Abstract
“Trojan horse” antibiotic albomycins are peptidyl nucleosides consisting of a highly modified 4′-thiofuranosyl cytosine moiety and a ferrichrome siderophore that are linked by a peptide bond via a serine residue. While the latter component serves to sequester iron from the environment, the seryl nucleoside portion is a potent inhibitor of bacterial seryl-tRNA synthetases, resulting in broad-spectrum antimicrobial activities of albomycin δ2. The isolation of albomycins has revealed this biological activity is only optimized following two unusual cytosine modifications, N4-carbamoylation and N3-methylation. We identified a genetic locus (named abm) for albomycin production in Streptomyces sp. ATCC 700974. Gene deletion and complementation experiments along with bioinformatic analysis suggested 18 genes are responsible for albomycin biosynthesis and resistance, allowing us to propose a potential biosynthetic pathway for installing the novel chemical features. The gene abmI, encoding a putative methyltransferase, was functionally assigned in vitro and shown to modify the N3 of a variety of cytosine-containing nucleosides and antibiotics such as blasticidin S. Furthermore, a ΔabmI mutant was shown to produce the descarbamoyl-desmethyl albomycin analog, supporting that the N3-methylation occurs before the N4-carbamoylation in the biosynthesis of albomycin δ2. The combined genetic information was utilized to identify an abm-related locus (named ctj) from the draft genome of Streptomyces sp. C. Cross-complementation experiments and in vitro studies with CtjF, the AbmI homolog, suggest the production of a similar 4′-thiofuranosyl cytosine in this organism. In total, the genetic and biochemical data provide a biosynthetic template for assembling siderophore-inhibitor conjugates, and modifying the albomycin scaffold to generate new derivatives.
Albomycins are broad-spectrum antibiotics that have been isolated several times from soildwelling actinomycetes in various drug discovery programs since its first report dating back sixty years.1 Its extreme antibacterial potency and extended spectrum has prompted numerous attempts to develop it into a commercial drug2–7 and likewise spawned a longstanding interest in elucidating the mechanism of action of albomycins.8 These studies have revealed that albomycins are Trojan horse antibiotics that consist of a ferrichrome-type siderophore component that is indiscriminately taken up by bacteria as a mechanism to scavenge iron from the environment. Once inside the cell, host peptidases hydrolyze albomycin into two components, the aforementioned siderophore and a nucleoside compound termed SB-217452.7 The latter was independently identified as an enzyme inhibitor of bacterial seryl-tRNA synthetase (SerRS), an essential component for all protein synthesis. Because of the high efficiency of bacterial iron transport system and specificity of the inhibitor to the SerRS target, minimum inhibitory concentration (MIC) of albomycin δ2 was shown to be as low as 10 ng/ml against Streptococcus pneumonia, and 5 ng/ml against Escherichia coli.9
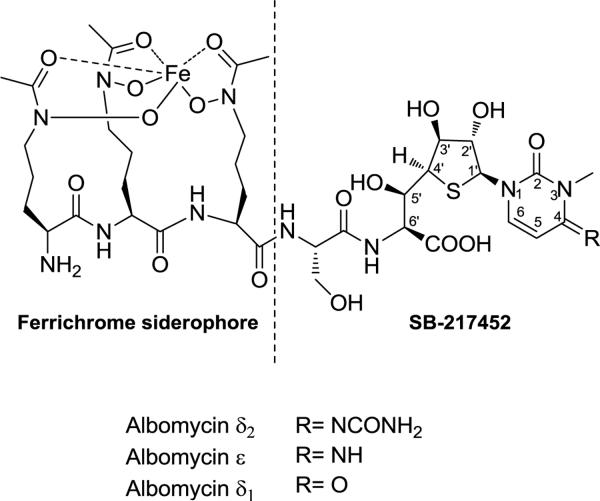
The exceptional antibiotic activity of albomycins stems from the many unusual chemical features (Figure 1). Structurally, albomycins belonging to the peptidyl nucleoside family consists of an N-terminus of hydroxymate siderophore composed of three tandem N5-acetyl-N5-hydroxy-l-ornithine residues. This siderophore is a chemical motif that is able to chelate a ferric iron with a high affinity and is often observed as a part of the structure of fungal ferrichromes. At the C-terminus of the siderophore tripeptide of albomycins is a serine residue linked by a peptide bond to a nonproteinogenic α-amino acid. This amino acid has a side chain consisting of a 4′-thioxylofuranose ring attached to a cytosine base through a C1′-N1 glycosidic bond that is reminiscent of the canonical ribonucleosides. In albomycin δ2, the major congener produced by Streptomyces sp. ATCC 700974, the cytosine base carries two modifications: one is methylation of the N3 and the other is carbamoylation of the N4 imino nitrogen. These modifications are known to significantly enhance the bioactivity of albomycins since two minor congeners that either lack the carbamoyl group (albomycin ε) or the N3-methylcytosine is substituted with a N3-methyluracil (albomycin δ1) have been isolated and assessed.2 While the C-terminal nonproteinogenic amino acid contains several uncommon features such as a thioether modification of the furanoside and N4-carbamoylation of cytosine, N3-methylated cytosine is only found in eukaryotic tRNAs and rRNAs and thought to finely modulate structures and functions of the RNAs.
Figure 1.
Structure of albomycins. The dashed line indicates the site of cleavage by bacterial peptidases to give a ferrichrome-type hydroxamate siderophore and the seryl thionucleoside compound SB-217452. The R in SB-217452 is identical to albomycin δ2.
The biosynthetic gene clusters for a number of nucleoside antibiotics have been recently identified and partially characterized.10–16 Most of the pyrimidine-containing nucleoside antibiotics are derived from uridyl ribonucleotide, and modification of the uracil base are infrequently encountered with only a few exceptions. In contrast, the cytosine-containing antibiotics often contain modifications of the canonical base; for example, blasticidins and mildiomycins are two cytosine-derived nucleoside antibiotics whose congeners can harbor a hydroxymethyl group at the C5 position. Amicetin, another cytosine-containing antibiotic for which the biosynthetic genes were recently identified, contains a paminobenzoic acid substituent at N4 of cytosine.17 In essence, modification by N3-methylation in conjunction with the N4-carbamoylation is so far only seen in albomycins. The totality of the chemical features and the possible biotechnological applications for drug discovery inspired us in part to search for the biosynthetic machinery responsible for albomycin assembly.
We previously identified a region on the chromosomal DNA of Streptomyces sp. strain ATCC 700974 that codes for a protein related to albomycin production. A gene, abmK (also known as serS2 or alb10), was shown to be a resistance gene of albomycin. Its encoded SerRS requires a high serine concentration for optimal activity.18 Contrastingly, at least one distinct SerRS homolog (SerS1) in this strain has a much lower serine requirement and was therefore concluded to be involved in housekeeping functions. Importantly, the effective in vitro inhibitory concentration of SB-217452 to SerS1 is at least ten times lower than to AbmK, consistent with a resistance role of AbmK. Given that selfresistance genes are often clustered with biosynthetic genes in actinomycetes, the region surrounding the abmK gene was hypothesized to harbor the structural genes for albomycin biosynthesis. We now report the strategy used to identify this region and demonstrate that the abmK resistance gene is indeed clustered with the albomycin biosynthetic genes by utilizing inactivation and complementation of abmKflanking genes along with the functional assignment and in vitro characterization of an S-adenosyl-l-methionine (SAM)-dependent N3-cytidine methyltransferase encoded by abmI. This data combined with bioinformatic analysis has allowed us to propose an albomycin biosynthetic pathway. Furthermore, a related biosynthetic gene cluster was identified by mining genome of Streptomyces sp. strain C. Of the eight abm homlogs residing within this locus, two were assigned functions identical to the respective abm genes, suggesting this strain may produce an albomycin-related metabolite. These findings set the stage to elucidate the mechanisms for installing the chemical features found in albomycins, which provide these Trojan horse antibiotics with such potent and intriguing biological activity.
RESULTS AND DISCUSSION
Cloning and Identification of the abm gene cluster in Streptomyces sp. ATCC 700974
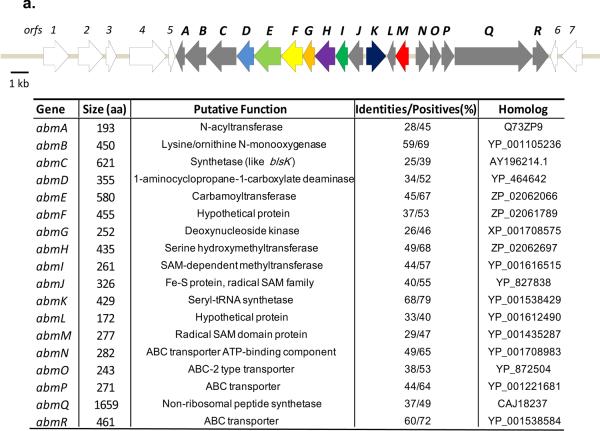
Ferrichrome siderophores contain hydroxamate bidentates that are essential for coordination of a ferric ion. Hydroxylation of the N5 of l-ornithine in hydroxamate bidentates is known to occur by a flavin-dependent monooxygenase termed SidA in Aspergillus.19 The hydroxamate functionality is also found in bacterial siderophores such as desferrioxamines and coelichelin produced by some Streptomyces species, and an identical enzymatic strategy involving a flavin-dependent monooxygenase has been described.20 We set out to identify sidA homologs encoded within the genome of the albomycin-producing strain. PCR with degenerate primers combined with genome walking yielded a ~1 kb DNA fragment encoding protein sequences with similarity to SidA. This fragment was used as a probe to screen a cosmid library, and shot-gun sequencing of two positive cosmids revealed two distinct gene clusters containing sidA homologs. One locus had a genetic organization that is highly similar to the cch cluster responsible for the biosynthesis of coelichelin in Streptomyces coelicolor (Supporting Figure S1).20 The other cosmid (8F8) contained many novel genes including homologs for a seryl-tRNA synthetase and a deoxynucleoside kinase. A DNA fragment residing distantly from the sidA homolog in 8F8 was used to re-screen the library, which led to the identification of eight more overlapping cosmids. Two cosmids were subsequently sequenced and assembled into ~38 kb continuous DNA sequences, and FRAMEPLOT analysis revealed the presence of 25 complete orfs including the sidA homolog (abmB) (Figure 2a).
Figure 2.
Biosynthetic gene clusters of albomycins and a putative albomycin-type analog. (a) Genetic organization of the albomycin (abm) gene cluster in Streptomyces sp. ATCC 700974 and (b) ctj gene cluster in Streptomyces sp. strain C. The homologous abm and ctj genes are filled with same colors. Genes unique to each cluster are filled in grey.
REDIRECT gene inactivation technology21 was utilized to generate a number of deletion mutant strains, of which abmB was the initial target. A ΔabmB mutant strain was successfully prepared based on PCR analysis (Supporting Figure S2a), and in contrast to the wild-type strain, the ΔabmB mutant strain did not produce any albomycins as determined using HPLC (Supporting Figure S2b). Additionally, agar diffusion bioassay using Escherichia coli BW25113 as an indicator strain revealed the ΔabmB mutant strain lost its antibiotic activity, thus confirming the involvement of abmB in albomycin biosynthesis and supporting the utility of the bioassay as a preliminary screen for albomycin production.7 Close examination of the 38-kb DNA locus revealed two open reading frames orf5 and orf6 are conserved as two adjacent genes, SGR_4620 and SGR_4621, respectively, in the genome of Streptomyces griseus subsp. griseus strain NBRC 13350, which does not produce albomycins. Furthermore, the protein encoded by orf6 appears to be missing ~~25% of the C-terminal residues found in SGR_4621. Additional upstream (orfs 1–4) and downstream (orf7) orfs are also highly conserved in S. griseus, suggesting these orfs are not essential for albomycin production. Nonetheless, we inactivated orf 4 and orf7 encoding a putative secreted protease and nicotinate phosphoribosyltransferase, respectively. The resulting mutants retained their ability to produce albomycins based on bioactivity tests. In contrast, abmE and I were individually inactivated (Supporting Figure S3), and the resulting mutants were unable to produce albomycin related bioactivities. Thus, the abm biosynthetic gene cluster is proposed to include 18 genes (abmA-R) required for albomycin biosynthesis and resistance.
While we performing the mutational analysis, draft genome sequences of Streptomyces sp. ATCC 700974 were obtained by Illumina DNA sequencing. Analysis of the genome revealed that ATCC 700974 and NBRC 13350 are closely related streptomycetes. More than 98.7% genetic contents are conserved between the two strains. The abm cluster appears to be one of a few genetic islands located in a ~1.5 Mbp supercontig. A construction of the metabolic network for the albomycin producing strain is in progress. In summary, we have outlined a strategy for identifying and cloning biosynthetic gene clusters of Trojan horse antibiotics produced by Streptomyces. Two of these orfs, abmI and abmE encoding a putative N-methyltransferase and carbamoyltransferase, respectively, were chosen for further functional characterization due to their potentially unusual role in nucleoside modification.
Identification of an abm-like antibiotic biosynthetic gene cluster in Streptomyces sp. C
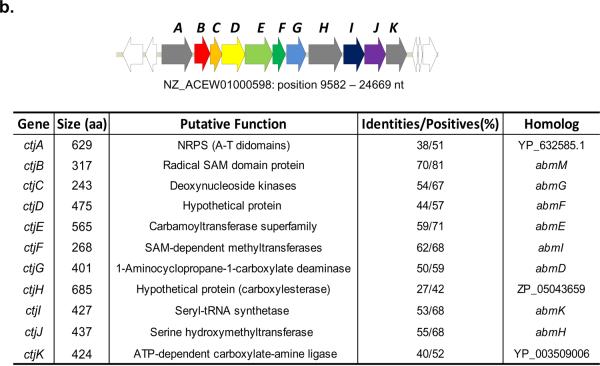
By searching GenBank with the abm genes, we identified a uni-directional gene cluster (Figure 2b) in Streptomyces sp. strain C whose genome was sequenced and released by the Broad Institute. In the 11-gene cluster, 8 genes are homologous to the abm genes (translated sequences identity ranging from 40% to 70%), particularly those likely involved in the biosynthesis of the SerRS inhibitor SB-217452. An abmK homologous gene was also identified in the gene cluster. It is speculated that the strain C might produce another Trojan horse antibiotic like albomycin targeting seryl-tRNA synthetase. The gene locus is thus named ctj. In contrast to the abm gene cluster, the locus contains an orf(annotated as ctjA) encoding an NRPS with an adenylation-thiolation didomain architecture showing homology to microcystin synthetase B. Other new orfs include ctjH encoding a hypothetic protein with low homology to proteins of carboxylesterase superfamily, and ctjK encoding a protein belonging to the ATP-dependent carboxylate-amine ligase family. As for the genes possibly involved in nucleoside modification, ctjE is a homolog of abmE, and ctjF is a homolog of abmI. Interestingly, excluding ctjC, D and E that correspond to abmG, F and E, the remaining ctj genes are architecturally rearranged compared to that observed in the abm cluster. This mosaic architecture is in sharp contrast to many homologous antibiotic biosynthetic gene clusters found in distinct actinomycetes,23 wherein the genetic organization between strains is nearly identical. Although the metabolic product for the ctj gene cluster remains unknown, the discovery does raise an question regarding the evolutionary history of albomycin, which has been isolated from a number of actinomycetes.24,25 Perhaps more importantly it suggests that the 4′-thioxylofuranosyl nucleoside core could be found in other bioactive secondary metabolites.
Genetic characterization of the genes encoding nucleoside modification enzymes
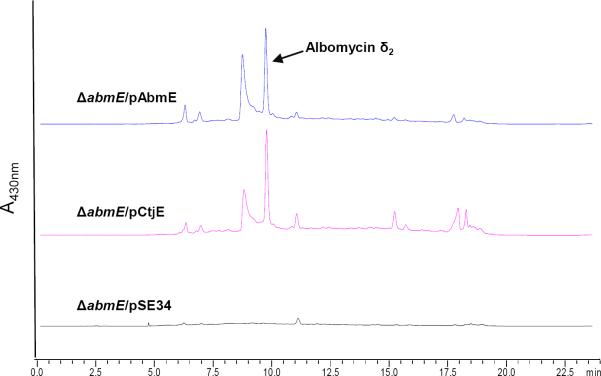
The abmE gene encodes a protein with sequence similarity to various carbamoyltransferases. We logically hypothesized that this gene product uses carbamoyl phosphate to modify the N4 nitrogen of the 4′-thiofuranosyl cytosine moiety. As previously mentioned, ΔabmE mutants were generated (Supporting Figure S3) and demonstrated not to produce any antibiotic activity measured with the bioassay. To exclude the possibility of a downstream polar effect, the genes abmA-D were PCR-amplified and cloned into a plasmid (pSE34) under the control of ermE* promoter for expression in the mutant. Following introduction of these constitutively expressed genes into the ΔabmE mutant strain, bioactivity was still not detected in liquid fermentation media. However, bioactivity was observed on solid agar plates after the ΔabmE host cells were freshly transformed. The activity was significantly less than the wild-type, and its production was highly unstable and ephemeral. We managed to isolate some active compound from agar plates that were streaked with fresh transformants. The purified active fraction yielded an [M+H]+ ion at m/z 1004.5, which is in agreement with the molecular formula of C36H55FeN10O18S for the descarbamoyl albomycin, albomycin δ1 (Supporting Figure S4). When abmE or ctjE was cloned into the abmA-D expression plasmid, the wild-type phenotype was restored within the ΔabmE mutant and albomycin δ2 was robustly produced in both cases (Figure 3) (Supporting Figure S5). Together, the results unambiguously demonstrate that the abmE gene product is responsible for N-carbamoylation of the cytosine nucleoside during albomycin biosynthesis and suggest Streptomyces sp. C produces a similar N-carbamoylated nucleoside antibiotic. In addition, since the major product of the ΔabmE mutant contains the N3-methyl group, N-carbamoylation is proposed to be the last step in the biosynthetic pathway.
Figure 3.
HPLC analysis of extracts of the ΔabmE mutant strain. The ΔabmE mutant strain was complemented with either abmE or homologous gene ctjE and shown to restore the albomycin δ2 production in contrast to the empty vector pSE34.
The gene abmI encodes an S-adenosyl-l-methionine (SAM)-dependent methyltransferase that was hypothesized to methylate the N3 of the cytosine in albomycins with cofactor SAM contributing a methyl group. To validate the proposed function, abmI was deleted in-frame in the wild-type to create a mutant strain (Supporting Figure S3). The ΔabmI strain produced a low level of bioactivity that was detected after ~20-fold concentration from the fermentation broth. After four chromatographic separations, an active fraction showed a major compound with an [M+H]+ ion of m/z 989.0 with the expected isotopic distribution which is in agreement with the molecular formula of C35H54FeN9O17S for descarbamoyl-desmethyl-albomycin (Figure 4). The MS peak was absent in a control sample that was extracted from an albomycin null mutant in which the NRPS gene abmQ was deleted. The bioactive metabolite was likely a new albomycin because its retention time was earlier than the known albomycin congeners under same reverse-phase HPLC conditions, which agrees with a more hydrophilic structure of descarbamoyl-desmethyl-albomycin (Supporting figure S6). The bioactivity was further confirmed to be a direct result of an albomycin analog by using different E. coli reporting strains in the bioassay including ΔfhuA (ferrichrome transporter gene inactivated) and abmK+ (albomycin resistant gene overexpressed) (Supporting Figure S7). The bioactivity of this biosynthetic intermediate was readily degraded with peptidases after being deferrated (Supporting Figure S7). An expression plasmid containing abmA-I under control of the ermE* promoter was subsequently introduced by conjugation into the ΔabmI mutant strain, but unlike our other complementation experiments, albomycin production was not restored despite screening a large number of colonies. The rationale for this finding is currently under investigation.
Figure 4.
ESI-MS analysis of a partially purified fraction from ΔabmI mutant that produces the descarbamoyl-desmethyl-albomycin compound. The insert is the simulated isotopic distribution of the MS ions according to the expected molecular formula. See Supporting Figure S6 for HPLC profile of the active fraction.
Biochemical characterization of a novel N3-cytosine modification enzyme
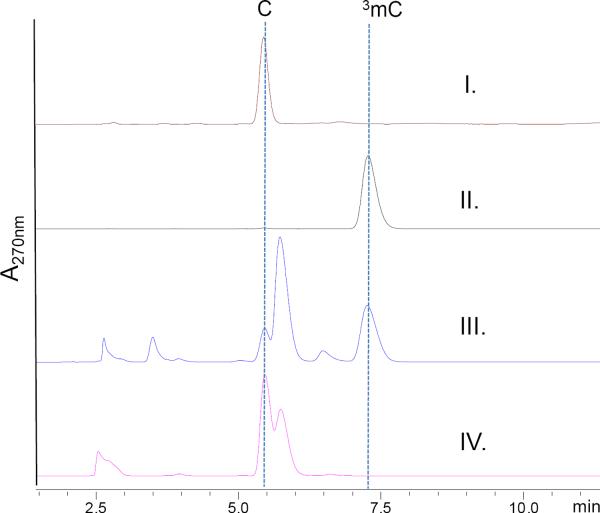
Due to the inability to complement abmI, we decided to finally turn our attention to characterization of the recombinant protein in vitro. We also targeted CtjF, the AbmI homolog, to provide additional evidence that an SB-217452-like compound is potentially produced within Streptomyces sp. C. Both proteins were soluble when produced in E. coli and purified to near homogeneity as judged by SDS-PAGE (Supporting Figure S8). Size exclusion chromatography using AbmI yielded a native molecular weight of ~60 kD suggesting AbmI functions as a dimer (calculated monomeric molecular weight is ~29 kD) (Supporting Figure S8). AbmI activity was initially tested with cytidine as a surrogate substrate along with SAM. After incubation at 30°C, a new peak appeared in an HPLC analysis with the same elution time as authentic N3-methylcytidine (3mC), which is absent in the control using boiled enzyme. The major [M+H]+ ion of this new peak was m/z 258.0, consistent with the molecular formula of C10H15N3O5 for 3mC and thus confirmed that 3mC was the product of the methylation reaction. S-Adenosyl-l-homocysteine (SAH) nucleosidase was added into the AbmI reactions to lessen potential product inhibition caused by SAH that has been observed in other methyltransferases.26 As expected, better conversion was revealed by the analysis (Figure 5) (Supporting Figure S9). Preliminary characterization of the enzyme showed optimal activity was obtained at pH 6.75 in phosphate buffer (Supporting Figure S10). The substrate specificity of AbmI was next explored to reveal that AbmI is able to methylate cytitidine derivatives including both the 2′- and 3′-deoxyribosyl cytidine nucleosides as detected by LCMS (Supporting Figure S11). Interestingly, besides being able to methylate the nucleoside base cytosine, AbmI also tolerates the fluoro- and methyl-substitutions on the C5 of cytidine nucleosides (Supporting Figure S11), suggesting a relaxed specificity for not only the ribose but also the nucleoside base. Individual kinetic parameters of these AbmI catalyzed reactions were not measurable as the rate relative to nucleoside substrate concentration was linear in the range of 100 μM to 10 mM substrates. Nonetheless, the specific activities of 2′-deoxyribosyl cytidine and 2′-deoxyribosyl-5-methylcytidine were calculated ~50 M−1 s−1 and ~48 M−1 s−1, respectively. Activity tests using CtjF revealed the identical methyltransferase activity with these nucleosides; however, with 2′-deoxy-5-fluoro-cytidine or 2′-deoxy-5-methyl-cytidine as the substrate, CtjF was apparently much less efficient than AbmI (Supporting Figure S12).
Figure 5.
HPLC analysis of AbmI in vitro reaction with cytidine as a substrate. I. Cytidine standard (C); II. N3-methylacytidine (3mC); III. AbmI reaction supplied with SAH nucleosidase after 4-hour incubation; IV. Reaction conditions are identical to III except that AbmI was heat-inactivated prior to the incubation. A major peak adjacent to cytidine in III and IV is adenine due to SAH degradation. Other minor peaks in III are also result of the SAH nucleosidase.
In contrast to the relaxed specificity with cytosine-containing nucleosides, all uracil-containing substrates including uracil, uridine or thioribouridine were not converted to product (e.g. uridine in Supporting Figure S11). Although this specificity can be explained by the necessity of the lone pair of electrons on N4 to contribute by formation of an imine concomitant with methyl transfer, this result appears inconsistent with the isolation of albomycin δ1 from either the wild-type or ΔabmE mutant strains. However, it was previously reported that albomycin δ1 could be made by mild alkaline hydrolysis of albomycin δ2.27 We repeated this experiment by incubating albomycin δ2 in 50mM Tris-HCl pH 9 for 3 hr at 37°C. LCMS analysis showed that the majority of the starting material was converted into a compounds with a [M+H]+ ion of m/z 1004.7 that agrees with the molecular formula of albomycin δ1 (Supporting Figure S13). Thus, albomycin δ1 is not a biosynthetic intermediate but instead a shunt product that is obtained under conditions in the absence of N-carbamoylation.
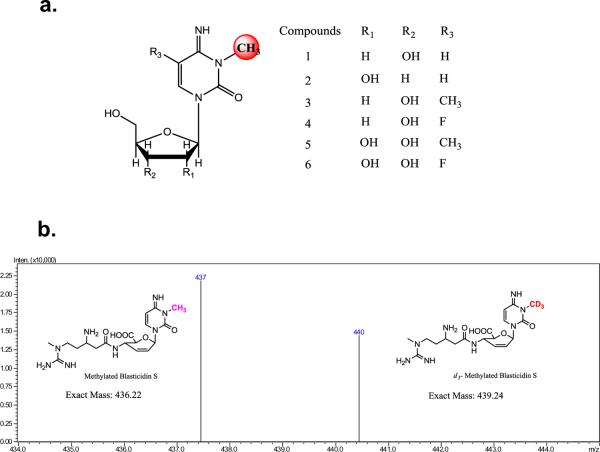
To further investigate the unusual substrate flexibility of AbmI methylation, we tested blasticidin S, the unmodified cytosine-containing congener of blasticidins that are isolated from Streptomyces griseochromogenes. It has been established that blasticidin S-resistant bacteria encode a cytosine deaminase that renders this antibiotic inactive.28 The methylation reaction was analyzed with a LCMS method that showed a new peak eluted immediately after the blasticidin S peak (Supporting Figure S14a). It was absent with boiled AbmI in a control. The new peak had a [M+H]+ ion of m/z 437.0, consistent with the molecular formula of C18H28N8O5 for the monomethylated blasticidin S (Supporting Figure S14b and c). To verify that the new species is indeed methylated blasticidin S, [methyl-d3]-SAM was used as a tracer with unlabeled SAM at a 2:3 ratio. LCMS analysis of the reaction revealed the product contained [M+H]+ ions of m/z 440 and 437 with the expected 2:3 ratio (Figure 6b). The results therefore demonstrate that AbmI can be used to modify other cytosine-containing antibiotics,29 which allows access to structural derivatives such as the monomethylated blasticidin S that we are currently testing for its inhibitory activity as well as for circumvention of deaminase-mediated resistance.
Figure 6.
AbmI specificity toward cytosine-containing compounds in in vitro reaction. (a) Structures of the cytosine-containing nucleosides. The red ball denotes the site of methylation. LCMS data are provided in Supporting Figure S11. (b) Positive ESI-MS analysis of the deuterium labeled methyl blasticidin S product of an AbmI in vitro reaction. Substrates d3-SAM and 1H3-SAM were pre-mixed in a 2:3 ratio. The HPLC profile of the reaction is provided in Supporting Figure S14.
The proposed albomycin biosynthetic pathway
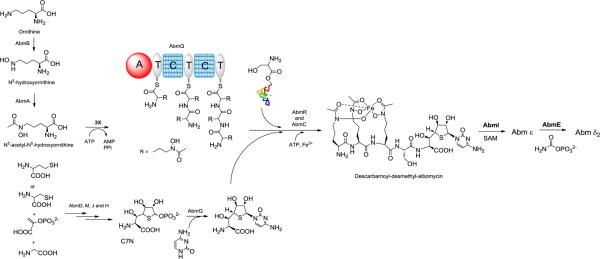
Based on the predicted functions of the proteins encoded by the abm and ctj genes and the cumulative data from this and other studies, we can now propose a biosynthetic pathway for albomycin δ2 (Figure 7). The biosynthesis of the ferrichrome siderophore moiety in albomycins is proposed to require AbmA, B and Q. AbmA, a flavin-dependent monooxygenase catalyzes the N5 hydroxylation of l-ornithine. AbmB, an N-acyltransferase acetylates N5 with acetyl-CoA to afford the N5-acetyl-N5-hydroxy-l-ornithine (AHO) building block that is used by AbmQ, a non-ribosomal peptide synthetase (NRPS) for iterative condensation. AbmQ include one adenylation (A) domain, two condensation (C) domains and three thiolation (T) domains. This domain organization is very similar to that found in Sib1, the Schizosaccharomyces pombe ferrichrome NRPS that utilizes AHO as a substrate.30 The N-terminal A domain of this bacterial ferrichrome NRPS is proposed to load the building blocks onto the three T domains. Interestingly, linear non-ribosomal peptides are typically released by a C-terminal thioesterase domain, but there is no such domain within AbmQ to catalyze this process. A nucleophilic molecule such as an activated serine must be in place to release the siderophore tripeptide anchored on the terminal T domain of AbmQ. We speculate two possible mechanisms for off-loading the tripeptide. The first intriguing mechanism is an off-loading utilizing a seryl-tRNA as a substrate, which is likely supplied in situ by the seryl-tRNA synthetase found in the cluster, AbmK. This mechanism is similar to a few relatively recent studies that have demonstrated tRNAs can serve as adaptors to shuttle in amino acids to antibiotic biosynthesis.31–33 But the activated serine can also possibly be a free seryl-adenylate. In either way, an intermediate in the biosynthetic pathway must be a siderophore-containing tetrapeptide. A second mechanism employs SB-217452 as the co-substrate since it has been isolated via fermentation.7 However, the peptide bonds are labile to proteases during the fermentation and SB-217452 is highly toxic to producing cells. SB-217452 unlikely reaches a micromolar concentration in the cells and may instead be due to degradation of albomycin δ2. Regardless, a protein candidate encoded within the cluster, coupling the siderophore to the Ser residue is not obvious. We hypothesize either off-loading by AbmR, an ABC transporter component of NTPase superfamily that is encoded just downstream of abmQ, or AbmC, which has low sequence similarity to lysyl-tRNA synthetase and other ATP-utilizing enzymes. These two enzymes are also candidates for catalyzing the formation of the remaining amide bond that links the unusual 4′-thioxylofuranosyl amino acid to the Ser residue. We are now analyzing the ΔabmC and ΔabmR mutants for production of ferrichrome-type siderophore and other biosynthetic intermediates, hence facilitating the elucidation of the mechanisms of peptide bond formation.
Figure 7.
The proposed albomycin biosynthetic pathway in Streptomyces sp. ATCC 700974. Three hypothetical pathways leading to the formation of the C7N amino acid are presented in Supporting Figure S15.
Biosynthesis of the unusual amino acid containing the 4′-thioxylofuranosyl cytidine moiety of albomycin is just as intriguing (Supporting Figure S15) and likely requires minimally five enzymes: AbmD, a pyridoxal-5'-phosphate (PLP)-dependent enzyme, AbmF, a hypothetical protein, AbmM, a radical SAM domain protein, AbmJ, a second radical SAM protein, and AbmH, a putative serine hydroxymethyltransferae (SHMT). AbmH may catalyze an aldol-type condensation to yield the final C7N scaffold as shown in Figure 7. This speculation is based on the discovery of SHMT homologs found within the gene cluster of caprazamycin and A-90289 ( 36/48, % identity /% similarity).12,34 These uridyl nucleoside antibiotics also contain a nonproteinogenic amino acid residue that has a C7N scaffold, proposed to be the product of a SHMT, which catalyzes the formation of a C-C bond between the α-carbon of a glycine and the C-5′ of uridine-5′-aldhyde. However, this activity has not been functionally demonstrated and the modest sequence identity may imply a chemically similar but unique function for AbmH. AbmD has homology to a number of enzymes capable of transforming simple sulfur-containing amino acids, including 1-aminocyclopropane-carboxylate deaminase (ACCD),35,36 cystathionine betasynthase (CBS)37 and cysteine desulfhydrase.38 AbmM and J, like other radical SAM family of proteins,39 may harness the chemistry to catalyze a range of reactions such as oxidation, isomerization and sulfur insertion to close the thiofuranosyl ring. Based on these observations and assumptions, we propose three hypothetical pathways that could lead to the formation of the 4′-thioxylofuranose in albomycins (Supporting Figure S15). Elucidation of the thioether group formation in albomycin is very important because substitution of the sulfur for an oxygen resulted in complete loss of the bioactivity of albomycins.4
Following the thiosugar formation, a cytosine base is likely attached to yield a typical N-glycosidic bond (Figure 7). This process would require that the anomeric carbon is activated for attack. We propose this occurs by generation of the sugar-1-phoshate through retention of a phosphate group derived from phosphoenolpyruvate in assembly of the C7N thiosugar scaffold. Interestingly, there is only one gene abmG within the cluster that appears to encode enzymes involved in nucleoside metabolism. AbmG is a homolog of human deoxycytidine kinase (dCK),40 which catalyzes 5′-phophorylation of both purine and pyrimdine nucleosides. Rather surprisingly, however, dCK has significant deoxyribonucleoside phosphorylase activity in phosphate buffer that yields the free base and 2-deoxy-α-d-ribofuranose-1-phosphate as products.40 Thus, AbmG may catalyze the reverse phosphorolysis reaction to yield the unusual cytosine-containing nucleoside, which is feasible given that nucleoside phosphorylases are known to catalyze readily reversible reactions. Finally, either prior to or following attachment to the ferrichrome siderophore—the latter of which is preferred given the isolation of the desmethyl- and descarbamoyl derivatives—the cytosine base is modified by N-methylation catalyzed by AbmI and N-carbamoylation catalyzed by AbmE.
While a majority of Abm proteins have been assigned biosynthetic roles, AbmN, O and P encoded by three consecutive genes may contribute to a second self-resistance mechanism of the albomycin producer in addition to AbmK. Homologous abmN to P are often found in an operon in other sequenced actinomycete genomes. These genes encode proteins with similarity to three components of an ABC transporter system with AbmN being the ATPase, and AbmO and P the permeases. Some AbmP homologs are annotated as multidrug exporters (e.g. YP_001221681). Unlike many other antibiotic biosynthetic gene clusters in streptomycetes, no pathway-specific regulatory protein gene was found in the abm cluster. However, production of albomycins is apparently regulated since the production is initiated in the late growth phase.41 Thus, an involvement of a global regulator or unidentified nucleic acid regulatory elements is probable.
Studies of albomycin biosynthetic enzymes will lead to discoveries of new potential for generating molecular diversity and more potent antibacterial agents. SAM-dependent methyltransferase like AbmI belongs to a few highly diverse families of enzymes that have remarkably flexible catalytic requirements.42 Enzyme catalyzing N3-methylation of cytosine in the nucleosides of nucleic acids versus the N3-methylation of cytosine in albomycins can be such an example. A methyltransferase has been recently identified responsible for 3mC modification of tRNAs in yeast.43,44 A thorough biochemical and structural analysis of these N3-methylpyrimidine methyltransferases is of general interest to gain insights into the reaction mechanisms, specificity and protein evolution. Additionally, nucleoside methyltransferases may be useful in chemo-enzymatic synthesis45 of small molecules46 and modified RNA oligonucleotides, the former of which was shown here with AbmI catalyzed modification of blasticidin S. Novel genes for nucleoside modification are one of many chemical biological facets uncovered in albomycin biosynthesis. Other aspects such as the peptidyl bond formation, glycosidic bond formation, thioether group formation and roles of aminoacyl-tRNA synthetases and tRNAs in secondary metabolism are the subjects in ongoing investigations.
Albomycin δ2 as a natural product antibacterial antibiotic has one of the lowest MICs reported to date. It is effective at clearing mice infected with Yersinia enterocolitica and Streptococcus pneumonia.24 Naturally occurring genetic elements conferring resistance to SB-217452 in human pathogens has yet to be found. One of the largest hurdles toward the development of albomycin is the limited supply of the structurally complex antibiotic. Now that the albomycin biosynthetic genes have been identified and the whole genome sequenced of a producing organism, we envision the implementation of engineering strategies to improve the production of albomycins or biosynthetic intermediates that can be used for semisynthetic purposes.
METHODS
Amplification of sidA homologous gene and screening of a fosmid library
SidA _F2_tag and SidA_R2_tag degenerate primers were used to amplify a 0.3-kb fragment with Streptomyces sp. ATCC 700974 genomic DNA. After being confirmed as homologous to sidA, its upstream ~0.7-kb was amplified with BIO S&T APA genomic walking kit. A re-amplifed 1-kb DNA was used as a probe to screen a fosmid library constructed in pCC1FOS vector with protocols suggested by Epicenter Biotechnology. DNA labeling was performed using instruction of Rediprime II dCTP random prime labeling system following the instruction (GE healthcare). From ~2,000 arrayed fosmid clones, about 60 positive colonies were identified in colony hybridization. 10 cosmids were rigorously mapped with restriction enzymes and totally 4 were sent to Functional Biosciences, Inc. for shot-gun sequencing. Inserts of 3 cosmids (8F8, 5C3, 22H10) were assembled into large contigs with the longest one ~38 kb. 5B9 cosmid apparently harbors a homologous cch cluster. DNA sequence analysis was performed in Vector NTI v.10 with the built-in functions and Frameplot 2.0. GenBank Accession Number for the analyzed region in ATCC 700974 is JN252488.
Targeted gene deletion and genetic complementation in Streptomyces sp. ATCC 700974
All targeted abm genes were inactivated by REDIRECT PCR-targeting strategy used for Streptomyces.21 Primer pairs were designed to amplify the apramycin resistance cassette from pIJ773. In-frame deletions of abm genes were constructed in the cosmids in E. coli and the resultant cosmids were conjugated into ATCC 700974 via E. coli BW12567/pUZ8002. Double-crossover mutants that were apramycin-resistant but chloramphenicol (or kanamycin) sensitive were chosen to be confirmed by PCR the targeted gene were deleted in frame in chromosome (e.g. ΔabmB, I and E in Supporting Figures S2 and S3). Gene oriT was amplified from pIJ773 with primers oriT-EcoRI-F/oriT-SacI-R to clone into plasmid pSE34 (pWHM3)47 to give pSE34-oriT. Genes used for the complementation experiments were cloned into this vector between XbaI and HindIII sites under the control of an ermE* promoter. The resulting plasmids were introduced into Streptomyces sp. ATCC 700974 by conjugation.
Isolation of wild-type and descarbamoyl-desmethyl albomycins
We were able to process 4-L cultures at one time using the method described below. Streptomyces spores/mycelia were used to inoculate a TSB seed culture that was grown for 20–30 hours. 5% inoculums were transferred to albomycin production liquid media in shaking flasks and the strains were fermented an additional 90–100 hours. One liter cell-free broth was passed through a 20cm × 2.6cm column filled with XAD4 or XAD7HP resins (Sigma). The column was washed with 200 ml 3% methanol and albomycin bioactive fraction was eluted with 300 ml 50% methanol. After removal of most of the methanol by a rotovap, the remaining was reverse-extracted with ethyl acetate, retaining the aqueous phase that was subsequently lyophilized. The dried sample was brought up in water to be separated by size exclusion chromatography and/or preparative HPLC. For size exclusion chromatography, a 95 × 2.6 cm column containing BioGel P-2 (Bio-Rad) was packed. The eluent of 25 mM NaCl was delivered by a peristaltic pump on a BioLogic (Bio-Rad) system. Flow rate was 15 ml/hr. Elution was collected by a fraction collector in 5 ml/tube. For preparative HPLC, a YMC CombiPrep ODS-A 5μm 20×50mm column (Waters) was used. The mobile phase was developed using 0.1% TFA in water (A) and methanol (B) at a flow rate of 8 ml/min with a linear increase between the times: 0–10 min, 0–40% B; 10–15 min, 40– 95% B. Fractions were collected manually and tested for bioactivities. The active fractions were dried and prepared to be further separated with a Phenomenex Sphereclone 10μ ODS (2) 250 × 4.6mm HPLC column. The mobile phase was water with 0.1% TFA (A) and acetonitrile (ACN) with 0.1% TFA (B). Flow rate 1.5 ml/min. The gradient was a linear increase between the times: 0 – 15 min, 0 – 35% B; 15 –18 min, 15 – 85% B. Fractions were collected by a fraction collector and assayed for bioactivities. At this point, the wild-type albomycin fraction was minimally 80% pure. Two additional HPLC purification steps were used for the albomycin analogs that were produced in low quantities. First, a hydrophilic interaction HPLC column polyHYDROXYETHYL Aspartamide 100 × 4.6 mm 100Å manufactured by PolyLC, Inc. was used. Dried sample was dissolved in 65% ACN and loaded into the column. A chromatogram was developed with an isocratic flow of 70% ACN and 30% 20 mM NH4Ac (flow rate 0.7 ml/min). Wild type albomycins were eluted at ~ 5 min and the descarbamoyl-desmethyl-albomycin were eluted at a time between7 and 9.5 min as detected by bioactivities. The bioactive fractions can be combined and dried for further separation on a Phenomenex Synergi 4μ Fusion-RP 80Å 150 × 4.6 mm column. The mobile phase consisted of water with 0.1% TFA (A) and ACN with 0.1% TFA (B). Flow rate 1.5 ml/min. A series of linear gradients was developed in the following manner: 0–4 min, 0–12% B; 4–24 min, 12–27% B; 24–26 min, 27–95% B. Fractions were collected by a fraction collector and assayed for bioactivities. It was estimated the best descarbamoyl-desmethyl-albomycin fraction had around 80% purity starting from a 4 liter culture. All the HPLC steps were performed with a Shimadzu Prominence system including a photodiode array detector (SPD-M20A) and a fraction collector (FRC-10A). Albomycins have absorbance maxima at 270 nm, 306 nm and 430 nm.
AbmI in vitro biochemical assays
The assay mixture for an optimized AbmI reaction consisted of 50mM NaH2PO4 pH 6.8, 3 mM SAM, 1 μM S-adenosylhomocysteine (AdoHcy) nucleosidase (G-Biosciences, St. Louis, MO), 100 – 4000 μM nucleoside substrate and 500 nM-50 μM AbmI. The reaction was incubated at 30°C for a varied time from 30 min to overnight. The reactions samples were analyzed with HPLC using the conditions as in the preparation of descarbamoyl-desmethyl-albomycin but the time program was changed to: 0–8 min, 0% B; 8–15 min, 0–5% B; 15 to 17 min, 5–85% B. For inline LC/MS analysis of the two enzyme reactions, the LC conditions were not changed. Electrospray ionization mass spectroscopy (ESI-MS) was performed on Shimadzu LCMS-2010EV that has a single stage quadrupole mass spectrometer which was set to analyze the compounds in positive mode in this work.
Methylation of blasticidin S
Blasticidin S (MP Biomedicals) was added into a 20-μl AbmI reaction at 1 mM final concentration with 3 mM SAM, 1 μM S-AdoHcy nucleosidase and 5 μM enzyme in 50mM NaH2PO4 pH 6.8 buffer. The control reaction was identical to the AbmI reaction except that prior to adding other components, the enzyme was mixed with ~15 μl water and boiled at 98°C for 10 min. The two samples were incubated at 30°C overnight and then analyzed with HPLC. The two elution solvents, water with 0.1% TFA (A) and ACN with 0.1% TFA (B) were mixed in a linear gradient between the times: 0–3 min, 0% B; 3–7 min, 0–15% B; 7–12 min, 15–30%; 12–14 min, 30–85%. For stable isotope labeling of the methylated product, S-Adenosyl-l-methionine-d3 (S-methyl-d3) (C/D/N Isotopes) was pre-mixed with 1H3-SAM in a 2:3 ratio and added to the AbmI reaction at 1.5 mM final concentration. The reaction was incubated at 30°C for 4 hours.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from American Heart Association (Beginning Grant-in-Aid 09BGIA2070029 to S.C.), American Cancer Society (IRG 85-001-22 to S. V. L.) and the National Institute of Health (AI087849 to S. V. L.). S.C. would also like to thank the research groups of Drs. Stephen Bergmeier and Hao Chen at Ohio University, Dr. Ying Huang at the Institute of Microbiology of CAS and Ohio University Genomics Facility.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Braznikova G. Iron-containing antibiotic produced by Actinomyces subtropicus. Nov. Med. 1951;23:3. [Google Scholar]
- (2).Maehr H, Berger J. The production, isolation and characterization of a grisein-like sideromycin complex. Biotechnology and Bioengineering XI. 1969:1111–1123. doi: 10.1002/bit.260110608. [DOI] [PubMed] [Google Scholar]
- (3).Garrod LP. Correspondence. British Medical Journal. 1956:1362. [Google Scholar]
- (4).Paulsen H, Brieden M, Benz G. Synthese des Sauerstofanalogs der Desferriforn von δ1-Albomycine. Liebigs Ann Chem. 1987:565–575. [Google Scholar]
- (5).Gause GF. Recent studies on albomycin, a new antibiotic. Brit. Med. J. 1955;12:1177–1179. doi: 10.1136/bmj.2.4949.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Stapley EO, Ormond RE. Similarity of Albomycin and Grisein. Science. 1957;125:587–589. doi: 10.1126/science.125.3248.587. [DOI] [PubMed] [Google Scholar]
- (7).Stefanska AL, Fulston M, Houge-Frydrych CSV, Jones JJ, Warr SR. A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. Journal of Antibiotics. 2000;53:1346–1353. doi: 10.7164/antibiotics.53.1346. [DOI] [PubMed] [Google Scholar]
- (8).Braun V, Pramanik A, Gwinner T, Koberle M, Bohn E. Sideromycins: tools and antibiotics. BioMetals. 2009;22:3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pramanik A, Braun V. Albomycin Uptake via a Ferric Hydroxamate Transport System of Streptococcus pneumoniae R6. J. Bacteriol. 2006;188:3878–3886. doi: 10.1128/JB.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Walsh CT, Zhang W. Chemical Logic and Enzymatic Machinery for Biological Assembly of Peptidyl Nucleoside Antibiotics. ACS Chemical Biology. 2011;6:1000–1007. doi: 10.1021/cb200284p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Funabashi M, Yang Z, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Chi X, van Lanen SG. An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat Chem Biol. 2010;6:581–586. doi: 10.1038/nchembio.393. [DOI] [PubMed] [Google Scholar]
- (12).Kaysser L, Lutsch L, Siebenberg S, Wemakor E, Kammerer B, Gust B. Identification and Manipulation of the Caprazamycin Gene Cluster Lead to New Simplified Liponucleoside Antibiotics and Give Insights into the Biosynthetic Pathway. Journal of Biological Chemistry. 2009;284:14987–14996. doi: 10.1074/jbc.M901258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rackham EJ, Grüschow S, Ragab AE, Dickens S, Goss RJM. Pacidamycin Biosynthesis: Identification and Heterologous Expression of the First Uridyl Peptide Antibiotic Gene Cluster. ChemBioChem. 2010;11:1700–1709. doi: 10.1002/cbic.201000200. [DOI] [PubMed] [Google Scholar]
- (14).McCarty RM, Bandarian V. Deciphering Deazapurine Biosynthesis: Pathway for Pyrrolopyrimidine Nucleosides Toyocamycin and Sangivamycin. Chemistry & biology. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li L, Xu Z, Xu X, Wu J, Zhang Y, He X, Zabriskie TM, Deng Z. The Mildiomycin Biosynthesis: Initial Steps for Sequential Generation of 5-Hydroxymethylcytidine 5′-Monophosphate and 5-Hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. ChemBioChem. 2008;9:1286–1294. doi: 10.1002/cbic.200800008. [DOI] [PubMed] [Google Scholar]
- (16).Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM. The Blasticidin S Biosynthesis Gene Cluster from Streptomyces griseochromogenes: Sequence Analysis, Organization, and Initial Characterization. ChemBioChem. 2003;4:821–828. doi: 10.1002/cbic.200300583. [DOI] [PubMed] [Google Scholar]
- (17).Zhang G, Zhang H, Li S, Xiao J, Zhang G, Zhu Y, Niu S, Ju J, Zhang C. Characterization of the Amicetin Biosynthesis Gene Cluster from Streptomyces vinaceusdrappus NRRL 2363 Implicates Two Alternative Strategies for Amide Bond Formation. Applied and Environmental Microbiology. 2012;78:2393–2401. doi: 10.1128/AEM.07185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zeng Y, Roy H, Patil PB, Ibba M, Chen S. Characterization of Two Seryl-tRNA Synthetases in Albomycin-Producing Streptomyces sp. Strain ATCC 700974. Antimicrob. Agents Chemother. 2009;53:4619–4627. doi: 10.1128/AAC.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Eisendle M, Oberegger H, Zadra I, Haas H. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N5- monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Molecular Microbiology. 2003;49:359–375. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- (20).Barona-Gomez F, Lautru S, Francou F-X, Leblond P, Pernodet J-L, Challis GL. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology. 2006;152:3355–3366. doi: 10.1099/mic.0.29161-0. [DOI] [PubMed] [Google Scholar]
- (21).Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proceedings of the National Academy of Sciences. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang W, Ostash B, Walsh CT. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proceedings of the National Academy of Sciences. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Baltz RH. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot. 2010;63:506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- (24).Pramanik A, Stroeher UH, Krejci J, Standish AJ, Bohn E, Paton JC, Autenrieth IB, Braun V. Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcus pneumoniae. International Journal of Medical Microbiology. 2007;297:459–469. doi: 10.1016/j.ijmm.2007.03.002. [DOI] [PubMed] [Google Scholar]
- (25).Baltz RH. Antimicrobials from Actinomycetes: Back to the Future. Microbe. 2007;2:125–131. [Google Scholar]
- (26).Lee J-H, Bae B, Kuemin M, Circello BT, Metcalf WW, Nair SK, van der Donk WA. Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proceedings of the National Academy of Sciences. 2010;107:17557–17562. doi: 10.1073/pnas.1006848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Benz G, Schröder T, Kurz J, Wünsche C, Karl W, Steffens G, Pfitzner J, Schmidt D. Constitution of the Deferri form of the Albomycins δ1, δ2 and ε. Angewandte Chemie International Edition in English. 1982;21:527–528. [Google Scholar]
- (28).Nawa K, Tanaka T, Kamakura T, Yamaguchi I, Endo T. Inactivation of blasticidin S by Bacillus cereus. VI. Structure and comparison of the bsr gene from a blasticidin S-resistant Bacillus cereus. Biol. Pharm. Bull. 1998;21:983–988. doi: 10.1248/bpb.21.893. [DOI] [PubMed] [Google Scholar]
- (29).Isono K. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J. Antibiot (Tokyo) 1988;41:1711–1739. doi: 10.7164/antibiotics.41.1711. [DOI] [PubMed] [Google Scholar]
- (30).Schwecke T, Göttling K, Durek P, Dueñas I, Käufer NF, Zock-Emmenthal S, Staub E, Neuhof T, Dieckmann R, von Döhren H. Nonribosomal Peptide Synthesis in Schizosaccharomyces pombe and the Architectures of Ferrichrome-Type Siderophore Synthetases in Fungi. ChemBioChem. 2006;7:612–622. doi: 10.1002/cbic.200500301. [DOI] [PubMed] [Google Scholar]
- (31).Zhang W, Ntai I, Kelleher NL, Walsh CT. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proceedings of the National Academy of Sciences. 2011;108:12249–12253. doi: 10.1073/pnas.1109539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: Elucidation of the role of seryl-tRNA. Proceedings of the National Academy of Sciences. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gondry M, Sauguet L, Belin P, Thai R, Amouroux R, Tellier C, Tuphile K, Jacquet M, Braud S, Courcon M, Masson C, Dubois S, Lautru S, Lecoq A, Hashimoto S.-i., Genet R, Pernodet J-L. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- (34).Yang Z, Chi X, Funabashi M, Baba S, Nonaka K, Pahari P, Unrine J, Jacobsen JM, Elliott GI, Rohr J, Van Lanen SG. Characterization of LipL as a Non-heme, Fe(II)-dependent α-Ketoglutarate:UMP Dioxygenase That Generates Uridine-5'-aldehyde during A-90289 Biosynthesis. Journal of Biological Chemistry. 2011;286:7885–7892. doi: 10.1074/jbc.M110.203562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Thibodeaux CJ, Liu H.-w. Mechanistic Studies of 1-Aminocyclopropane-1-carboxylate Deaminase: Characterization of an Unusual Pyridoxal 5'-Phosphate-Dependent Reaction. Biochemistry. 2011;50:1950–1962. doi: 10.1021/bi101927s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Walsh C, Pascal RA, Johnston M, Raines R, Dikshit D, Krantz A, Honma M. Mechanistic studies on the pyridoxal phosphate enzyme 1-aminocyclopropane-1-carboxylate deaminase from Pseudomonas sp. Biochemistry. 1981;20:7509–7519. doi: 10.1021/bi00529a028. [DOI] [PubMed] [Google Scholar]
- (37).Aitken SM, Kirsch JF. The enzymology of cystathionine biosynthesis: strategies for the control of substrate and reaction specificity. Archives of Biochemistry and Biophysics. 2005;433:166–175. doi: 10.1016/j.abb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- (38).Soutourina J, Blanquet S, Plateau P. Role of d-Cysteine Desulfhydrase in the Adaptation of Escherichia coli to d-Cysteine. Journal of Biological Chemistry. 2001;276:40864–40872. doi: 10.1074/jbc.M102375200. [DOI] [PubMed] [Google Scholar]
- (39).Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- (40).Usova E, Maltseva T, Földesi A, Chattopadhayaya J, Eriksson S. Human Deoxycytidine Kinase as a Deoxyribonucleoside Phosphorylase. Journal of Molecular Biology. 2004;344:1347–1358. doi: 10.1016/j.jmb.2004.10.016. [DOI] [PubMed] [Google Scholar]
- (41).Fiedler HP, Walz F, Dohle A, Zahner H. Albomycin: Studies on fermentation, isolation and quantitative determination. Applied Microbiology and Biotechnology. 1985;21:341–347. [Google Scholar]
- (42).Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends in biochemical sciences. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Noma A, Yi S, Katoh T, Takai Y, Suzuki T, Suzuki T. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA. 2011;17:1111–1119. doi: 10.1261/rna.2653411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).D'Silva S, Haider SJ, Phizicky EM. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA. 2011;17:1100–1110. doi: 10.1261/rna.2652611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lee BWK, Sun HG, Zang T, Kim BJ, Alfaro JF, Zhou ZS. Enzyme-Catalyzed Transfer of a Ketone Group from an S-Adenosylmethionine Analogue: A Tool for the Functional Analysis of Methyltransferases. Journal of the American Chemical Society. 2010;132:3642–3643. doi: 10.1021/ja908995p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang C, Weller RL, Thorson JS, Rajski SR. Natural Product Diversification Using a Non-natural Cofactor Analogue of S-Adenosyl-l-methionine. Journal of the American Chemical Society. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- (47).Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J. Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.