.Kiefer T, Yuan L, Spriggs LL, Ram P, Hill SM. Melatonin inhibits estrogen receptor transactivation and cAMP levels in breast cancer cells. Breast Cancer Res Treat. 2002;71:37–45. doi: 10.1023/a:1013301408464. [DOI] [PubMed] [Google Scholar]
- 28.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–20. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 29.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian cock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancer. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 31.Kuo SJ, Chen ST, Yeh KT, et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454:467–74. doi: 10.1007/s00428-009-0761-7. [DOI] [PubMed] [Google Scholar]
- 32.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–7. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 play an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 34.Hogenesch JB. It’s all in a day’s work: regulation of DNA excision repair by the circadian clock. Proc Natl Acad Sci U S A. 2009;6:2481–2. doi: 10.1073/pnas.0813323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua H, Wang Y, Wan C, et al. Circadian gene mPer2 overexpression induces cell apoptosis. Cancer Sci. 2006;97:589–96. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azmi S, Sun H, Ozog A, Taneja R. mSharp1/DEC2, a helix-loop-helix protein functions as a transcriptional repressor of E-box activity and Str13 expression. J Biol Chem. 2003;278:20098–109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Sato F, Kawamoto T, et al. Anti-apoptotic effect of the basic helix-loop- helix (bHLH) transcription factor DEC2 in human breast cancer cells. Genes Cells. 2010;15:315–25. doi: 10.1111/j.1365-2443.2010.01381.x. [DOI] [PubMed] [Google Scholar]
- 38.Cho Y, Noshiro M, Choi M, et al. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC)1 and DEC2 function as corepressors of retinoid X receptors. Mol Pharmacol. 2009;76:1369–9. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
- 39.Széles L, Póliska S, Nagy G, et al. Research resource: transcriptome profiling of genes regulated by RXR and its permissive and nonpremissive partners in differentiating monocyte-derived dendritic cells. Mol Endocrinol. 2010;24:2218–31. doi: 10.1210/me.2010-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eck K, Yuan L, Spriggs LL, Hill SM. Pathways through which a regimen of melatonin and retionic acid induces apoptosis in MCF-7 human breast cancer cells. Breast Cancer Res Treat. 2000;61:229–39. doi: 10.1023/a:1006442017658. [DOI] [PubMed] [Google Scholar]
- 41.Hill SM, Blask DE, Xiang S, et al. Melatonin and associated signaling pathways that control normal breast epithelium and breast cancer. J Mammary Gland Biol Neoplasia. 2011;3:235–45. doi: 10.1007/s10911-011-9222-4. [DOI] [PubMed] [Google Scholar]
- 42.Sun XH, Copeland NG, Jenkins N, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Bio. 1991;11:5603–11. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–8. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrinello S, Lin CQ, Murata K, et al. ITF-2 and Id2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. J Biol Chem. 2001;276:39213–9. doi: 10.1074/jbc.M104473200. [DOI] [PubMed] [Google Scholar]
- 45.Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–9. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Shen Q, Kim HT, et al. The rexinoid bexarotene represses cyclin D1 transcription by inducing DEC2 transcriptional repressor. Breast Cancer Res Treat. 2010;128:667–77. doi: 10.1007/s10549-010-1083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang S, Mao L, Yuan L, et al. Impaired mouse mammary gland growth and development is mediated by melatonin and its MT1 G protein-coupled receptor via repression of ERa, Akt1, and Stat5. J Pineal Res. 2012 doi: 10.1111/j.1600-079X.2012.01000.x. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]