Abstract
RNA silencing, or RNA interference (RNAi) in metazoans mediates development, reduces viral infection and limits transposon mobility. RNA silencing involves 21–30 nucleotide RNAs classified into microRNA (miRNA), exogenous and endogenous small interfering RNAs (siRNA), and Piwi-interacting RNA (piRNA). Knock-out, silencing and mutagenesis of genes in the exogenous siRNA (exo-siRNA) regulatory network demonstrate the importance of this RNAi pathway in antiviral immunity in Drosophila and mosquitoes. In Drosophila, genes encoding components for processing exo-siRNAs are among the fastest evolving 3% of all genes, suggesting that infection with pathogenic RNA viruses may drive diversifying selection in their host. In contrast, paralogous miRNA pathway genes do not evolve more rapidly than the genome average. Silencing of exo-siRNA pathway genes in mosquitoes orally infected with arboviruses leads to increased viral replication, but little is known about the comparative patterns of molecular evolution among the exo-siRNA and miRNA pathways genes in mosquitoes. We generated nearly complete sequences of all exons of major miRNA and siRNA pathway genes dicer-1 and dicer-2, argonaute-1 and argonaute-2, and r3d1 and r2d2 in 104 Aedes aegypti mosquitoes collected from six distinct geographic populations and analyzed their genetic diversity. The ratio of replacement to silent amino acid substitutions was 1.4 fold higher in dicer-2 than in dicer-1, 27.4 fold higher in argonaute-2 than in argonaute-1 and similar in r2d2 and r3d1. Positive selection was supported in 32% of non-synonymous sites in dicer-1, in 47% of sites in dicer-2, in 30% of sites in argonaute-1, in all sites in argonaute-2, in 22% of sites in r3d1 and in 55% of sites in r2d2. Unlike Drosophila, in Ae. aegypti, both exo-siRNA and miRNA pathway genes appear to be undergoing rapid, positive, diversifying selection. Furthermore, refractoriness of mosquitoes to infection with dengue virus was significantly positively correlated for nucleotide diversity indices in dicer-2.
Introduction
RNA silencing, or RNA interference (RNAi), in plants and animals mediates normal growth and development [1], [2], controls or eliminates viral infection [3] and limits transposon mobility in both germ line [4] and somatic cells [5], [6]. RNA silencing involves small RNAs that are 21–30 nucleotides (nt) in length and are divided into three main classes: microRNAs (miRNAs), exogenous and endogenous small interfering RNAs (exo- and endo-siRNAs), and Piwi-interacting RNAs (piRNAs).
Much of what we know about RNAi in insects has been elucidated in Drosophila melanogaster, where the biogenesis and regulatory functions of each of the small RNA classes have been separated into distinct pathways [7]. The exo-siRNA pathway has a central role in Drosophila antiviral immunity [8],[9] and is initiated by Dicer-2 (Dcr2). Dcr2 is an RNase III family protein that recognizes cytoplasmic long dsRNA and cleaves it into ∼21 bp siRNAs [10], [11]. The siRNAs, in association with Dcr2 and the dsRNA-binding protein R2D2, are loaded into a multi-protein RNA-induced silencing complex (RISC), which contains Argonaute-2 (Ago2) [12], [13], [14]. In the effector stage of the pathway, the RISC unwinds and degrades one of the siRNA strands and retains the other strand as a guide for recognition and sequence-specific cleavage of viral mRNA, mediated by the “slicer” endonuclease activity of Ago2 [15], [16], [17], [18].
MicroRNAs (miRNAs) are 22–23 nt RNA guides that regulate cellular functions such as differentiation and development and metabolic homeostasis. Although only invertebrates have siRNAs, both vertebrates and invertebrates have miRNAs, which are transcribed from the cellular genome as primary miRNAs by RNA polymerase II and are processed sequentially by two distinct endonucleases in the RNase III family, nuclear Drosha and cytoplasmic Dicer 1 (Dcr1), the only ortholog of the dcr gene family in mammals. Dcr1 processes pre-miRNA to imperfectly base-paired duplex miRNA with ∼23 nt strands and acts with the dsRNA-binding protein R3D1 to load the miRNA guide strand into an Argonaute-1 (Ago1)-containing RISC [13], [19]. Typically miRNAs recognize targets in the 3′ non-coding region of cellular mRNAs by imperfect complementarity and inhibit their translation [20].
There is currently a great deal of interest in identifying genes that condition the ability of arthropods to transmit RNA viruses that are pathogenic to humans and domestic animals. Of particular interest is the mosquito Aedes aegypti, which is an important vector of a number of pathogenic arthropod borne viruses (arboviruses), including the dengue viruses (DENV1-4), yellow fever virus and chikungunya virus, and is also a tractable genetic system with which to identify candidate genes [21]. Aedes aegypti is distributed in all subtropical and tropical regions of the world. Most importantly, Ae. aegypti populations demonstrate a great deal of variation in their susceptibility to arboviral infection [22].
Several lines of evidence suggest the importance of the exo-siRNA pathway in antiviral immunity in Drosophila and mosquitoes. Drosophila with mutations in or depletion of known exo-siRNA pathway components are hypersensitive to RNA virus infections and develop a dramatically increased viral load [9], [23], [24]. Increases in arboviral replication occur after knock-down of one or more genes in the exo-siRNA pathway [25], [26]. siRNAs derived from the infecting virus genome (viRNAs) have been discovered and characterized in infected insects [27], [28], [29], [30]. Many insect pathogenic viruses encode suppressors of RNAi that counteract insect immunity [31].
Noting that interaction between RNA viruses that encode suppressors of RNAi and their insect hosts may lead to a co-evolutionary “arms race” and directional selection on RNAi genes, Obbard et al. [32] undertook a comparative study of the rates of amino acid evolution in exo-siRNA and miRNA pathway components in three species of Drosophila. They showed that among Drosophila species, the ratio of replacement to silent amino acid substitutions (w = KA/KS) among the exo-siRNA genes dcr2, r2d2, and ago2 is much greater than w among their miRNA-pathway counterparts dcr1, r3d1, and ago1. In fact it was shown that Dcr2, R2D2, and Ago2 are among the fastest evolving 3% of all Drosophila proteins [32]. Recent selective sweeps in ago2 have reduced genetic variation across a region of more than 50 kb in the genomes of Drosophila melanogaster, D. simulans, and D. yakuba, and it was estimated that selection has fixed adaptive substitutions in this gene every 30–100 thousand years [33]. The rapid evolution of exo-siRNA pathway genes compared with miRNA pathway genes supported a hypothesis for directional selection on host antiviral RNAi genes driven by evolution of virus-encoded suppressors of RNAi (VSRs) that enable viruses to evade RNA silencing [32], [34].
More than 50 VSRs encoded by plant and insect pathogenic viruses have been described. The Flock House virus (Nodaviridae) protein B2 is one of the best characterized animal VSRs [35]. B2 binds both siRNA duplexes and long dsRNA, thereby preventing dsRNA binding by proteins in the exo-siRNA pathway. No arbovirus VSRs have been identified [31], [36], [37]; however, mosquito-borne alphaviruses were engineered to express B2 and then used to infect mosquitoes orally or by injection [28] [38]. These mosquitoes had reduced pools of viRNAs, increased infectious virus titers and, most importantly, greatly decreased survival rates relative to mosquitoes infected with non-recombinant viruses.
The ability of arboviruses to cause persistent, non-cytopathic infections in both mosquito cells and mosquitoes despite the RNAi response has led to speculation about arboviral mechanisms of immune suppression or evasion in insect cells. Entomopathogenic VSRs are generally virulence factors that increase the likelihood of virus transmission by killing the insect hosts; however, pathogenic rather than persistent infections of mosquitoes by arboviruses would be detrimental to transmission and maintenance in nature. Thus, arboviral mechanisms of evasion are unlikely to involve VSRs that increase virulence, but could involve strategies such as rapid evolution of genome ‘decoy’ regions [39] or RNAi-escape mutations [29].
A recent review concluded that mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission in mosquitoes in a similar way to Drosophila antiviral immunity [40]. However, there has been no characterization and comparison of the molecular evolution of miRNA and exo-siRNA genes in a mosquito. Herein we describe the intraspecific patterns of molecular evolution of ago1, ago2, dcr1, dcr2, r3d1, and r2d2 within and among collections of Ae. aegypti from throughout its geographic range. We tested whether the interspecific evolutionary patterns of small RNA pathway genes among Drosophila species [32] are also apparent intraspecifically in Ae. aegypti. We compared nearly complete sequences of all exons in each of the six genes from 104 individual Ae. aegypti collected in six geographically distinct sites throughout the mosquito's range. We determined if amino acids encoded by the six major genes in the two pathways appear to be under positive selection by performing a fixed-site phylogenetic analysis by maximum likelihood (PAML) [41] on each of the six genes. Identified variable sites were further characterized as to the likelihood that they would alter protein structure or function using the program SIFT (Sorting Intolerant From Tolerant) [42]. Finally, we sought to determine if sequence diversity in the six genes is correlated with susceptibility to infection with DENV2 by calculating Pearson's correlation coefficients between vector competence data gathered in previous studies for four of the six mosquito collections [43], [44], [45] and various measures of genetic diversity.
Materials and Methods
Mosquito strains and DNA extraction
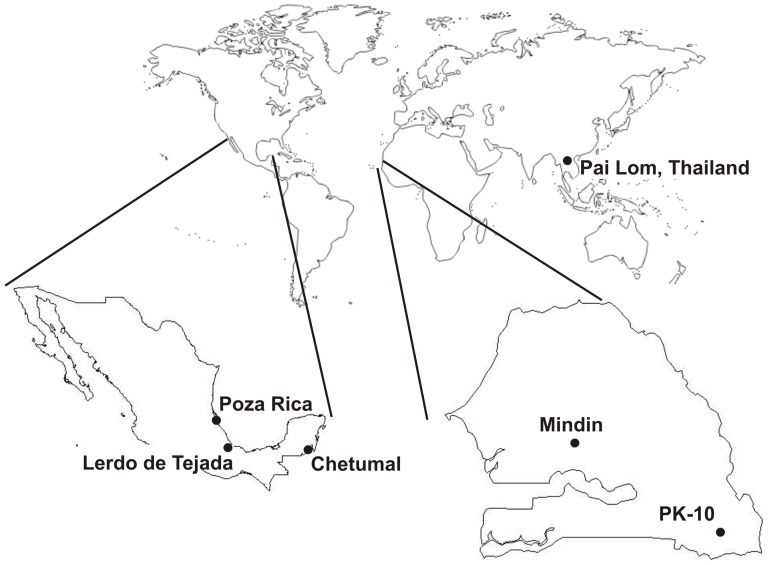
Six geographically distinct Ae. aegypti populations were analyzed in this study (Figure 1). DNA was analyzed from 18 Ae. aegypti individuals collected from Poza Rica, 18 from Lerdo de Tejada, and 18 from Chetumal in Mexico. Details on these collection sites and determination of their vector competence for DENV2 were published previously [43], [44]. DNA was analyzed from 20 mosquitoes from PK-10 (near Kedouguo) and ten from Mindin, Senegal [45]. All mosquitoes from PK-10 and five from Mindin were the formosus subspecies of Ae. aegypti as determined by the absence of silver scales on the first abdominal tergite [46]. Vector competence in Ae. aegypti formosus for flaviviruses tends to be lower than in subspecies Ae. aegypti aegypti [47], [48], [49]. The 20 Ae. aegypti from Pai Lom, Thailand were from a laboratory colony provided by Dr. L. C. Harrington at Cornell University. DNA was extracted from all 104 mosquitoes using the Qiagen DNeasy Blood and Tissue Kit (Valencia, CA).
Figure 1. Aedes aegypti collection sites.
PCR amplification
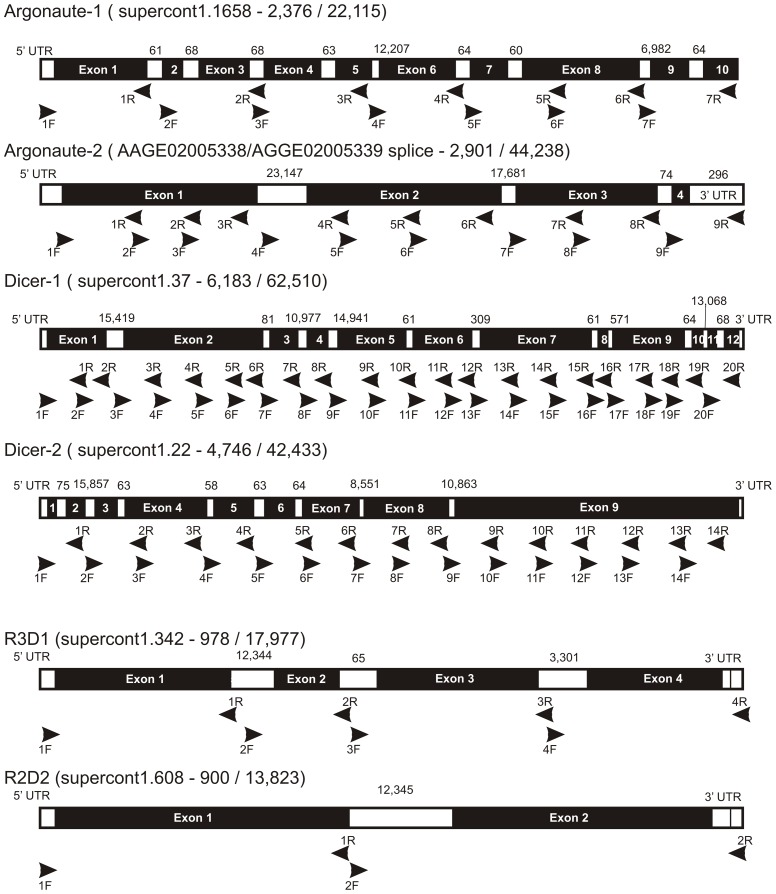
Primers for PCR (Table 1) were designed to amplify most of the exon regions of dcr1, dcr2, ago1, ago2, r2d2 and r3d1 genes. Primer locations and regions amplified with respect to supercontigs in the Ae. aegypti genome project in VectorBase (http://www.vectorbase.org/) are listed in Table 1 and shown in Figure 2. There were 56 amplicons analyzed in each of the 104 mosquitoes: dcr1 (20 amplicons), dcr2 (14), ago1 (7), ago2 (9), r3d1 (4), and r2d2 (2). In dcr1, 94% (6,183/6,581) of nucleotides were sequenced while in dcr2, 95% (4,746/4,976) of nucleotides were sequenced. In ago1 and ago2, 96% (2,376/2,477) and 97% (2,901/2,979) of nucleotides were sequenced, respectively. In r3d1, 99% (978/989) of nucleotides and 94% (900/956) of r2d2 nucleotides were sequenced.
Table 1. Primers for PCR amplification of most of the exon regions of dcr1, dcr2, ago1, ago2, r2d2 and r3d1.
| Gene | Amplicon ID | Forward/Reverse Primers | Start location | PCR product size | Ta |
| Ago1 | Ago1#1 | TCGTGCTGCGTGCCAATCACTTCCA* | 32 | 455 | 55.8 |
| AATRACTTAYCCATCGGGCGAACTG* | |||||
| Ago1#2 | CCTGGGCGGAGGTCGTG | 487 | 440 | 55.8 | |
| AGGAAATAGAATCAAGAAGGGGAGT* | |||||
| Ago1#3 | GCTTCCCTCTGCAGCTAGAAA | 928 | 455 | 53.0 | |
| GCGTCCTCCGTACTGTAACTTC | |||||
| Ago1#4 | GTGGCACGTTAAAATCAGC* | 13,585 | 402 | 53.0 | |
| TACCAGGAACACCCCAAT* | |||||
| Ago1#5 | TTCAACAGCGGAAGTCAA | 14,007 | 428 | 53.0 | |
| GCAACTCCCGAACCATAC | |||||
| Ago1#6 | GCAGCAGCACCGCCAGG | 14,379 | 403 | 55.8 | |
| CTTTCCCCGCATCAAACTCA* | |||||
| Ago1#7A | CATTTGCTTCTAATTTCAG* | 21,724 | 203 | 48.2 | |
| TTATTTGTCCTTACCTGG* | |||||
| Ago1#7B | CGCCCATTTGGTGGCATTC | 21,886 | 236 | 50.4 | |
| CTTTCTTTAAGCGAAGTACA* | |||||
| Ago2 | Ago2#1 | ACTGTATGATACTAAACGC* | 5 | 428 | 50.4 |
| CGACGGTGACGACGAAT | |||||
| Ago2#2 | GCATTCGTCGTCACCGTCGCA | 405 | 337 | 53.0 | |
| AGCCGCATAGGCATTTTT | |||||
| Ago2#3 | TAGCGAGTTCACCAAGC | 689 | 345 | 48.2 | |
| AACGACTAAGGTTATCAT* | |||||
| Ago2#4 | TTCATATTTCTCTCTATTGC* | 23,994 | 427 | 48.2 | |
| AGGATACCGAAGTTGTTTGT | |||||
| Ago2#5 | CAACTTCGGTATCCTTCT | 24,406 | 399 | 50.4 | |
| TTCCCGTCTTGTAATCTCC | |||||
| Ago2#6 | GTACGGAGATTACAAGACGGGAAT | 24,782 | 412 | 58.5 | |
| CCAACAAAACTTACCTTGAGCCA* | |||||
| Ago2#7 | GCATTTTACAGATCAACGCCA* | 42,911 | 368 | 48.2 | |
| ACGAAGTTCTATGGTCAGTA | |||||
| Ago2#8 | CTGACCATAGAACTTCGTGC | 43,261 | 428 | 53.0 | |
| GCTACTCACTCTTTGATGTAGACGC* | |||||
| Ago2#9 | CGTCCTCTGAACATGAACAACCT | 43,756 | 389 | 48.2 | |
| AGTATCCTAGAGCCAACAA* | |||||
| Dcr1 | Dcr1#1 | GTCCTACGACGAGAATGGCTTAC* | 14 | 430 | 53.0 |
| ATCACCAGCAGATTTACTT | |||||
| Dcr1#2 | TTGTGGCTATTTGGATCT | 375 | 292 | 48.2 | |
| AACCTGTTAGTGGACCTTA* | |||||
| Dcr1#3 | ATTTGCTACCAGCCCTAA* | 16,012 | 444 | 48.2 | |
| CATTGGAATACTGTTGGAAA | |||||
| Dcr1#4 | TTTTCCAACAGTATTCCAAT | 16,435 | 453 | 48.2 | |
| CATTCTGGTTGTAATAGTTTG | |||||
| Dcr1#5 | CAACCAGAATGATCCAGATGCTTTA | 16,877 | 448 | 53.0 | |
| CATGCGTTCGCTCAGATCCTCAC | |||||
| Dcr1#6 | CTTCCACCGCATATCATATTCTC | 17,236 | 315 | 48.2 | |
| TCTCCATATAGATAGC | |||||
| Dcr1#7 | TCGTTAGCGTAATTTGATAACAC* | 17,603 | 347 | 55.8 | |
| TCTTACTTACCACAATGTCTTCCT* | |||||
| Dcr1#8 | GGCTTGCCCATGCCTACGGAGAT | 28,917 | 264 | 48.2 | |
| GATGTTACAAGAATAGGTACT* | |||||
| Dcr1#9 | AAACTCATTTCTTTCCCTCAGATC* | 44,077 | 442 | 55.8 | |
| TTCCAATCAACGGTAACAACACT | |||||
| Dcr1#10 | ACGAGAAAGTGTTGTTACCG | 44,490 | 448 | 48.2 | |
| CTTGTAGGAAGAGCC* | |||||
| Dcr1#11 | GTGAATCGGAAAGGGGTGGCTCT | 44,906 | 428 | 55.8 | |
| ATCGCAGCAGATTTGTCA | |||||
| Dcr1#12 | ATCTGCTGCGATTGAGGAA | 45,322 | 259 | 48.2 | |
| TAGTGTAAACTTGTCATGTATAAAT* | |||||
| Dcr1#13 | GCCAGTGTTTCTCAACCTGTATT* | 45,814 | 450 | 55.8 | |
| GTCCCACAACTCCTAATGCGTT | |||||
| Dcr1#14 | AACGCATTAGGAGTTGTGGGACG | 46,242 | 450 | 53.0 | |
| GGTTGTCGGTCAGATTGTTGAGA | |||||
| Dcr1#15 | TCTCAACAATCTGACCGACAACC | 46,669 | 410 | 50.4 | |
| CGGAGTTCACTTACCTT* | |||||
| Dcr1#16 | GTATGCTTGTAGTTTGCTAGTCC* | 47,080 | 199 | 50.4 | |
| TTCACTTTTAACCATGTAGA* | |||||
| Dcr1#17 | GACAATGAAGAGGGCGAAAC | 47,815 | 425 | 55.8 | |
| AAGGCACTGTAACCATCCAAGA | |||||
| Dcr1#18 | TCTTGGATGGTTACAGTGCCTTC | 48,218 | 297 | 53.0 | |
| GGTGCCTGAAATACTTGTGG | |||||
| Dcr1#19 | GATTTCCACAAGTATTTCAG | 48,490 | 306 | 53.0 | |
| CTTACCTTCCACACAGCGTCCA* | |||||
| Dcr1#20 | ACCGAAGTATGATGGGACC | 61,863 | 348 | 50.4 | |
| GTGATATTTCAACGACGTTTGTT* | |||||
| Dcr2 | Dcr2#1 | CCGCTTGACAAATTTTTCAG* | 53 | 312 | 48.2 |
| GCCCATACTCACTTATCCAG* | |||||
| Dcr2#2 | CCATTAACTGAAGGTG | 16,101 | 441 | 48.2 | |
| ATAGAGTAACATTCCAGAAAGACCG | |||||
| Dcr2#3 | GTGTAATCGGTCTTTCTG | 16,510 | 394 | 53.0 | |
| ACGCCAGTCTTAGCATTG | |||||
| Dcr2#4 | GCAGTTTGCGAAAGCCTG* | 17,047 | 342 | 48.2 | |
| AATGAAAGACATCACCCTTCTATCC* | |||||
| Dcr2#5 | TAAAAAGAATGAAACAAACG | 17,451 | 405 | 48.2 | |
| CCTTGGCGGTGAAAAACGGCG | |||||
| Dcr2#6 | ATTCCGCCGTTTTTCA | 17,831 | 365 | 50.4 | |
| AAATCCTTCCAATGACG | |||||
| Dcr2#7 | TCAGACCTCGGCAAACTA* | 26,751 | 398 | 50.4 | |
| AGAAATTCCTTCCACAGT | |||||
| Dcr2#8 | CCGAAATAGAACTTGCTC | 27,070 | 373 | 48.2 | |
| CGTAACATAACTTACCCGTAC* | |||||
| Dcr2#9 | CATTTGCTTTTCTTTTCAGTTCCTA* | 38,272 | 394 | 53.0 | |
| TGCTTTCCTTTCCGCTCCTTG | |||||
| Dcr2#10 | GAGCGGAAAGGAAAGCAG | 38,649 | 394 | 50.4 | |
| CTACGGGTACATCCAAGA | |||||
| Dcr2#11 | CAATCTTGGATGTACCCGTAG | 39,022 | 357 | 55.8 | |
| GAGGTGGTTGCCAGTCGT | |||||
| Dcr2#12 | CAACCACCTCTAGCAACG | 39,369 | 411 | 48.2 | |
| ATCTACTTCACGAATATCAA | |||||
| Dcr2#13 | CGCACAATGTCCTGAAGC | 39,700 | 456 | 53.0 | |
| GATCGGTAATTTCGTGTT | |||||
| Dcr2#14 | TTACCGATCAGGTGAAC | 40,147 | 435 | 50.4 | |
| AAACGAAACATTACTTAGCAC* | |||||
| R3D1 | R3D1#1 | AAAATCATTTCAGATGAGTA* | 14 | 343 | 50.4 |
| GACTTACCATCCTTGTCCTG* | |||||
| R3D1#2 | GAAATATGTCATGTTTAAGG* | 12,625 | 182 | 48.2 | |
| CGCAGATGGAATAACTTACT* | |||||
| R3D1#3 | GTTCAATATGCTCACTATCA* | 12,815 | 357 | 48.2 | |
| TAAATCCCTACCACC* | |||||
| R3D1#4 | AGCGGGAAAACTATTACCAA* | 16,389 | 360 | 50.4 | |
| CCAGACAAGTACATAGACAT* | |||||
| R2D2 | R2D25′ | CTTGTGGTGTAGAATAATGG | 16 | 603 | 48.2 |
| TAATCATCTCGTTGC | |||||
| R2D2 | ATACTTTAGATACCTCCCATTGACA* | 13,713 | 659 | 50.4 |
Primer locations are numbered with respect to supercontigs in the Ae. aegypti genome project in Vector Base (Figure 2). An * indicates that all or part of that primer is located in an intron.
Figure 2. PCR primer locations on miRNA and siRNA pathway genes.
Positions are numbered with respect to supercontigs in the Ae. aegypti genome project in VectorBase. Start position of each primer and lengths of amplicons are given in Table 1. Lengths of exons are given above each gene.
Single strand conformational polymorphism (SSCP) analysis was performed on all PCR products [50]. Gels were scored based on the SSCP banding patterns observed for each of the 56 amplicons for each individual mosquito in the study. Unique haplotypes were identified and recorded from each SSCP gel. Each unique haplotype for each of the 56 amplicons was sequenced on both strands on the ABI 3130×L Genetic Analyzer at the Proteomics and Metabolomics facility at Colorado State University. Sequences were obtained from at least two mosquitoes to test the sensitivity of the SSCP technique when a haplotype pattern occurred more than once.
Sequence analysis
Forward and reverse strand sequences were assembled for each unique haplotype with SeqMan 2 (DNAStar, Madison, WI). Amino acid sequences for each locus were aligned in MAFFT ver. 6.606 [51] using the iterative refinement method (≥1,000 iterations) with consistency and weighted sum-of pairs scores (G-INS-i). The corresponding nucleotide sequence alignments were derived from amino acid alignments in MacClade ver. 4.03 [52]. No indels were inferred for any of the six sequence sets. DNAsp 5.10 (http://www.ub.es/dnasp) was used to determine the number of haplotypes (h), numbers of segregating sites that were synonymous (Ss) or caused amino acid replacements (SA), and the average number of nucleotide differences per site between all sequence pairs (pi) [53]. DNAsp also estimated K, the average number of nucleotide differences between all sequence pairs [54] (equation A3) and among replacement sites (KA) and synonymous sites (KS) and their ratio (w = KA/KS). The effective recombination rate between adjacent sites (R), the PHI test for recombination and Fu and Li's F* test were also calculated. The w ratio was used to infer the action of natural selection from comparative sequence data [41]. If replacements/substitutions are deleterious and therefore subject to purifying selection, KA<KS and w<1. Alternatively, when replacement substitutions are favored by natural selection KA>KS and w>1.
Phylogenetic analyses
Maximum likelihood (ML) analyses of nucleotide characters from each of the molecular data matrices were performed using RAxML ver. 7.0.3 [55]. Because RAxML only implements general time reversible (GTR) Q-matrices for nucleotide characters, more restrictive variants of the GTR matrix were not used when selected by the Akaike Information Criterion. Optimal likelihood trees were searched for using 1,000 independent searches starting from randomized parsimony trees with the GTR-GAMMA model and four discrete rate categories. Likelihood bootstrap (BS) analyses [56] were conducted with 2,000 replicates with ten searches per replicate using the “–f i” option, which “refine[s] the final BS tree under GAMMA and a more exhaustive algorithm” [57].
Tests for positive selection
CodeML [41] in the PAML package was used to perform a maximum likelihood analysis of protein-coding DNA sequences using codon substitution models [58]. CodeML estimated KA and KS and performed likelihood ratio tests (LRTs) of positive selection along lineages based on w to identify amino acid sites potentially under positive selection. We loaded the ML tree topology derived from RAxML and enforced that as a constraint. PAML was set to explicitly account for the nucleotide content at each codon position when calculating KA and KS. PAML optimized the branch lengths based on the particular model that it employed for each analysis. Five models (M0, M1a, M2a, M7 and M8) were compared. These five were reported to be the most effective models for detecting positive selection based on both simulations and empirical data [41]. Model M0 assumes and calculates one ω for all codons. Model M1a is a neutral model that assumes two site classes: w 0<1 (estimated empirically from the data) and w 1 = 1. Model M2a is a selection model that is compared with M1a by a LRT. It adds a third site class to M1a, with w 2>1 estimated empirically. Model M7 (beta) is a flexible null model, in which ω for a codon is a random draw from the beta-distribution, with 0<w<1. Model M8 (beta & w) is compared with M7 (beta) by a LRT. It adds an extra site class to M7 (beta), with w s>1 estimated empirically. Positive selection was inferred at individual amino acids using the Bayes empirical Bayes method [59].
A star tree is commonly used as a null model in phylogenetic comparative methods. All branches emerge from a single common ancestor (no topology) rather than emerging internally from one another and all branches are of equal length (no differential stasis). All branches in a star tree evolve independently of one another. Recombination among nuclear genes can homogenize haplotypes and thus create a star phylogeny. If a phylogeny derived from a dataset is resolved as a star phylogeny or if the data fit a star phylogeny as well as they fit any alternative phylogeny then the branches are said to evolve independently. Independence is desirable when testing for evidence of selection because specific hierarchies in which branches evolve in a dependent manner may lead to false detection of sites under selection because the hierarchy may be incorrect for sites under selection [59]. Results obtained from the analysis of star trees were highly similar to those obtained from the estimated gene tree (results not shown), suggesting that positive selection results were not seriously affected by possible recombination events.
Phenotype correlation analysis
Phenotypic data consisted of DENV2 midgut infection barriers (MIB = proportion of orally exposed female mosquitoes that fail to develop a midgut infection) and midgut escape barriers (MEB = proportion of females with a midgut infection that fail to develop a disseminated infection). Phenotypic data were not available for the Thailand or Mindin collections. Pearson correlation coefficients and Fisher exact tests were performed to compare the average number of nucleotide differences per site between all sequence pairs (pi), the average number of nucleotide differences between all sequence pairs (K), among replacement sites (KA), among synonymous sites (KS) and w between each phenotype (i.e. MIB, MEB) using R v2.11.1 (http://cran.r-project.org/).
Results
Intraspecific patterns of siRNA and miRNA gene variation in Ae. aegypti
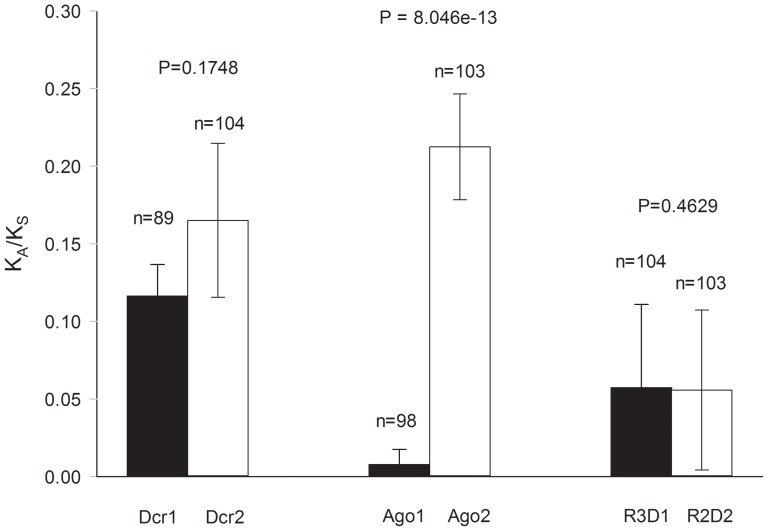
Table 2 lists, for each gene and each mosquito collection, sample sizes (N), numbers of segregating sites encoding synonymous (Ss) and replacement (Sa) substitutions, numbers of haplotypes (h), average numbers of nucleotide differences per site between all sequence pairs (pi), average numbers of nucleotide differences between all sequence pairs (k), average numbers of nucleotide differences among replacement sites (KA), average numbers of nucleotide differences among synonymous sites (KS), KA/KS = w, effective recombination rates between adjacent sites (R), and the PHI tests for recombination alongside Fu and Li's F* test. Graphs of w for all six genes appear in Figure 3. In Ae. aegypti, w was 1.4-fold higher in dcr2 than in dcr1, as compared to 5.4-fold higher in an interspecific study in Drosophila [32]. The Ae. aegypti ω ratio in ago2 was 27.4 fold higher than in ago1 (the KA for ago1 in all Drosophila spp. was zero [32]) . The nearly six-fold higher w ratios found in r2d2 as compared to r3d1 in Drosophila spp. [32] were not found among Ae. aegypti collections. Instead w was approximately the same in both genes. The probability values from Fisher's exact test of Sa and Ss appear above the bar graphs for the three gene pairs in Figure 3; only the ago1 vs. ago2 comparison in mosquitoes was significantly different. In contrast, among Drosophila spp. all three comparisons were significant.
Table 2. Intraspecific patterns in miRNA and siRNA pathway genes of Ae. aegypti.
| Site | N | Ss | Sa | h | p | k | KS | KA | KA/KS | R | PHI | F* |
| Dcr1 | ||||||||||||
| All Sites | 103 | 158 | 103 | 206 | 0.0068 | 37.94 | 0.0212 | 0.0025 | 0.1163 | 0.0125 | 1.11E-16 | 0.31 |
| Poza Rica | 18 | 88 | 44 | 36 | 0.0067 | 40.08 | 0.0210 | 0.0022 | 0.1050 | 0.0087 | 1.31 | |
| Chetumal | 18 | 84 | 30 | 36 | 0.0045 | 28.02 | 0.0148 | 0.0013 | 0.0908 | 0.0036 | 0.82 | |
| L.d.Tejada | 18 | 59 | 24 | 36 | 0.0041 | 25.46 | 0.0129 | 0.0014 | 0.1053 | 0.0123 | 1.13 | |
| Thailand | 20 | 67 | 33 | 40 | 0.0041 | 24.49 | 0.0121 | 0.0016 | 0.1326 | 0.0051 | 0.66 | |
| PK10 | 19 | 54 | 21 | 38 | 0.0042 | 23.54 | 0.0126 | 0.0016 | 0.1234 | 0.0062 | 1.42 | |
| Mindin | 10 | 82 | 43 | 20 | 0.0065 | 40.41 | 0.0183 | 0.0026 | 0.1424 | 0.0154 | 0.99 | |
| Dcr2 | ||||||||||||
| All Sites | 104 | 109 | 85 | 190 | 0.0050 | 23.87 | 0.0141 | 0.0023 | 0.1650 | 0.0080 | 1.63E-14 | −0.27 |
| Poza Rica | 18 | 32 | 24 | 27 | 0.0023 | 10.92 | 0.0063 | 0.0010 | 0.1651 | 0.0007 | 0.13 | |
| Chetumal | 18 | 20 | 12 | 32 | 0.0021 | 9.82 | 0.0060 | 0.0009 | 0.1426 | 0.0072 | 1.25 | |
| L.d.Tejada | 18 | 32 | 28 | 33 | 0.0036 | 16.98 | 0.0083 | 0.0021 | 0.2503 | 0.0114 | 0.82 | |
| Thailand | 20 | 26 | 14 | 39 | 0.0028 | 13.41 | 0.0088 | 0.0010 | 0.1129 | 0.0090 | 2.06* | |
| PK10 | 20 | 47 | 25 | 40 | 0.0047 | 22.19 | 0.0135 | 0.0019 | 0.1421 | 0.0240 | 1.45 | |
| Mindin | 10 | 71 | 39 | 20 | 0.0060 | 28.44 | 0.0156 | 0.0028 | 0.1772 | 0.0151 | 0.31 | |
| Ago1 | ||||||||||||
| All Sites | 104 | 76 | 10 | 136 | 0.0076 | 14.91 | 0.0310 | 0.0002 | 0.0077 | 0.0127 | 6.42E-13 | −0.23 |
| Poza Rica | 18 | 37 | 1 | 28 | 0.0057 | 13.52 | 0.0230 | 0.0001 | 0.0026 | 0.0085 | 1.65* | |
| Chetumal | 18 | 26 | 0 | 8 | 0.0016 | 3.71 | 0.0063 | 0.0000 | 0.0000 | 0.0000 | 0.17 | |
| L.d.Tejada | 18 | 29 | 0 | 22 | 0.0040 | 9.49 | 0.0162 | 0.0000 | 0.0000 | 0.0050 | 1.68* | |
| Thailand | 20 | 40 | 2 | 38 | 0.0062 | 14.68 | 0.0244 | 0.0002 | 0.0094 | 0.0118 | 1.87** | |
| PK10 | 20 | 37 | 5 | 38 | 0.0074 | 14.41 | 0.0277 | 0.0007 | 0.0268 | 0.0480 | 1.4 | |
| Mindin | 10 | 63 | 4 | 20 | 0.0084 | 19.62 | 0.0321 | 0.0002 | 0.0065 | 0.1044 | 0.26 | |
| Ago2 | ||||||||||||
| All Sites | 104 | 52 | 75 | 165 | 0.0053 | 13.31 | 0.0134 | 0.0029 | 0.2124 | 0.0041 | 2.42E-13 | 0.97 |
| Poza Rica | 18 | 15 | 14 | 30 | 0.0032 | 7.86 | 0.0076 | 0.0017 | 0.2224 | 0.0096 | 1.36 | |
| Chetumal | 18 | 13 | 18 | 32 | 0.0030 | 7.37 | 0.0068 | 0.0017 | 0.2442 | 0.0063 | 0.47 | |
| L.d.Tejada | 18 | 12 | 18 | 34 | 0.0034 | 8.48 | 0.0071 | 0.0019 | 0.2630 | 0.0164 | 1.07 | |
| Thailand | 20 | 8 | 11 | 19 | 0.0020 | 4.90 | 0.0041 | 0.0012 | 0.2995 | 0.0039 | 0.57 | |
| PK10 | 20 | 7 | 9 | 34 | 0.0020 | 5.05 | 0.0046 | 0.0012 | 0.2604 | 0.0464 | 1.68* | |
| Mindin | 10 | 33 | 23 | 19 | 0.0076 | 21.91 | 0.0186 | 0.0039 | 0.2086 | 0.0063 | 1.71* | |
| R3D1 | ||||||||||||
| All Sites | 104 | 21 | 9 | 87 | 0.0046 | 4.47 | 0.0163 | 0.0009 | 0.0572 | 0.0131 | 2.64E-03 | 0.69 |
| Poza Rica | 18 | 14 | 2 | 17 | 0.0041 | 3.98 | 0.0131 | 0.0011 | 0.0861 | 0.0091 | 1.23 | |
| Chetumal | 18 | 9 | 2 | 9 | 0.0023 | 2.20 | 0.0064 | 0.0009 | 0.1415 | 0.0009 | 0.96 | |
| L.d.Tejada | 18 | 13 | 3 | 16 | 0.0040 | 3.92 | 0.0152 | 0.0004 | 0.0270 | 0.0075 | 0.64 | |
| Thailand | 20 | 4 | 0 | 4 | 0.0012 | 1.14 | 0.0048 | 0.0000 | 0.0000 | 0.0000 | 1.02 | |
| PK10 | 20 | 12 | 4 | 37 | 0.0048 | 4.67 | 0.0159 | 0.0012 | 0.0763 | 0.1607 | 1.56 | |
| Mindin | 10 | 12 | 2 | 16 | 0.0053 | 5.20 | 0.0194 | 0.0006 | 0.0325 | 0.0675 | 1.33 | |
| R2D2 | ||||||||||||
| All Sites | 104 | 6 | 5 | 19 | 0.0064 | 2.81 | 0.0243 | 0.0014 | 0.0557 | 0.0162 | 6.65E-01 | 0.46 |
| Poza Rica | 18 | 3 | 2 | 6 | 0.0012 | 1.04 | 0.0037 | 0.0004 | 0.1005 | 0.0000 | 0.79 | |
| Chetumal | 18 | 7 | 1 | 16 | 0.0026 | 2.32 | 0.0089 | 0.0006 | 0.0683 | 0.0263 | 0.21 | |
| L.d.Tejada | 18 | 4 | 1 | 14 | 0.0022 | 2.00 | 0.0074 | 0.0006 | 0.0823 | 0.0436 | 1.52 | |
| Thailand | 20 | 6 | 2 | 8 | 0.0019 | 1.73 | 0.0056 | 0.0008 | 0.1441 | 0.0006 | 0.97 | |
| PK10 | 20 | 0 | 2 | 3 | 0.0012 | 0.51 | 0.0000 | 0.0015 | 0.0000 | 0.0000 | 0.69 | |
| Mindin | 10 | 10 | 5 | 15 | 0.0066 | 5.90 | 0.0198 | 0.0022 | 0.1122 | 0.0290 | 1.18 |
Sample size (N), numbers of segregating sites that were synonymous (Ss) or led to amino acid replacements (Sa), the number of haplotypes (h), the overall pi(the average number of nucleotide differences per site between two sequences) are listed for each gene and collection. Also listed are the average number of nucleotide differences, k and K among synonymous sites (Ks) and among replacement sites (Ka) and their ratio (Ka/Ks). The effective recombination rate between adjacent sites (R) and the PHI test for recombination are listed alongside Fu and Li's F* test and associated significance tests (*P<0.05, **P<0.01).
Figure 3. The ratio (w = Ka/Ks) for miRNA (ago1, dcr1, and r3d1) and siRNA (ago2, dcr2, and r2d2) pathway genes among Ae. aegypti collections.
The average number of nucleotide differences among replacement sites (Ka) relative to the average number of nucleotide differences among synonymous sites (Ka).
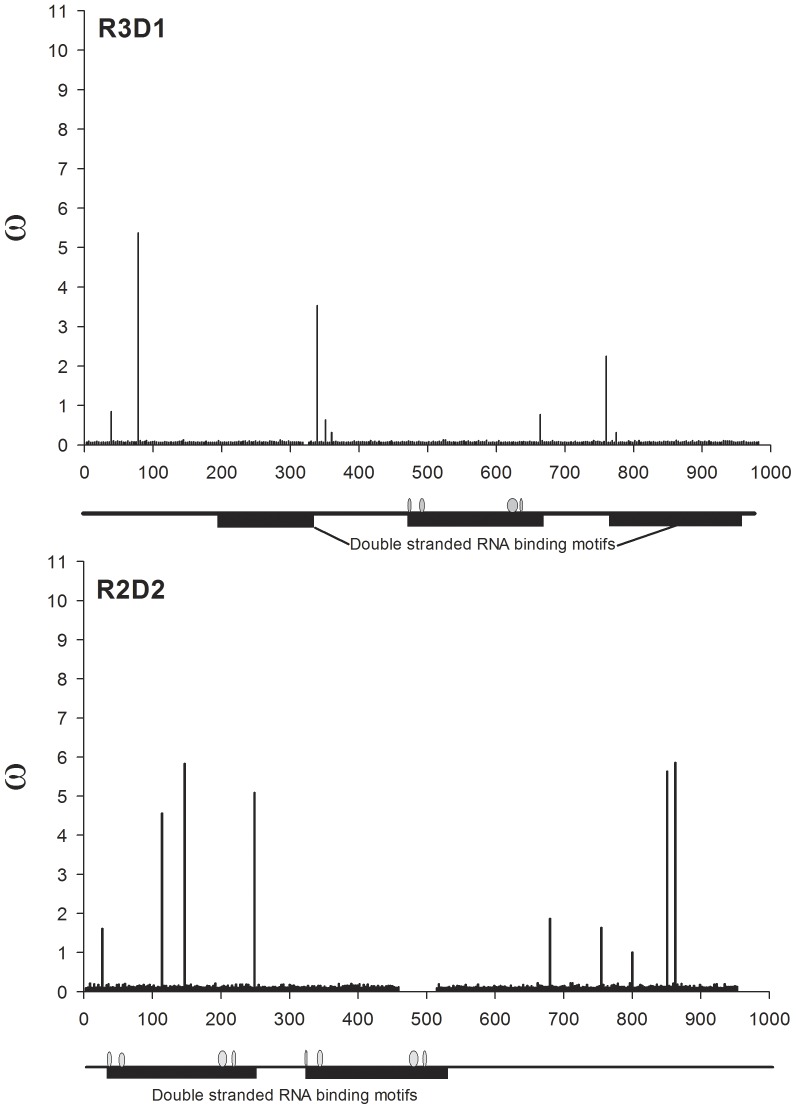
Obbard et al. [32] found no replacement substitutions in ago1 among Drosophila spp. In contrast, we found 10 nucleotide substitutions that caused amino acid replacements in Ae. aegypti ago1. In ago2, ω was 2.5 fold higher among Drosophila spp. than among Ae. aegypti populations. In dcr1, w was approximately the same among Drosophila spp. as among Ae. aegypti populations, while w in dcr2 was 3.5 fold higher among Drosophila spp. than among Ae. aegypti populations. In r3d1, w was 2.2 fold higher among Drosophila spp. than among Ae. aegypti populations while in r2d2, ω was 13.2 fold higher among Drosophila spp. The same trends in w occurred in all six Ae. aegypti collections (Table 2).
While not significant, comparison of w between Dcr1 and Dcr2 among Ae. aegypti populations reveals the same trend as among Drosophila spp, with a higher ω in the siRNA pathway gene. In contrast, replacement substitutions in ago1 were detected in four of the six Ae. aegypti collections while no replacement substitutions were found among four species of Drosophila. The same large disparity in ω between Drosophila Ago1 and Ago2 was evident among Ae. aegypti populations, but the same was not true of the Ae. aegypti dsRNA-binding proteins R3D1 and R2D2, amongst which w was approximately identical.
Functional domain analysis
Functional domains on each of the six proteins were defined by annotations in GenBank: Dcr1: AAW48724 and Dcr2: AAW48725 (DExD/H-like helicases, helicase superfamily c-terminal domains, dsRNA-binding, PAZ dicer-like, and ribonuclease domains); Ago1: XP_001662554 and Ago2: ACR56327 (conserved domains of unknown function (DUF), PAZ argonaute-like and PIWI domains); R3D1: XP_001659426 and R2D2: XP_001655660 (double-stranded RNA-binding motifs) (Figures 4–6 and Table 3). The average numbers of nucleotide differences per site between all sequence pairs (pi) were compared for each gene. The proportion of segregating sites in regions of known function versus regions where no function has been assigned to date were not significantly different for any of the six genes as determined by Fisher's Exact Test. Average values of pi were compared between regions of known versus unassigned function using a Student's t-test and a significant difference was only seen for R3D1 in which pi was greater in regions of known function (Table 3).
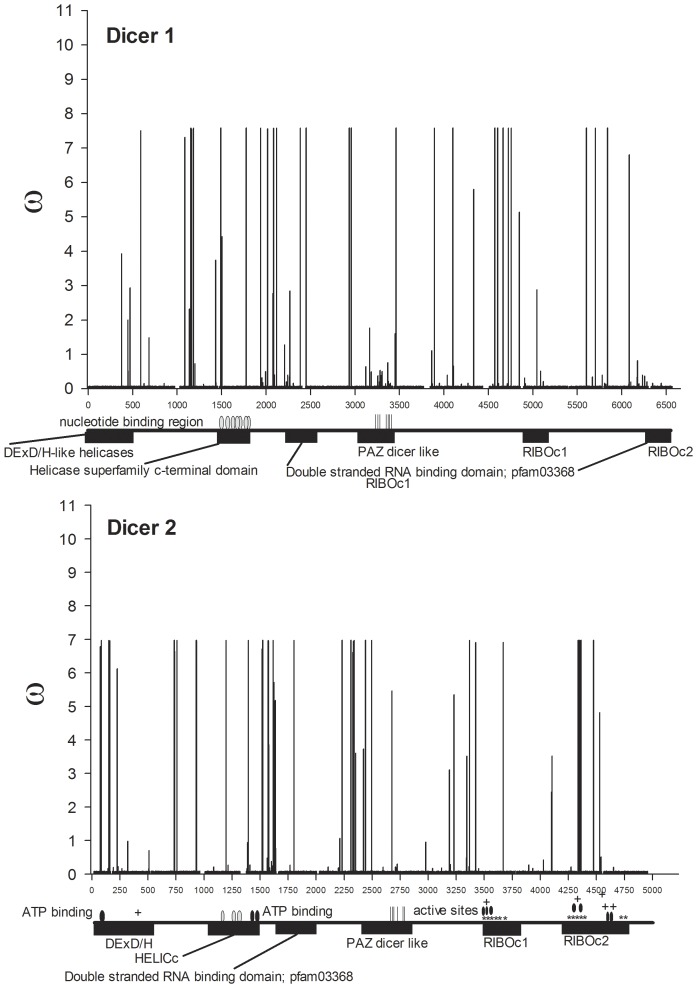
Figure 4. w ratios for all nucleotides mapped across annotated Dicer genes.
The amino-terminal domains of most Dicer enzymes contain a DExH-box RNA or ERCC4-like helicase domain followed by members of helicase superfamily c-terminal domains containing a number of nucleotide binding regions (gray ovals) and an ATP binding domain on Dcr 2 (black ovals). One or two double stranded RNA-binding domains (dsRBDs) consisting of ∼100 amino acid residues occur in the center and the carboxyl end of Dicers. The first of these dsRBDs is followed by the oligonucleotide-binding (indicated with vertical lines) PAZ domain located in the center of Dicer where it binds the 5′ phosphates and 2 nt 3′ hydroxyl overhangs. Cleavage is accomplished by dimerized RNase III domains (labeled RIBOc). Active sites (black ovals) are shown on Dcr2. The areas of dimerization are labeled with ‘*’ while regions of metal ion-binding are labeled with a ‘+.’
Figure 6. w ratios for all nucleotides mapped across annotated R3D1 (cognate binding protein for Dcr1) and R2D2 (binding protein for Dcr2).
Table 3. Functional domain analysis.
| Gene | Function | Sites | SNPs | Prob. | pi | pi(known function) | Prob. |
| FET | pi(unassigned) | t-test | |||||
| Dcr1 | Unassigned | 39 | 0 | 0.0000 | |||
| DExD/H-like helicase | 474 | 11 | 0.0109 | ||||
| Unassigned | 972 | 50 | 0.0102 | ||||
| Helicase superfamily c-terminal domain | 327 | 9 | 0.0030 | ||||
| Unassigned | 465 | 32 | 0.0085 | ||||
| Double stranded RNA binding domain; pfam03368 | 288 | 10 | 0.0063 | ||||
| Unassigned | 513 | 9 | 0.0030 | ||||
| PAZ_dicer_like | 360 | 28 | 0.0070 | ||||
| Unassigned | 1524 | 39 | 0.0049 | ||||
| RIBOc1 | 255 | 3 | 0.0050 | ||||
| Unassigned | 633 | 35 | 0.0099 | 0.0063 | |||
| RIBOc2 | 333 | 19 | 0.9448 | 0.0056 | 0.0061 | 0.9109 | |
| Dcr2 | Unassigned | 63 | 6 | 0.0126 | |||
| DExD/H-like helicase | 426 | 28 | 0.0054 | ||||
| Unassigned | 519 | 14 | 0.0048 | ||||
| HELICc | 321 | 13 | 0.0029 | ||||
| Unassigned | 208 | 24 | 0.0074 | ||||
| Double stranded RNA binding domain; pfam03368 | 266 | 10 | 0.0046 | ||||
| Unassigned | 483 | 23 | 0.0081 | ||||
| PAZ | 351 | 13 | 0.0058 | ||||
| Unassigned | 684 | 25 | 0.0055 | ||||
| RIBOc1 | 246 | 3 | 0.0006 | ||||
| Unassigned | 465 | 12 | 0.0042 | ||||
| RIBOc2 | 485 | 15 | 0.0046 | 0.0040 | |||
| Unassigned | 229 | 8 | 0.6061 | 0.0024 | 0.0064 | 0.1322 | |
| Ago1 | Domain of Unknown Function DUF1785; Pfam 08699 | 159 | 0 | 0.0000 | |||
| PAZ_argonaute_like | 273 | 3 | 0.0015 | ||||
| Piwi_ago-like | 1257 | 62 | 0.0081 | ||||
| Piwi domain; pfam02171 | 885 | 41 | 0.0088 | ||||
| 5′ RNA guide strand anchoring site | 12 | 1 | 0.0008 | 0.0038 | |||
| Unassigned | 204 | 8 | 1.0000 | 0.0093 | 0.0093 | - | |
| Ago2 | Unassigned | 933 | 22 | 0.0059 | |||
| Domain of Unknown Function DUF1785; Pfam 08699 | 147 | 6 | 0.0087 | ||||
| PAZ_argonaute_like | 312 | 12 | 0.0069 | ||||
| Unassigned | 513 | 18 | 0.0081 | ||||
| Piwi domain; pfam02171 | 912 | 14 | 0.0027 | 0.0061 | |||
| Unassigned | 78 | 2 | 0.1482 | 0.0014 | 0.0052 | 0.7332 | |
| R3D1 | Unassigned | 195 | 4 | 0.0036 | |||
| Double-stranded RNA binding motif | 135 | 3 | 0.0069 | ||||
| Unassigned | 138 | 6 | 0.0010 | ||||
| Double-stranded RNA binding motif | 201 | 8 | 0.0061 | ||||
| Unassigned | 96 | 2 | 0.0030 | ||||
| Double-stranded RNA binding motif | 198 | 6 | 0.0061 | 0.0063 | |||
| Unassigned | 15 | 0 | 0.7084 | 0.0000 | 0.0019 | 0.0099 | |
| R2D2 | Unassigned | 30 | 2 | 0.0010 | |||
| Double-stranded RNA binding motif | 210 | 4 | 0.0009 | ||||
| Unassigned | 66 | 3 | 0.0155 | ||||
| Double-stranded RNA binding motif | 153 | 1 | 0.0014 | 0.0011 | |||
| Unassigned | 441 | 11 | 0.1775 | 0.0064 | 0.0076 | 0.2665 |
The proportions of segregating sites in regions of known function versus regions where functions have not been assigned were compared by Fisher's Exact Test (FET) and the resulting probability is listed. Average values of pi(average number of nucleotide differences per site between sequence pairs) were compared between regions of known versus unassigned function using a student's t-test and a significant difference was only seen for R3D1, in which pi was greater in regions of known function.
Tests for positive selection
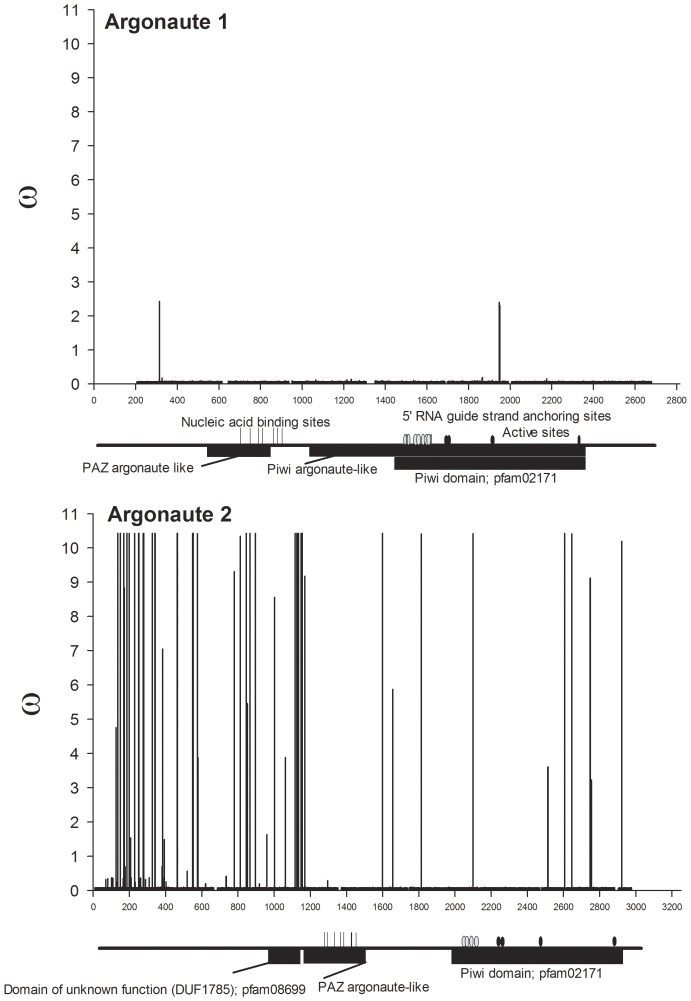
Maximum likelihood analysis of protein-coding DNA sequences was used to estimate the average number of nucleotide differences among replacement sites (KA) and synonymous sites (KS) and perform LRTs for positive selection. The ML tree topologies derived from RAxML were enforced as a constraint in CodeML from the PAML package [41]. Five models (M0, M1a, M2a, M7 and M8) were compared and results for each of the five models and each of the six genes are shown in Table S1. For each gene the model with the greatest likelihood score is highlighted in grey. In each case the M2a model had a significantly better fit than the M1a model and the M8 model had a significantly better fit than the M7 model, implying that models of positive selection had a better fit than neutral models. All of the amino acids identified using the naïve empirical Bayes (NEB) method were also identified using the Bayes empirical Bayes (BEB) method, while a few additional sites were identified with BEB.
The alternative amino acids found to be under positive selection using BEB are listed in Table S2 alongside w from the M8 model and its standard error. All replacement to synonymous substitution ratios (w) are mapped across dicer genes in Figure 4, across argonaute genes in Figure 5 and across genes encoding dsRNA-binding proteins in Figure 6. Even though clusters are visually evident, the distances between positively selected sites (PSSs) were formally subjected to a chi-square goodness-of-fit test to determine if they were distributed in a random (Poisson) fashion. Only ago2, dcr1, and dcr2 had enough sites to perform this test and the PSSs all three of these genes were clustered rather than randomly distributed (analyses available on request).
Figure 5. w ratios for all nucleotides mapped across annotated Argonaute genes.
Eukaryotic argonaute proteins are characterized by having both PAZ and Piwi domains. Piwi domains are found only in Ago proteins and are structurally related to the RNase H family of ribonucleases. Nucleic acid binding sites are indicated by vertical lines. The 5′ ends of siRNAs and miRNAs are important for mRNA target recognition and definition of the site of RNA cleavage. These binding sites are indicated by grey ovals. Active sites (black ovals) in the Ago1 ribonuclease correspond to Tyr684, Glu686, Pro757 and Gly895 and in Ago 2 correspond to Asp740, His742, Asp812, and His950.
An additional test was conducted on PSSs using the program Sorting Intolerant from Tolerant (SIFT) [42] to determine whether the amino acid changes in Table S2 are predicted to affect protein function. SIFT scores replacement substitutions on a scale from 0–1, where scores at or below 0.05 are likely to change protein function, whereas higher scores are not. Thirty PSSs were detected in Dcr1, eight (27%) of which were likely to change protein function; in Dcr2, 16 of 38 (42%) PSSs were likely to change protein function (Figure 4). In Dcr1, a single cluster consisting of positively-selected sites 384, 385, 388 and 395 was detected in a region without assigned function between the DExD/H-like helicase and helicase superfamily C-terminal domains. Replacements at PSSs 497, 502 and 592 were in the helicase superfamily C-terminal domain and two of these were predicted to change function. Replacements at PSSs 795, 817 and 978 were in the dsRNA-binding domain but were not predicted to change function. In Dcr2, one cluster of PSSs occurred in the ribonuclease III carboxyl-terminal domain and included amino acid sites 1446, 1450, and 1454, which are among the residues responsible for dimerization. Missense mutations in an RNase III-encoding domain of Drosophila dcr2 resulted in a profound loss of dsRNA processing activity and destabilization of the protein [24]. A second Dcr2 PSS cluster consisted of 8 PSSs, six with significant SIFT scores, in a region of unassigned function between the ribonuclease III C terminal domain and the dsRNA binding domain. A cluster also occurred in a region of unassigned function amino-terminal to the PAZ domain.
Replacement substitutions in ago1 were detected in four of the six Ae. aegypti collections even though no replacement substitutions were found in ago1 among four species of Drosophila. It is unclear why diversifying selection would occur within and among collections of Ae. aegypti while only purifying selection is evident among Drosophila spp. Three PSSs were identified in Ago1 but none of these was predicted to change protein function. In strong contrast, 16 of 38 PSSs were likely to change protein function in Ago2 (Figure 5). Four clusters of PSSs were found in Ago2; one of the clusters occurred in the amino-terminal region of the PAZ domain at residues 372, 375, 378, 383, 385, and 390, and the 383 and 390 replacements had significant SIFT scores. However, the oligonucleotide binding domain of PAZ occurs in the carboxyl-terminus in residues 418–472 according to GenBank annotation ACR56327. The amino-terminus of ago2 encodes a series of poly-glutamines, however the function(s) of these residues are unknown. One intriguing PSS occurred in the 5′ RNA guide strand anchoring site in the amino-end of the Piwi domain. This corresponds to amino acid residue 700 and involves a threonine-methionine replacement. Of two PSSs identified in R3D1, one was likely to change protein function. In R2D2, four of five PSSs were predicted to alter function.
Phenotype correlation analysis
Phenotype correlation analysis was performed to identify potential correlations of mosquitoes with DENV2 midgut infection barrier (MIB) and escape barrier (MEB) phenotypes with the number of nucleotide differences between all sequence pairs (k), nucleotide differences per site between all sequence pairs (pi) and and/or the number of nucleotide differences among replacement sites (KA) and among synonymous sites (KS) and/or the KA/KS ratio (w). Phenotype data were available for Poza Rica (MIB: 21%, MEB: 18%), Lerdo de Tejada (MIB: 25%, MEB: 36%), and Chetumal, Mexico (MIB: 9%, MEB: 8%) [43],[44] and PK10, Senegal (MIB: 8%, MEB: 76%) [45] collections. Pearson correlation coefficients are displayed in Table 4. No significant MIB correlations were found. No significant MEB correlations with these variables were observed for the argonaute, r3d1 or r2d2 genes. However, w in dcr1 was significantly positively correlated with MEB and pi, k, and KS were significantly correlated with MEB for dcr2. This suggests the possibility that diversity in Dicers may increase the frequency of mosquitoes with MEBs.
Table 4. Phenotype correlation analysis.
| Gene | p | k | Ks | Ka | Ka/Ks |
| Ago1 | |||||
| MIB | 0.000 | 0.031 | 0.003 | 0.343 | 0.338 |
| MEB | 0.629 | 0.439 | 0.564 | 0.837 | 0.832 |
| Ago2 | |||||
| MIB | 0.640 | 0.646 | 0.489 | 0.580 | 0.019 |
| MEB | 0.585 | 0.580 | 0.773 | 0.581 | 0.398 |
| Dcr1 | |||||
| MIB | 0.108 | 0.152 | 0.105 | 0.085 | 0.017 |
| MEB | 0.203 | 0.338 | 0.304 | 0.020 | 0.900* |
| Dcr2 | |||||
| MIB | 0.026 | 0.026 | 0.157 | 0.064 | 0.723 |
| MEB | 0.956* | 0.957* | 0.984** | 0.608 | 0.002 |
| R3D1 | |||||
| MIB | 0.050 | 0.051 | 0.153 | 0.386 | 0.493 |
| MEB | 0.642 | 0.640 | 0.574 | 0.066 | 0.216 |
| R2D2 | |||||
| MIB | 0.000 | 0.041 | 0.072 | 0.410 | 0.543 |
| MEB | 0.299 | 0.520 | 0.633 | 0.829 | 0.718 |
To identify potential correlations of Ae aegypti susceptibility to dengue virus infection with measures of nucleotide and amino acid diversity in RNAi genes, Pearson correlation analyses were performed between DENV-2 MIB (the proportion of orally virus-exposed female mosquitoes that fail to develop a midgut infection) and MEB (the proportion of females with a midgut infection that fail to develop a disseminated infection) with p (number of nucleotide differences per site between all sequence pairs), k (number of nucleotide differences between all sequence pairs), KA (number of nucleotide differences among replacement sites), KS (number of nucleotide differences among synonymous sites) and KA/KS. Pearson correlation coefficients are shown.
P≤0.05,
P≤0.01.
Discussion
Intraspecific patterns of variation between miRNA and siRNA pathway genes in Ae. aegypti
In contrast to Drosophila spp., we found that in Ae. aegypti mosquitoes both exo-siRNA and miRNA pathway genes appear to be undergoing rapid, positive, diversifying selection. However, in similarity to Drosophila, most positively selected sites occurred in protein regions without defined functions (Table S2 and Figs. 3–5). Our observations are consistent with a hypothesis that diversifying selection acts on both dcr1 and dcr2 to maintain the intraspecific w ratios among dcr1 genes of Ae. aegypti populations at the same level as among Drosophila species. Mandibulate arthropods are unique in having two Dicers [34]. With the exception of Cnidarians and Porifera, the remainder of animals so far examined, including vertebrates, possess a single Dicer that produces miRNAs and, in the case of invertebrates such as C. elegans, also siRNAs. It is possible that Dcr1 retains some role in antiviral RNAi in mosquitoes but not in Drosophila. Mosquitoes (Culicidae) are members of the primitive fly suborder Nematocera while Drosophilidae are one of the most recently evolved of fly families [60]. In addition, replication of some mammalian viruses has been shown to be indirectly inhibited by host miRNAs and some viruses exploit host miRNAs during replication [61]; however, potential roles of mosquito miRNAs in arbovirus replication have not been explored. It is possible that components that have been assigned functions in distinct RNA silencing pathways, including the miRNA pathway, interact with or serve as redundant or backup antiviral mechanisms for the exo-siRNA pathway in insects. Evidence of interactions between components of RNA silencing pathways in Drosophila was provided by the demonstration that R3D1-Dcr2 heterodimers, rather than the canonical R2D2-Dcr2 complexes, are involved in Ago2-RISC-loading of endo-siRNAs [5], [6]. The endo-siRNA pathway, which has been shown to function in suppressing transposon activity in somatic cells of Drosophila, can also be triggered by transient transfection of exogenous dsRNA, suggesting a potential role in antiviral defense [62]. Potential interactions between siRNA and miRNA pathways in mosquitoes, particularly in antiviral defense or control of transposon activity, remain to be examined.
Evidence for a role of the piRNA pathway in insect antiviral defense also has emerged recently. In our examination of antiviral RNAi in Anopheles gambiae mosquitoes we found that dsRNA-mediated silencing of the ago3 gene concurrent with O'nyong-nyong virus infection resulted in increased virus titers, hinting at a possible role for Ago3 in antiviral immunity [25]. Furthermore, RNA virus-specific piRNAs, in addition to viral siRNAs, were recently described in Drosophila ovary cells [63] and other studies showed that piwi-family mutants of Drosophila were more susceptible to Drosophila virus X infection than wild-type flies [8]. We also found that in cultured C6/36 (Aedes albopictus) cells a single-nucleotide deletion in dcr2 causes a defect in the exo-siRNA pathway-mediated antiviral defense that was apparently compensated by the piRNA pathway [30], [64]. The redundant role of the piRNA pathway in antiviral defense in mosquito somatic cells and its particular relevance in dcr2-null cell culture lines has recently been confirmed [65].
Sources of diversifying selection
Obbard et al. [34] suggested that a “molecular arms race” with pathogenic virus suppressors of RNAi drives siRNA pathway gene diversity in Drosophila spp. There are various reasons to believe that infections with arboviruses are not the drivers of diversifying selection that we have documented in the miRNA and siRNA pathways in Ae aegypti. A hallmark of arboviruses is that they have little, if any, effect on survivorship or fecundity in their insect hosts [66]. Two studies further suggest that selection has, in fact, prevented the evolution of potent VSRs in arboviruses [28], [38]. Even if infection by arboviruses imposed a minimal fitness effect, a number of field studies have demonstrated that very few mosquitoes (10−3 to 10−4) collected in areas endemic for certain arboviruses are actually infected at any given time [67], [68], [69] thus providing only rare opportunities for selection. An interesting alternative possibility for selection may involve the recently discovered high prevalence of persistent insect-only flaviviruses in natural populations [70], [71], [72]. These viruses are maintained through vertical transmission from one generation to the next without obvious pathogenesis and without requiring horizontal transmission through infected vertebrates.
We suggest that infections with entomopathogenic viruses or transposon invasion and movement are more likely causes of the diversifying selection detected in this study. Unfortunately, few mosquito-pathogenic RNA viruses have been identified or well-studied. One such virus, Nodamura virus (Nodaviridae), was isolated from Culex tritaeniorhynchus in Japan [73] and can experimentally infect and produce pathogenesis in Ae. aegypti [74]. It has a bipartite, positive-strand RNA genome that replicates through a dsRNA intermediate. Unique features of viruses in this family are that their replication complexes are sequestered in membrane-enclosed spherules within the mitochondrial outer membrane during replication and they encode B2-type VSRs [75]. Small RNAs that appeared to be derived from the RNA genome of a previously-undescribed nodavirus were recently discovered by deep sequencing analysis of a small RNA library from Ae. aegypti [63], suggesting that other potentially pathogenic mosquito viruses remain to be found.
Phenotype correlation analysis
Our a priori hypothesis was that Ae. aegypti collections that were more refractory for DENV2 disseminated infection would also have higher rates of evolution in genes encoding components of the siRNA pathway compared to DENV2 susceptible populations. The trends shown in correlation of vector competence with certain measures of genetic diversity in RNAi pathway genes in Table 4 need to be tested in more Ae. aegypti populations and possibly in other Aedes species. Furthermore, we need to perform quantitative trait loci mapping and association studies to test for a correlation between miRNA and siRNA pathway alleles and arbovirus susceptibility. If a correlation is detected it could suggest that RNA silencing evolved in mosquitoes as a means to combat entomopathogenic virus infection or genome invasion by transposons but that this evolution may have indirectly provided a regulatory mechanism for replication and transmission of arboviruses.
Supporting Information
CodeML results for each of the five models for detection of positive selection and each of the six genes. For each gene, the ML model is highlighted in grey. The number of positively selected sites (PSS) identified using the naive empirical Bayes (NEB) and Bayes empirical Bayes (BEB) methods are listed for each gene. l = −log likelihood ratio. The likelihood ratio test was computed between Models M2a and M1a (c2[1 d.f.] = 2ΔL) and between Models M7 and M8 (χ2 [1 d.f.] = 2ΔL) in all 12 comparisons, P<0.0001.
(DOCX)
Alternative amino acids found to be under positive selection in CodeML using BEB. The locations of the replacements are indicated in parentheses (U = region with unassigned function, Pw = Piwi, Pz = PAZ, DUF = domain of unknown function, Hc = Helicase superfamily c-terminal domain, Hcd = Helicase dimerization domain, Dc = DExD/H-like helicase, Db = double stranded RNA binding domain). Sites are listed alongside w ratios from the M8 model and its standard error. SIFT scores <0.05 indicate the replacement amino acid is likely to change protein function and are underlined and appear in bold. Sites that are clustered (<10 nt apart) appear in boxes.
(DOCX)
Funding Statement
This work received financial support from the UNICEF/UNDP/WorldBank/WHO Special Programme for Research and Training in Tropical Disease (TDR): A50363. This work was also supported by NIH/NIAID contract N01-AI 25489 and R01 AI034014 to CDB and AI083368 to WCB4. SAB was supported by the FTP Training Grant CCT 822307 from the Centers for Disease Control and Prevention. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 2. Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38: S31–S36. [DOI] [PubMed] [Google Scholar]
- 3. Ding S-W, Voinnet O (2007) Antiviral Immunity Directed by Small RNAs. Cell 130: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zamore PD (2007) RNA silencing: Genomic defence with a slice of pi. Nature 446: 864–865. [DOI] [PubMed] [Google Scholar]
- 5. Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, et al. (2008) An endogenous small interfering RNA pathway in Drosophila . Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, et al. (2008) Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science 320: 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zambon RA, Vakharia VN, Wu LP (2006) RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster . Cellular Microbiology 8: 880–889. [DOI] [PubMed] [Google Scholar]
- 9. Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler J-L (2006) Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila . Nat Immunol 7: 590–597. [DOI] [PubMed] [Google Scholar]
- 10. Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- 11. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, et al. (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila . Nat Immunol 9: 1425–1432. [DOI] [PubMed] [Google Scholar]
- 12. Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, et al. (2003) R2D2, a Bridge Between the Initiation and Effector Steps of the Drosophila RNAi Pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- 13. Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rand TA, Ginalski K, Grishin NV, Wang X (2004) Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci USA 101: 14385–14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwarz DS, Hutvágner G, Haley B, Zamore PD (2002) Evidence that siRNAs Function as Guides, Not Primers, in the Drosophila and Human RNAi Pathways. Molecular Cell 10: 537–548. [DOI] [PubMed] [Google Scholar]
- 16. Schwarz DS, Tomari Y, Zamore PD (2004) The RNA-Induced Silencing Complex Is a Mg2+-Dependent Endonuclease. Current Biology 14: 787–791. [DOI] [PubMed] [Google Scholar]
- 17. Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Development 19: 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, et al. (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes & Development 20: 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Förstemann K (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. (2007) Genome Sequence of Aedes aegypti, a Major Arbovirus Vector. Science 316: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Black WCt, Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury CV, Fernandez-Salas I, et al. (2002) Flavivirus susceptibility in Aedes aegypti . Arch Med Res 33: 379–388. [DOI] [PubMed] [Google Scholar]
- 23. Wang X-H, Aliyari R, Li W-X, Li H-W, Kim K, et al. (2006) RNA Interference Directs Innate Immunity Against Viruses in Adult Drosophila . Science 312: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim DH, Kim J, Kim S, Carthew RW, Lee YS (2008) Functional analysis of dicer-2 missense mutations in the siRNA pathway of Drosophila . Biochemical and Biophysical Research Communications 371: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, et al. (2004) RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae . Proceedings of the National Academy of Sciences of the United States of America 101: 17240–17245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, et al. (2009) Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog 5: e1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aliyari R, Wu Q, Li H-W, Wang X-H, Li F, et al. (2008) Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila . Cell Host & Microbe 4: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Myles KM, Wiley MR, Morazzani EM, Adelman ZN (2008) Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proceedings of the National Academy of Sciences 105: 19938–19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brackney DE, Beane JE, Ebel GD (2009) RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification. PLoS Pathog 5: e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, et al. (2010) Comparison of Dengue Virus Type 2-Specific Small RNAs from RNA Interference-Competent and -Incompetent Mosquito Cells. PLoS Negl Trop Dis 4: e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li F, Ding SW (2006) Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60: 503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Obbard DJ, Jiggins FM, Halligan DL, Little TJ (2006) Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol 16: 580–585. [DOI] [PubMed] [Google Scholar]
- 33. Obbard DJ, Jiggins FM, Bradshaw NJ, Little TJ (2011) Recent and Recurrent Selective Sweeps of the Antiviral RNAi Gene Argonaute-2 in Three Species of Drosophila . Molecular Biology and Evolution 28: 1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM (2009) The evolution of RNAi as a defence against viruses and transposable elements. Philosophical Transactions of the Royal Society B: Biological Sciences 364: 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Li WX, Ding SW (2002) Induction and Suppression of RNA Silencing by an Animal Virus. Science 296: 1319–1321. [DOI] [PubMed] [Google Scholar]
- 36. Attarzadeh-Yazdi G, Fragkoudis R, Chi Y, Siu RWC, Ulper L, et al. (2009) Cell-to-Cell Spread of the RNA Interference Response Suppresses Semliki Forest Virus (SFV) Infection of Mosquito Cell Cultures and Cannot Be Antagonized by SFV. J Virol 83: 5735–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, et al. (2007) La Crosse Bunyavirus Nonstructural Protein NSs Serves To Suppress the Type I Interferon System of Mammalian Hosts. J Virol 81: 4991–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE (2009) Suppression of RNA Interference Increases Alphavirus Replication and Virus-Associated Mortality in Aedes aegypti Mosquitoes. BMC Microbiol 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siu RWC, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, et al. (2011) Antiviral RNA Interference Responses Induced by Semliki Forest Virus Infection of Mosquito Cells: Characterization, Origin, and Frequency-Dependent Functions of Virus-Derived Small Interfering RNAs. J Virol 85: 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blair CD (2011) Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol 6: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Z (2007) PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 42. Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bennett K, Olson K, Munoz Mde L, Fernandez-Salas I, Farfan-Ale J, et al. (2002) Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg 67: 85–92. [DOI] [PubMed] [Google Scholar]
- 44. Lozano-Fuentes S, Fernandez-Salas I, de Lourdes Munoz M, Garcia-Rejon J, Olson KE, et al. (2009) The Neovolcanic Axis Is a Barrier to Gene Flow among Aedes aegypti Populations in Mexico That Differ in Vector Competence for Dengue 2 Virus. PLoS Negl Trop Dis 3: e468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sylla M, Bosio C, Urdaneta-Marquez L, Ndiaye M, Black WC (2009) Gene Flow, Subspecies Composition, and Dengue Virus-2 Susceptibility among Aedes aegypti Collections in Senegal. PLoS Negl Trop Dis 3: e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mattingly PF (1957) Genetical aspects of the Aedes aegypti problem. I. Taxonomy and bionomics. Ann Trop Med Parasitol 51: 392–408. [PubMed] [Google Scholar]
- 47. Lorenz L, Beaty BJ, Aitken THG, Wallis GP, Tabachnick WJ (1984) The Effect of Colonization Upon Aedes aegypti - Susceptibility to Oral Infection with Yellow-Fever Virus. American Journal of Tropical Medicine and Hygiene 33: 690–694. [DOI] [PubMed] [Google Scholar]
- 48. Tabachnick WJ, Wallis GP, Aitken THG, Miller BR, Amato GD, et al. (1985) Oral Infection of Aedes aegypti with Yellow-Fever Virus - Geographic-Variation and Genetic Considerations. American Journal of Tropical Medicine and Hygiene 34: 1219–1224. [DOI] [PubMed] [Google Scholar]
- 49. Wallis GP, Aitken THG, Beaty BJ, Lorenz L, Amato GD, et al. (1985) Selection for Susceptibility and Refractoriness of Aedes aegypti to Oral Infection with Yellow-Fever Virus. American Journal of Tropical Medicine and Hygiene 34: 1225–1231. [DOI] [PubMed] [Google Scholar]
- 50.Black WCI, DuTeau NM (1997) RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors: a Methods Manual. London, New York: Chapman and Hall. pp. 361–373.
- 51. Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298. [DOI] [PubMed] [Google Scholar]
- 52.Maddison DR, Maddison WP (2001) MacClade: Analysis of phylogeny and character evolution. [DOI] [PubMed]
- 53.Nei M (1987) Molecular Evolutionary Genetics. New York: Columbia University Press. 512 p.
- 54. Tajima F (1983) Evolutionary Relationship of DNA Sequences in Finite Populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 56. Felsenstein J (1985) Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 57.Stamatakis A (2008) The RAxML 7.0.4 Manual. Ludwig-Maximilians-Universitat Munchen: The Exelixis Lab, Teaching & Research Unit, Bioinformatics Department of Computer Science.
- 58. Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution 11: 725–736. [DOI] [PubMed] [Google Scholar]
- 59. Yang Z, Wong WSW, Nielsen R (2005) Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection. Molecular Biology and Evolution 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 60. Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, et al. (2011) Episodic radiations in the fly tree of life. Proceedings of the National Academy of Sciences 108: 5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schütz S, Sarnow P (2006) Interaction of viruses with the mammalian RNA interference pathway. Virology 344: 151–157. [DOI] [PubMed] [Google Scholar]
- 62. Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K (2009) Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu Q, Luo Y, Lu R, Lau N, Lai EC, et al. (2010) Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proceedings of the National Academy of Sciences 107: 1606–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, et al. (2010) C6/36 Aedes albopictus Cells Have a Dysfunctional Antiviral RNA Interference Response. PLoS Negl Trop Dis 4: e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM (2012) Production of Virus-Derived Ping-Pong-Dependent piRNA-like Small RNAs in the Mosquito Soma. PLoS Pathog 8: e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lambrechts L, Scott TW (2009) Mode of transmission and the evolution of arbovirus virulence in mosquito vectors. Proceedings of the Royal Society B: Biological Sciences 276: 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chow VT, Chan YC, Yong R, Lee KM, Lim LK, et al. (1998) Monitoring of dengue viruses in field-caught Aedes aegypti and Aedes albopictus mosquitoes by a type-specific polymerase chain reaction and cycle sequencing. The American Journal of Tropical Medicine and Hygiene 58: 578–586. [DOI] [PubMed] [Google Scholar]
- 68. Chung YK, Pang FY (2002) Dengue virus infection rate in field populations of female Aedes aegypti and Aedes albopictus in Singapore. Tropical Medicine & International Health 7: 322–330. [DOI] [PubMed] [Google Scholar]
- 69. Urdaneta L, Herrera F, Pernalete M, Zoghbi N, Rubio-Palis Y, et al. (2005) Detection of dengue viruses in field-caught Aedes aegypti (Diptera: Culicidae) in Maracay, Aragua state, Venezuela by type-specific polymerase chain reaction. Infection, Genetics and Evolution 5: 177–184. [DOI] [PubMed] [Google Scholar]
- 70. Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, et al. (2007) Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology 359: 405–414. [DOI] [PubMed] [Google Scholar]
- 71. Hoshino K, Isawa H, Tsuda Y, Sawabe K, Kobayashi M (2009) Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology 391: 119–129. [DOI] [PubMed] [Google Scholar]
- 72. Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD (2012) Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 427: 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Scherer WF, Hurlbut HS (1967) Nodamura Virus from Japan: A New and Unusual Arbovirus Resistant to Diethyl Ether and Chloroform. Am J Epidemiol 86: 271–285. [DOI] [PubMed] [Google Scholar]
- 74. Tesh RB (1980) Infectivity and Pathogenicity of Nodamura Virus for Mosquitoes. J Gen Virol 48: 177–182. [Google Scholar]
- 75. Venter P, Schneemann A (2008) Recent insights into the biology and biomedical applications of Flock House virus. Cellular and Molecular Life Sciences (CMLS) 65: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CodeML results for each of the five models for detection of positive selection and each of the six genes. For each gene, the ML model is highlighted in grey. The number of positively selected sites (PSS) identified using the naive empirical Bayes (NEB) and Bayes empirical Bayes (BEB) methods are listed for each gene. l = −log likelihood ratio. The likelihood ratio test was computed between Models M2a and M1a (c2[1 d.f.] = 2ΔL) and between Models M7 and M8 (χ2 [1 d.f.] = 2ΔL) in all 12 comparisons, P<0.0001.
(DOCX)
Alternative amino acids found to be under positive selection in CodeML using BEB. The locations of the replacements are indicated in parentheses (U = region with unassigned function, Pw = Piwi, Pz = PAZ, DUF = domain of unknown function, Hc = Helicase superfamily c-terminal domain, Hcd = Helicase dimerization domain, Dc = DExD/H-like helicase, Db = double stranded RNA binding domain). Sites are listed alongside w ratios from the M8 model and its standard error. SIFT scores <0.05 indicate the replacement amino acid is likely to change protein function and are underlined and appear in bold. Sites that are clustered (<10 nt apart) appear in boxes.
(DOCX)