Abstract
An acoustophoresis-based microfluidic flow-chip is presented as a novel platform to facilitate analysis of proteins and peptides loosely bound to the surface of beads or cells. The chip allows for direct removal of the background surrounding the beads or cells, followed by sequential treatment and collection of a sequence of up to five different buffer conditions. During this treatment, the beads/cells are retained in a single flow by acoustic radiation force. Eluted peptides are collected from the outlets and subsequently purified by miniaturized solid-phase extraction and analyzed with matrix assisted laser desorption mass spectrometry. Fundamental parameters such as the system fluidics and dispersion are presented. The device was successfully applied for wash and sequential elution of peptides bound to the surface of microbeads and human spermatozoa, respectively.
INTRODUCTION
Proteins and peptides that are loosely attached to the surface of cells have been the focus of several recent studies, given their importance for the understanding of human biology and treatment of disease. Such studies have, e.g., investigated the sperm surface,1, 2 where malfunction in any of a number of subsequent biochemical interactions can cause infertility. Another important example where surface bound peptides and proteins have very important regulatory functions is the major histocompatibility complex, where MHC:peptide complexes serve to regulate the innate immune responses.3, 4 The interaction between these peptides and proteins and the cell surface can be of varying nature, e.g., hydrophobic, lipid anchored, or electrostatic interactions.5, 6
Proteomics has been used as means for exploring the membrane bound portion of the cell surface biology.7, 8, 9 However, analysis and isolation of loosely bound surface molecules and peripheral membrane proteins require new methods that allow for collection of the proteins or peptides from the cell surface, while avoiding the background of the unbound molecules. Commonly employed procedures involve numerous centrifugation steps for washing or surface labeling, e.g., biotinylation of the cells prior to elution and subsequent purification. Many of these methods are labor intensive, complex, and introduce many sources of analytical uncertainty. A particularly important aspect of any method used to harvest surface bound molecules is the short time from wash to elution, as living cells continuously generate new background in the surrounding medium, e.g., by leakage of various cytoplasmic molecules.
Microfluidic sample preparation of cells is an attractive tool for sample preparation of cells with special relevance for analysis of the cell surface. Currently, the most widespread microfluidic technologies used for cell surface analysis are miniaturized patch clamp systems,10, 11 fluorescence-activated cell sorting (FACS),12 and surface plasmon resonance systems.13, 14 These microfluidic systems are very good for investigating known interactions of peptides and proteins with cells, but are less suited for use in a discovery mode.
A promising area of microfluidic cell handling is that of continuous flow separation, where cells or micrometer sized particles are subjected to forces acting perpendicular to a laminar flow, thus inducing a deflection of the particle trajectories inside a microchannel.15, 16 An interesting feature of this technique is the possibility to continuously transfer a stream of particles or cells between different liquids in a flow channel. It has previously been shown that acoustophoresis,17 where acoustic forces are utilized, is highly effective for buffer medium transfer of particles or cells from complex biofluids.18, 19, 20, 21, 22, 23, 24 Some benefits of this technique are: the rapid transverse motion of cells induced by the acoustic force (up to 1 mm s−1),25 that the force depends on the intrinsic acoustic properties of the cell or particle, and that the microfluidic system design is straightforward and flexible.
In this study, we present an acoustophoresis microchannel for rapid multiplexed buffer/medium exchange that offers the unique possibility to challenge cells in a sequence of different buffer conditions and also to extract the molecular material from each buffer condition as separate samples. We present fundamental data on the fluidic performance of the device and demonstrate that the system can be used to elute surface-bound peptides from beads and cells by exposure to a pH gradient. The eluted peptides were concentrated and purified by reversed-phased solid-phase extraction (RP-SPE) in our miniaturized integrated selective enrichment target (ISET) platform26, 27, 28 and the final analysis read-out was made by matrix assisted laser desorption mass spectrometry (MALDI) mass spectrometry.
MATERIALS AND METHODS
Chemicals
Unless otherwise specified all chemicals were purchased from Sigma-Aldrich. Co. (St. Louis, MO, USA) and used without any further purification.
Ion exchange beads used (strong anion exchange, SAX and strong cation exchange, SCX), Reprosil 5 μm, were from Dr. Maisch HPLC GmbH (Beim Brückle, Germany) and Poros R2 50 μm beads were from Applied Biosystems (Carlsbad, CA, USA). High-performance liquid chromatography (HPLC) grade water was used for all aqueous preparations.
Samples
Alcohol dehydrogenase 1 (ADH) from yeast and bovine serum albumin (BSA) were digested with trypsin from Promega (Madison, WI, USA) in a 1:100 ratio for 4 h at 37 °C. The digested peptide mixture was diluted 100 times with 10 mM phosphate buffered saline (PBS), pH 7.4, and frozen to stop the reaction. A stock solution of 1 μM ADH was used to prepare standard samples by dilution and spiking with antigen. The universal proteomics standard (UPS 1) containing 48 different proteins was reconstituted and digested with trypsin according to the manufacturer’s instructions. Frozen aliquots of 2.5 pmol were used to prepare samples immediate before use. Human semen (spermatozoa, head size approximately 5 by 3 μm) from a healthy volunteer was allowed to liquefy for 1 h prior to dilution in pH 7 phosphate buffer (PB). A set of PB with different pH range for peptide elution were prepared with HPLC grade water. For investigation of microchip flow patterns, Evans Blue dye 100 μg ml−1 was dissolved in PB, pH 7.
Incubation of ion exchange beads with samples
Beads were washed, conditioned, and diluted to 1 mg ml−1 in sample binding buffer. A defined amount of peptides was added and allowed to bind for 2 h, prior to injection into the chip.
Chip design
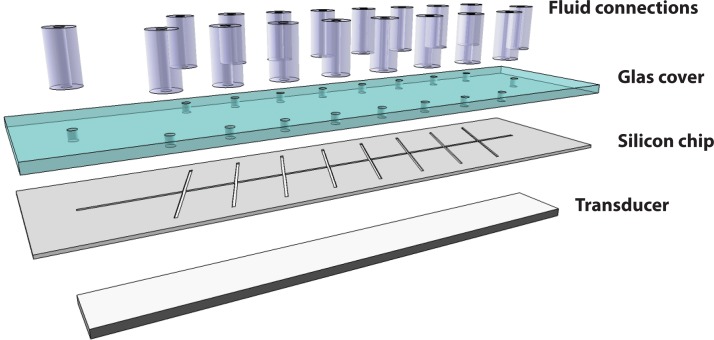
All experiments were performed on a silicon/glass chip previously described in detail by Augustsson et al.29 Briefly, a microchannel structure was KOH wet etched in 〈110〉 silicon, consisting of a main channel (60 mm long, 375 μm wide, and 150 μm deep) intersected by eight equidistant channel segments. These side branches incidence at an angle of ∼70° with respect to the main channel due to the orientation of the 〈111〉 stop etch planes constituting the channel walls. The channel was sealed by anodic bonding of a borosilicate glass cover having drilled holes for the liquid interconnects (Figure 1).
Figure 1.
Schematic of chip assembly. The flow channels are manufactured by wet etching in 〈110〉 silicon and sealed by anodic bonding of a glass cover having drilled holes for liquid interconnects. The ultrasonic transducer is attached to the underside of the silicon chip.
Ultrasonic actuation
For ultrasonic actuation, a piezoceramic transducer (PZT26, 60 mm long and 7 mm wide) resonant at 2 MHz was glued to the backside of the silicon chip. The actuation frequency was set to 1.950 MHz in all experiments. This frequency fulfills a half wavelength resonance criterion in the aqueous media flowing in the 375 μm wide main channel. The signal to the piezoceramic transducer was a sinusoidal varying voltage from a function generator (HP 3325B, Hewlett-Packard Inc., Palo Alto, CA, USA) and a power amplifier (AG 1020, T&C Power conversion Inc., Rochester, NY, USA). The voltage and power to the transducer were monitored by an oscilloscope (TDS 210, Tektronix UK Ltd., Bracknell, UK) and a digital power meter (5000-EX and 5010B, Bird Electronic Corp., Ceveland, OH, USA), respectively. The voltage was set to ∼7 Vpp in the bead based experiments, delivering a net power of ∼0.5 W to the device. In the experiments involving cells, the voltage was increased to ∼15 Vpp corresponding to a net delivered power of ∼2 W.
Setup of the chip flow-system
A schematic of the system is shown in Figure 2. Beads or cells were infused in the main channel from a plastic syringe (1 ml) (BD Plastipak, Becton Dickinson S.A., Madrid, Spain) mounted in a syringe pump (WPI sp260p, World Precision Instruments Inc., Sarasota, FL, USA). To minimize the effect of sedimentation of beads or cells during sample injection, this syringe pump was mounted vertically with the syringe pointing downwards. Elution buffers were supplied to the intersecting side channels via 8 plastic syringes (1 ml) mounted in parallel in a modified syringe pump. From the side outlets, fluids could either be withdrawn directly into 8 syringes mounted in parallel on a syringe pump or each outlet stream was sampled individually by 100 μl sample loops inserted between the outlets and the syringes.
Figure 2.
Schematic of the system. (a) The beads or cells are added at a specified flow rate from one syringe (cell/bead injection). In parallel, 8 syringes supply the fluids for treatment at a flow rate that is one third of the main flow. On the outlet side, 8 syringes collect the samples. Beads or cells are retained in the main flow channel by an acoustic force potential. (b) Beads/cells are pre-aligned in the inlet channel. (c) In the eight consecutive flow junctions, buffer is injected on one side of the channel and removed on the opposing side. Liquid solution injected through inlet 2 is brought into contact with the band of particles exposing them to, e.g., an increase in pH and thus trigging a release of peptides that will remain in the buffer and exit through outlet 5. The beads can thereafter be exposed to yet another increase in pH (e.g., inlet 4–outlet 7), where another set of peptides are released.
The flow rate of the main channel, defined by the sample injection syringe, was set to 60 μl min−1 and the side channel flow rate was set to 20 μl min−1, i.e., one third of the main channel flow rate, in all experiments.
Acoustophoresis sample preparation of cells
Samples of 1:5 diluted semen was injected directly into the chip. The sperm cells were exposed to a side channel 50 mM PB-buffer exchange sequence of pH 7, pH 9, pH 7, pH 11, and pH 7. Neutral buffer (pH 7) was used in-between to increase the spatial separation between the two elution buffers (pH 9 and pH 11). From each outlet, approximately 300–500 μl liquid was collected in individual syringes at each outlet. The syringes were emptied into individual 1 ml test tubes directly after each run.
MALDI MS analysis of samples
The samples eluted from the side outlets were first acidified with 1% TFA and then subjected to RP-SPE for purification and concentration using a previously described microfluidic solid phase extraction device, ISET,27, 28 and analyzed with MALDI-MS on a M@ldi LR (Waters/Micromass, Milford, MA, USA) and/or a MALDI Orbitrap XL (Thermo Scientific, Waltham, MA, USA) instrument. Prior to data acquisition, the MALDI settings were optimized to provide the best possible resolution, and mass accuracy. For the MALDI TOF MS analysis, a spectrum of 100 summed laser shots was acquired for each sample spot and for MALDI Orbitrap analysis FT-MS20 scans/spot were acquired. In some cases, peptides were identified by IT MS/MS performed with collision energy of 50% during an activation time of 30 ms and activation Q of 0.250. Resulting spectra were processed by xcalibur software v2.0.7 (Thermo Scientific, Waltham, MA, USA) and MS/MS spectra were manually inspected for confirmation of identity.
The concentrations of microbeads used in the experiments were analyzed using a Coulter counter (Multisizer 3, Beckman Coulter Inc., Fullerton, CA, USA). Concentration of Evans blue dye was analyzed using a FLUOstar Omega instrument (BMG LABTECH GmbH, Ortenberg, Germany) in absorbance mode for readouts at 595 nm.
Fluorescence measurements were made using samples spiked with fluorescein isothiocyanate (FITC) labeled angiotesin II, collected samples were pH adjusted to pH 11 and analyzed with the FLUOstar Omega in black 96 well plates (Corning, Lowell, MA, USA).
RESULTS AND DISCUSSION
Sequential buffer exchange
In the chip, particles or cells are introduced via an 11 mm long acoustophoresis pre-alignment segment. Particles are confined to a narrow region in the vertical center plane of the main channel flow by the acoustic force (Fig. 2b). The acoustic force on particles in acoustophoresis microchannels have been investigated in detail in previous publications.30, 31 At the first intersecting flow junction, a fraction of the main flow is withdrawn from one side while, at the same flow rate new carrier fluid is infused from the opposing side of the main channel. This causes the band of particles to experience a small shift towards the outlet side of the flow although still being retained in the main channel (Fig. 2c). After passing the flow junction, the particles will again be re-focused towards the vertical center plane by the acoustic force. The process of withdrawing fluid from one side and infusing new fluid from the other is repeated in 8 consecutive steps. With an exchange rate of one third of the main flow at each intersection, a fluid injected through, e.g., side inlet 2 will exit the main channel via side outlet no 5. During that passage, the fluid has been in direct contact with the suspended particles or cells for one third of the passage time (e.g., 0.35 s at 60 μl min−1).
In this fashion, the beads or cells are first washed and can then rapidly be challenged with a sequence of different buffer conditions at short exposure times, allowing molecules to be eluted from the surface. The eluates are collected via the side outlets for subsequent analysis. The first three side outlets will always contain the complex supernatant of the input particulate samples that needs to be removed in order to enable the analysis of extracted surface material.
System characterisation
In order to confirm the laminar flow condition in the chip, i.e., the ability to inject and collect liquid solutions while minimizing dispersion, the buffer was spiked with two different peptides, both, at a concentration of 1 pmol μl−1 and injected through side inlets 1 and 3. Consequently, these peptides should be detected in outlet 4 and 6, respectively. No cells or microbeads were used in the system during the experiment. A direct MALDI MS analysis of 1 μl samples collected from outlets 4-6 revealed that the two peptides were recovered in the anticipated outlets 4 and 6, with only a minor cross-talk detectable in outlet 5 (Fig. 3). This indicates a laminar flow and that neither flow-instabilities nor diffusion did induce any detrimental dispersion in the chip under these conditions.
Figure 3.
Low dispersion owing to the laminar flow conditions. The chip drawing to the right shows the pattern of injected samples and the laminar flow path. Mass spectra show MALDI MS analysis result of liquid solutions collected from outlets 4–6. Peptides were injected at a concentration of 1 μM. P1 and P2 denote peptide injected in inlet 1 and 3, respectively. A passage through the chip places P1 in outlet 4 and P2 in outlet 6. Very little cross-contamination was observed (outlet 5). The y-axis scaling is normalized to 15 000 counts.
A quantitative analysis of the dispersion in the acoustophoresis chip was also made using absorbance measurements of an injected Evans blue dye solution while focusing beads in the chip. A suspension of 5 μm polystyrene microbeads was continuously injected via the pre-focusing channel and a plug of dye solution was injected through side inlet 1. Figure 4 shows the mode of injection and the distribution of dye as measured by absorbance from the side outlet fractions, more than 70% of the Evans blue dye injected in inlet 1 was transferred to the intended outlet 4. While in this case 15%–20% of the dye had spread into each of outlets 3 and 5, only 0.1% of the molecules could escape all the way to outlets 2 or 6. The observed carry-over into neighboring side outlets is most likely a result of perturbations to the flow and bead associated entrapment of dye solution, as described in more detail by Augustsson et al.18 The effect of diffusion is expected to be miniscule for Evans blue molecules (diffusion constant D ≈ 3 × 10−10 m2/s).
Figure 4.
Dispersion in the system as measured by injection of a buffer containing Evans blue dye injected via side inlet 1, while all remaining side inlets were supplied with water. The collected outlet fractions were analyzed by absorbance at 595 nm, showing that ∼70% of the extracted dye molecules was collected from outlet 4. N = 1.
To determine the impact of particle loss in the device, the outlet fractions were analyzed using a Coulter counter. From these measurements, it was clear that some loss of particles occur, in total 8% of the input amount, were found in outlets 1 and 2. For outlet 3, the bead loss was 1% and for outlets 4 to 8 the particle loss was below 0.5% per outlet, as compared to the input amount. Here it is important to note that the first 3 outlets always will contain “all” the surrounding media (washes) of the originally input suspension and thus in any case has a limited analytical value. From an analytical perspective, the bead loss in the outlets 4–8 (<0.5%) will only be a very minor contribution to the final read-out in outlets 4–8.
Elution of peptides by a pH gradient
The initial experiments showed that a sample injected in one inlet could be recovered in a corresponding outlet, which greatly facilitates protocols treating cells with different solutions. Elution of molecules bound to a cell surface can be expected to be considerably more complicated, due to parameters such as the kinetics of elution-binding and the diversity of binding interactions. Microbeads with ion-exchange properties (SCX and SAX) were used to simulate cells with peptides bound to the cell surface by electrostatic interactions. Different peptide mixtures were absorbed and subjected to both discrete pH steps and pH gradients generated in the chip, for comparative purpose magnetic beads were used. These initial experiments showed that pH gradients could be created and that elution of simple samples in the pH gradient followed the predicted behavior (exemplified in Supplementary Fig. S1(a)-S1(c)32).
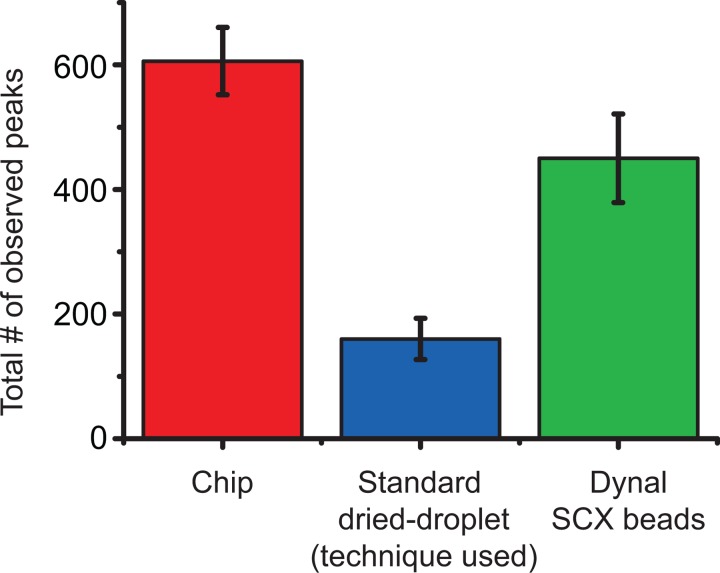
In one experiment, SCX beads (volume 1 ml, concentration 1 mg ml−1) were incubated with a total of 10 pmol of digested UPS1, in pH 6 PB buffer. This tryptic digest is a mixture of 48 proteins and yields roughly 2000 peptides of known mass and pI in the mass range 900–3000 Da. The SCX beads were then subjected to a pH gradient in the chip generated from 50 mM PB with adjusted pH. The theoretical gradient as seen from the outlet side was pH 7, 7, 7, 8, 9, 10, 11, 12 (pH at the inlets was 8, 9, 10, 11, 12, 12, 12, 12). In this case, the MALDI analysis resulted in a total of 606 different monoisotopic peaks being observed. This can be compared to only 160 peaks observed when 250 fmol of the same sample was analyzed directly using a standard dried droplet MALDI sample preparation. Using magnetic beads and five different pH’s (8, 9, 10, 11, 12) for elution, 450 peaks could be observed, Figure 5.
Figure 5.
The number of monoisotopic peaks observed with the same sample processed in triplicate using the acoustophoretic chip (red), a standard dried-droplet sample preparation (blue), and dynal magnetic SCX beads using five increasing pH steps. The error bars show the max and min number of peptides observed (N = 3).
As the pH increased along the channel, there was a trend that peptides of higher pI were eluted from the beads (supplementary Fig. S2 (Ref. 32)). Comparing the observed masses (10 ppm) revealed that 355 unique peptides were found (supplementary Fig. S3 (Ref. 32)), and 251 peptides were observed in more than one outlet. Although the number of peptides that were found in more than 4 outlets was only 71, this also includes matrix peaks and eventual contaminations. The observed cross-talk between the outlets is most likely down to slow elution kinetics, dispersion, and the chemical diversity of the peptides. For the first outlets 1–3, some background is likely to be caused by beads lost to the outlet; for the later outlets, the bead loss <0.5% into the collected sample would contribute to a very low amount of peptides. As only 100 μl of sample was collected from each outlet and from this volume approximately 1/3 was used for each MALDI analysis, the theoretical maximum of analytes in one spot is 333 fmol (10 pmol loaded).
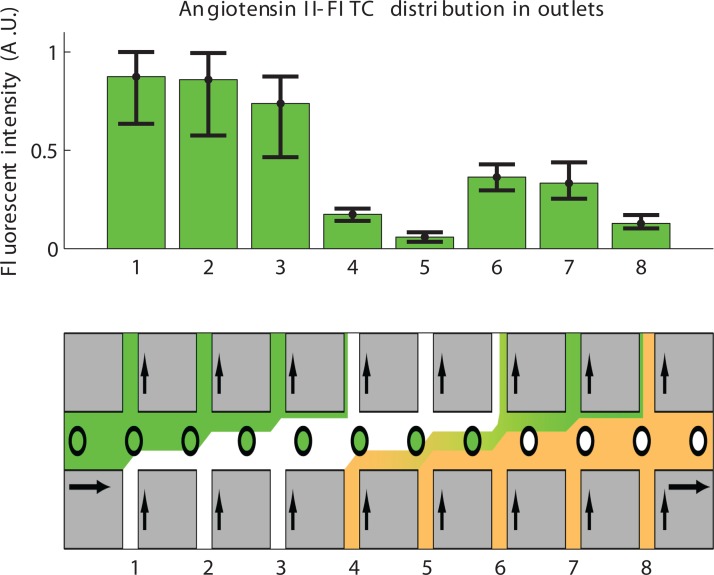
The dispersion in the acoustophoresis chip during pH induced elution of peptides was also investigated using fluorescence. Samples of FITC-labeled angiotensin II were incubated with SCX beads and subjected to a pH-increase induced elution by adding 2% NH4OH in inlet 4–8. The analysis revealed that the sample was considerably more spread out among the outlets (expected outlet 7) than would be expected from the laminar flow experiments (Fig. 6). This can be attributed to the slow kinetics of the elution from the SCX beads and the quick diffusion of OH−-ions in the chip. The fast diffusion of OH−-ions was confirmed by adding the pH indicator Thymol blue to all inlet buffers, and injecting NaOH (0.01 M) to one of the inlets (data not shown). This is not surprising since small molecules, such as an OH−-ion, diffuses more than five times faster than peptides and Evans blue dye molecules.
Figure 6.
Elution of FITC-labeled angiotensin II from 5 μm SCX beads (N = 4), beads with bound and free peptide was injected. The first 3 outlets comprise the washings of unbound peptide and the pH elution was injected at inlet 4–8 with the main part of eluted peptides being expected in outlet 7. The broadening of the elution is larger than was observed in the chip when no elution was applied (compare Figures 45). N = 4, error bars indicate min and max.
Analysis of cells
To test the acoustophoretic system with real cells, human spermatozoa were subjected to a pH7-pH9-pH7-pH11 sequence in order to test if any difference in the eluted fractions could be found, indicating that surface bound peptides could be recovered. Mass spectra of the side outlets 1 to 3 revealed high levels of unbound peptides in the seminal plasma being washed away, while outlets 5 (pH9) and 7 (pH11) revealed a slightly different population of eluted peptides, Figure 7. Note that Figure 7 shows linear mode (1.5–8 kDa) spectra of relatively small samples (10 μl). Using a high resolution Maldi Orbitrap instrument in the range 900–3000 Da (FT MS) and larger sample volumes (100 μl), many more differential peaks could be found in the fractions from the outlets (supplementary Fig. S4 (Ref. 32)). A number of possible peptide candidates from different proteins (SEMG1_HUMAN (P04279), SEMG2_HUMAN (Q02383), ALBU_HUMAN (P02768), PIP_HUMAN (P12273), KLK3_HUMAN (P07288), PPAP_HUMAN (P15309), CLUS_HUMAN (P10909) were found by the use of MALDI MS/MS, but the data cannot unequivocally prove that any of these peptides where in fact bound to the cell surface. This uncertainty of origin is a problem in all cell surface analysis techniques and will be addressed in future work. It is clear that differential peaks can be observed as a result of treatment in the acoustophoretic chip (supplementary Fig. S4 (Ref. 32)).
Figure 7.
Outlets 2 (pH7 wash), 5 (pH9 elution), and 7 (pH11 elution) after ISET sample preparation and MALDI-TOF MS in linear mode, range 1500–8000 Da of the spermatozoa samples. Red arrows indicate peaks that either increase in intensity or show up when the spermatozoa are exposed to increased pH in the chip.
Even though the laminar flow conditions help preserve a distinct readout of collected fluid fractions, it is clear from experimental data that a pH elution protocol will result in more of a gradient elution than distinct fractions. Though with the integrated wash and the sequential elution scheme offered by the chip, it is very likely that a later elution time indicates a stronger binding peptide.
Further refinements of the system design by implementation of a pressure driven flow in order to minimize the flow instabilities in system and allowing for injection and collection of buffers directly in test tubes instead of syringes will be a large improvement. Together with new elution protocols including detergents and perhaps most importantly implementation of liquid chromatography–mass spectrometry in the final analysis step, this should increase the value and usefulness of this acoustophoretic chip for investigation of biomolecules bound to cell surfaces.
CONCLUSION
The presented acoustophoretic microfluidic chip fulfills many important criteria for efficient extraction of surface associated molecules from microbeads or cells in a flow-through manner. By using buffers with different pH levels, it was possible to elute surface bound peptides from beads and human spermatozoa. Advantages of the chip design include the ability to perform multiplexed probing and the in-chip wash and removal of contaminating background increases the possibility of detecting true interactions. Also, cells are retained by acoustic forces with minimal destructive influence. Other important features are the ability to process samples without sample pre-treatment, such as centrifugation or swim-up of sperm cells, and that acoustophoresis in the MHz-range has been repeatedly reported as being a gentle method that inflicts minimal cell damage.33, 34, 35, 36, 37, 38
The chip may be suitable for assays involving, e.g., antibody screening, drug testing or inhibition of enzymatic reactions on the cell surface. While the platform is still under development, the presented acoustophoretic chip can provide unique opportunities for investigation of biomolecules bound to cell surfaces.
ACKNOWLEDGMENTS
The authors are grateful for support from the Swedish Research Council, the Royal Physiographic Society, the Crafoord Foundation, the Carl Trygger Foundation, the SSF Strategic Research Centre (Create Health), Fundacion Federico S.A., and the ELFA Foundation.
References
- Brewis I. A. and Gadella B. M., Mol. Hum. Reprod. 16(2 ), 68–79 (2010). 10.1093/molehr/gap077 [DOI] [PubMed] [Google Scholar]
- Jonsson M., Frohm B., and Malm J., J. Androl. 31(6 ), 560–565 (2010). 10.2164/jandrol.109.008672 [DOI] [PubMed] [Google Scholar]
- Blanchard N. and Shastri N., Curr. Opin. Immunol. 20(1 ), 82–88 (2008). 10.1016/j.coi.2007.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic B., Srinivasan P., Ueda Y., Tomita Y., and Maric M., PLoS ONE 5(5 ), E10599 (2010). 10.1371/journal.pone.0010599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulgrew-Nesbitt A., Diraviyam K., Wang J., Singh S., Murray P., Li Z., Rogers L., Mirkovic N., and Murray D., Biochim. Biophys. Acta 1761(8 ), 812–826 (2006). 10.1016/j.bbalip.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Cho W. and Stahelin R. V., Annu. Rev. Biophys. Biomol. Struct. 34, 119–151 (2005). 10.1146/annurev.biophys.33.110502.133337 [DOI] [PubMed] [Google Scholar]
- Cordwell S. J. and Thingholm T. E., Proteomics 10(4 ), 611–627 (2010). 10.1002/pmic.200900521 [DOI] [PubMed] [Google Scholar]
- Leth-Larsen R., Lund R. R., and Ditzel H. J., Mol. Cell. Proteomics 9(7 ), 1369–82 (2010). 10.1074/mcp.R900006-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger R. R. and Jensen O. N., Proteomics 10(22 ), 3997–4011 (2010). 10.1002/pmic.201000312 [DOI] [PubMed] [Google Scholar]
- Bao N., Wang J., and Lu C., Anal. Bioanal. Chem. 391(3 ), 933–42 (2008). 10.1007/s00216-008-1899-x [DOI] [PubMed] [Google Scholar]
- Andersson H. and van den Berg A., Curr. Opin. Biotechnol. 15(1 ), 44–9 (2004). 10.1016/j.copbio.2004.01.004 [DOI] [PubMed] [Google Scholar]
- Huh D., Gu W., Kamotani Y., Grotberg J. B., and Takayama S., Physiol. Meas. 26(3 ), R73–98 (2005). 10.1088/0967-3334/26/3/R02 [DOI] [PubMed] [Google Scholar]
- Maynard J. A., Lindquist N. C., Sutherland J. N., Lesuffleur A., Warrington A. E., Rodriguez M., and Oh S. H., Biotechnol. J. 4(11 ), 1542–58 (2009). 10.1002/biot.200900195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K. S. and Cheng Q., Anal. Bioanal. Chem. 387(5 ), 1831–1840 (2007). 10.1007/s00216-006-1052-7 [DOI] [PubMed] [Google Scholar]
- Lenshof A. and Laurell T., Chem. Soc. Rev. 39(3 ), 1203–1217 (2010). 10.1039/b915999c [DOI] [PubMed] [Google Scholar]
- Pamme N., Lab Chip 7(12 ), 1644–1659 (2007). 10.1039/b712784g [DOI] [PubMed] [Google Scholar]
- Laurell T., Petersson F., and Nilsson A., Chem. Soc. Rev. 36(3 ), 492–506 (2007). 10.1039/b601326k [DOI] [PubMed] [Google Scholar]
- Augustsson P., Åberg L. B., Swärd-Nilsson A. M. K., and Laurell T., Microchim. Acta 164(3–4 ), 269–277 (2009). 10.1007/s00604-008-0084-4 [DOI] [Google Scholar]
- Augustsson P., Persson J., Ekström S., Ohlin M., and Laurell T., Lab Chip 9(6 ), 810–818 (2009). 10.1039/b811027a [DOI] [PubMed] [Google Scholar]
- Persson J., Augustsson P., Laurell T., and Ohlin M., FEBS J. 275(22 ), 5657–5666 (2008). 10.1111/j.1742-4658.2008.06691.x [DOI] [PubMed] [Google Scholar]
- Petersson F., Nilsson A., Jönsson H., and Laurell T., Anal. Chem. 77(5 ), 1216–21 (2005). 10.1021/ac048394q [DOI] [PubMed] [Google Scholar]
- Hawkes J. J., Barber R. W., Emerson D. R., and Coakley W. T., Lab Chip 4(5 ), 446–452 (2004). 10.1039/b408045a [DOI] [PubMed] [Google Scholar]
- Liu Y., Hartono D., and Lim K. M., Biomicrofluidics 6(1 ), 12802–1280214 (2012). 10.1063/1.3671062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustsson P. and Laurell T., Lab Chip 12(10 ), 1742–52 (2012). 10.1039/c2lc40200a [DOI] [PubMed] [Google Scholar]
- Petersson F., Åberg L., Swärd-Nilsson A. M., and Laurell T., Anal. Chem. 79(14 ), 5117–23 (2007). 10.1021/ac070444e [DOI] [PubMed] [Google Scholar]
- Ekström S., Wallman L., Helldin G., Nilsson J., Marko-Varga G., and Laurell T., J. Mass Spectrom. 42(11 ), 1445–1452 (2007). 10.1002/jms.1278 [DOI] [PubMed] [Google Scholar]
- Ekström S., Wallman L., Hök D., Marko-Varga G., and Laurell T., J. Proteome Res. 5(5 ), 1071–1081 (2006). 10.1021/pr050434z [DOI] [PubMed] [Google Scholar]
- Ekström S., Wallman L., Malm J., Becker C., Lilja H., Laurell T., and Marko-Varga G., Electrophoresis 25(21–22 ), 3769–77 (2004). 10.1002/elps.200406094 [DOI] [PubMed] [Google Scholar]
- Augustsson P., Laurell T., and Ekström S., “ Flow-through chip for sequential treatment and analyte elution from beads or cells,” in Twelfth International Conference on Miniaturized Systems for Chemistry and Life Sciences on Micro Total Analysis Systems, San Diego, CA, USA, October 12–16, 2008, edited by Locascio L. E., Gaitan M., Paegel B. M., Ross D. J., Vreeland W. N. (Chemical and Biological Microsystems Society, San Diego, CA, USA, 2008), pp. 161–163.
- Augustsson P., Barnkob R., Wereley S., Bruus H., and Laurell T., Lab Chip 11, 4152–4164 (2011). 10.1039/c1lc20637k [DOI] [PubMed] [Google Scholar]
- Barnkob R., Augustsson P., Laurell T., and Bruus H., Lab Chip 10(5 ), 563–570 (2010). 10.1039/b920376a [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4749289 for further analysis of the pH gradient and mass spectra.
- Evander M., Johansson L., Lilliehorn T., Piskur J., Lindvall M., Johansson S., Almqvist M., Laurell T., and Nilsson J., Anal. Chem. 79(7 ), 2984–2991 (2007). 10.1021/ac061576v [DOI] [PubMed] [Google Scholar]
- Hultström J., Manneberg O., Dopf K., Hertz H. M., Brismar H., and Wiklund M., Ultrasound Med. Biol. 33(1 ), 145–151 (2007). 10.1016/j.ultrasmedbio.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Pui P. W. S., Trampler F., Sonderhoff S. A., Groeschl M., Kilburn D. G., and Piret J. M., Biotechnol. Prog. 11(2 ), 146–152 (1995). 10.1021/bp00032a005 [DOI] [PubMed] [Google Scholar]
- Radel S., McLoughlin A. J., Gherardini L., Doblhoff-Dier O., and Benes E., Ultrasonics 38(1–8 ), 633–637 (2000). 10.1016/S0041-624X(99)00211-5 [DOI] [PubMed] [Google Scholar]
- Yasuda K., Haupt S. S., Umemura S., Yagi T., Nishida M., and Shibata Y., J. Acoust. Soc. Am. 102(1 ), 642–645 (1997). 10.1121/1.421009 [DOI] [PubMed] [Google Scholar]
- Dykes J., Lenshof A., Åstrand-Grundström I. B., Laurell T., and Scheding S., PLoS ONE 6(8 ), e23074 (2011). 10.1371/journal.pone.0023074 [DOI] [PMC free article] [PubMed] [Google Scholar]