Abstract
The recent spate of discoveries of novel acyl-CoA mutases has engendered a growing appreciation for the diversity of 5′-deoxyadenosylcobalamin-dependent rearrangement reactions. The prototype of the reaction catalyzed by these enzymes is the 1,2 interchange of a hydrogen atom with a thioester group leading to a change in the degree of carbon skeleton branching. These enzymes are predicted to share common architectural elements: a Rossman fold and a TIM barrel domain for binding cofactor and substrate, respectively. Within this family, methylmalonyl-CoA mutase (MCM) is the best studied and is the only member found in organisms ranging from bacteria to man. MCM interconverts (2R)-methylmalonyl-CoA and succinyl-CoA. The more recently discovered family members include isobutyryl-CoA mutase (ICM), which interconverts isobutyryl-CoA and n-butyryl-CoA; ethylmalonyl-CoA mutase, which interconverts (2R)-ethylmalonyl-CoA and (2S)-methylsuccinyl-CoA, and 2-hydroxyisobutyryl-CoA mutase, which interconverts 2-hydroxyisobutyryl-CoA and (3S)-hydroxybutyryl-CoA. A variant in which the two subunits of ICM are fused to a G-protein chaperone, IcmF, has been described recently. In addition to its ICM activity, IcmF also catalyzes the interconversion of isovaleryl-CoA and pivalyl-CoA. This review focuses on the involvement of acyl-CoA mutases in central carbon and secondary bacterial metabolism and on their biotechnological potential for applications ranging from bioremediation to stereospecific synthesis of C2-C5 carboxylic acids and alcohols, and for production of potential commodity and specialty chemicals.
Novel B12-dependent Acyl-CoA Mutases and their Biotechnological Potential
AdoCbl (or 5'-deoxyadenosylcobalamin), a derivative of cobalamin, is used as a cofactor by enzymes that catalyze chemically challenging isomerization reactions using radical mechanisms (Fig. 1) (1–4). The cofactor’s distinguishing feature is its cobalt-carbon bond, which with a bond dissociation energy of ~31 kcal/mol (5), is key to its reactivity and its role as a radical reservoir (6). B12 is generally used to refer to the cobalamin cofactor in which the upper axial ligand and cobalt oxidation state are not specified. A generalized scheme of the overall reaction catalyzed by AdoCbl-dependent isomerases, which is reversible, is shown in Fig. 1 and begins with homolysis of the cobalt-carbon bond upon substrate binding, yielding the 5′-deoxyadenosyl (5′-dAdo•) and cob(II)alamin radicals. The 5′-dAdo• radical is the working radical and initiates the isomerization reaction by abstracting a hydrogen atom from the substrate, thereby propagating the carbon-centered radical from the cofactor to the substrate. Intramolecular migration of a variable group from a vicinal carbon generates a product-centered radical. The latter re-abstracts a hydrogen atom from 5′-deoxyadenosine generating the cofactor-derived radical pair, which recombines to complete a catalytic cycle. The carbon-centered radical intermediates formed during catalytic turnover are highly reactive and also susceptible to oxidative inactivation. Hence, it is interesting that AdoCbl-dependent enzymes are widely distributed in aerobic organisms in addition to being found in anaerobic ones (3).
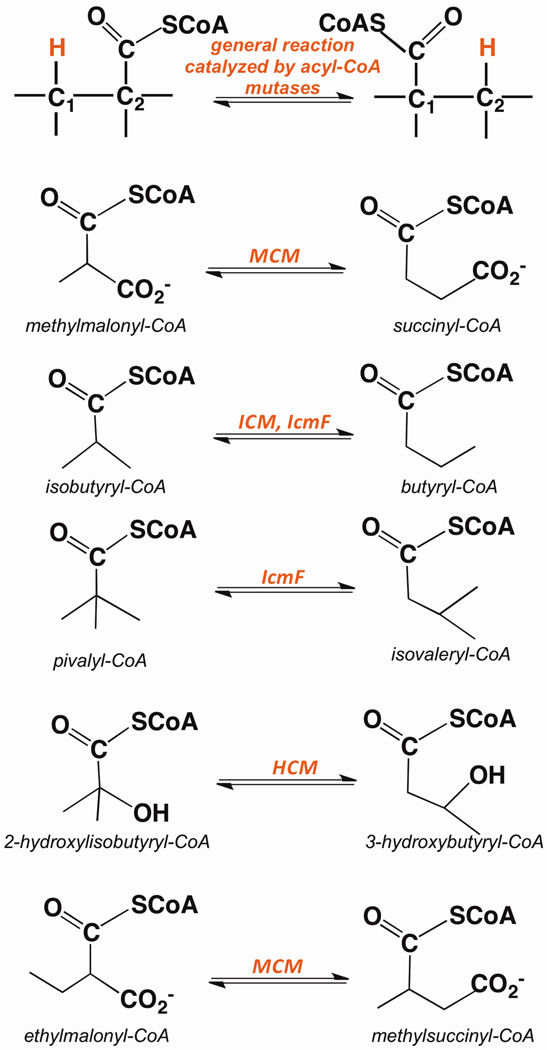
Figure 1.
Reactions catalyzed by acyl-CoA mutases. A generalized 1,2-rearrangement reaction catalyzed by AdoCbl-dependent mutases is shown in the top row followed by reactions catalyzed by MCM, ICM, IcmF, HCM and ECM.
Within the family of AdoCbl-dependent isomerases, two subfamilies can be distinguished: those that catalyze the migration of a carbon skeleton versus those that catalyze the migration of a heteroatom (e.g. an amino or hydroxyl group) (2, 7). Well-characterized members of the former subfamily include MCM (methylmalonyl-CoA mutase) and glutamate mutase (2) and of the latter, diol dehydratase and ethanolamine-ammonia lyase (8). The carbon-skeleton mutases in which an acyl-CoA group undergoes intramolecular migration, is the fastest growing group of AdoCbl-dependent mutases. A number of recently discovered AdoCbl-dependent enzymes belong to this subfamily (9–11) and are the subject of this article. Not included in this review are recently described putative AdoCbl-dependent isomerases involved in the anaerobic degradation of indoleacetate (12) and n-alkanes (13).
Organization of Acyl-CoA Mutases
A variety of subunit and oligomeric organizations are seen with acyl-CoA mutases (Fig. 2). MCM is the best characterized of the acyl-CoA mutases and is primarily organized either as an α2 homodimer (14) or an αβ heterodimer (15), in which the α chain houses the determinants for cofactor and substrate binding. In the Propionibacterium shermanii MCM, which has been characterized extensively, the β subunit displays sequence similarity to the α subunit but does not bind either substrate or cofactor (15). Hence, αβ MCMs contain a single active site per heterodimer. In contrast, human MCM, an α2 dimer, houses two active sites per homodimeric unit (14). The α subunit consists of an N-terminal triose phosphate isomerase (TIM) barrel constituting the substrate-binding domain and a C-terminal Rossman fold constituting the AdoCbl-binding domain (4). The two domains, connected by a linker, are juxtaposed such that the cofactor is sandwiched between them and sequestered from solvent (Fig. 3A) (15). Although the structures of other acyl-CoA mutases are not yet available, they are expected to share MCM’s architectural organization.
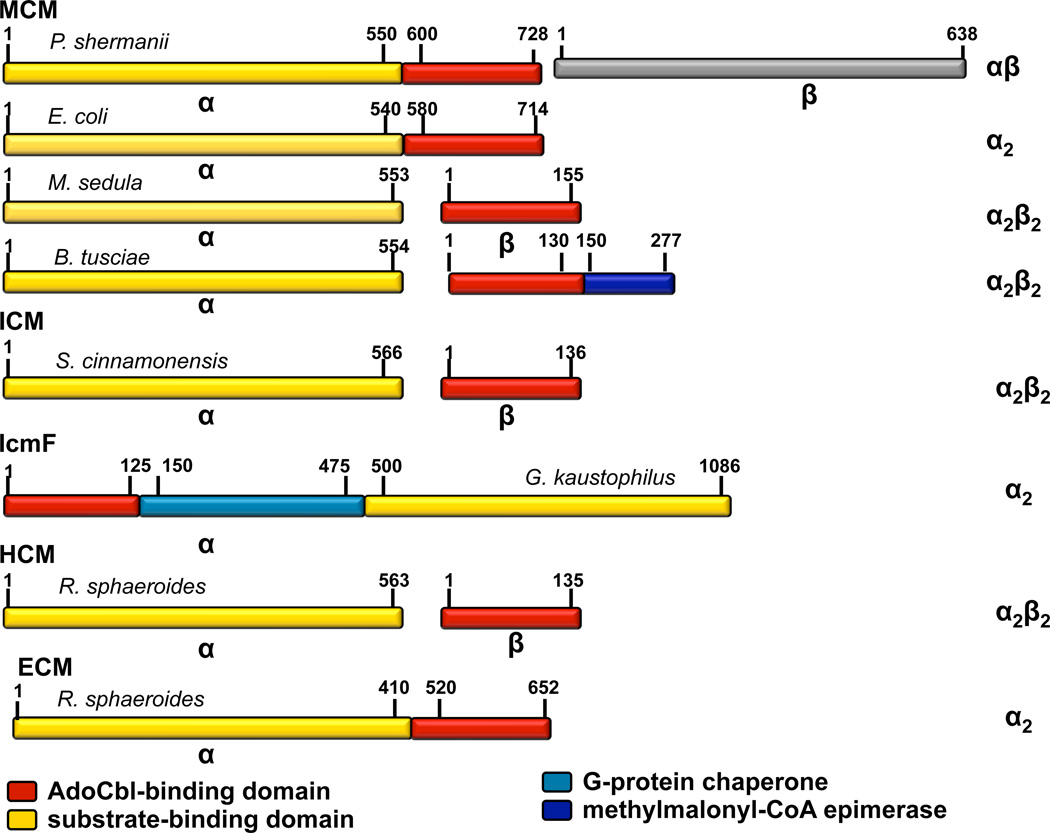
Figure 2.
Domain organization of acyl-CoA mutases. The domains/subunits are color-coded as follows: AdoCbl-binding domain (red), substrate-binding domain (yellow), methylmalonyl-CoA epimerase (navy) and G-protein chaperone (cyan). The oligomeric state of the proteins is shown on the right. The following proteins were used as examples: MCMs (Propionibacterium shermanii (αβ)(P11653, P11652), E. coli (α2)(AAA69084), Metallosphaera sedula (α2β2)(Msed_0638, Msed_2055) and Bacillus tusciae (α2β2)(YP_003589181)); ICM (Streptomyces cinnamonensis (α2β2)(AAC08713, CAB59633)), IcmF (Geobacillus kaustophilus (α2)(YP_149244)), HCM (Rhodobacter sphaeroides (α2β2)(Rsph17029_3657, Rsph17029_3654)), and ECM (Rhodobacter sphaeroides (α2)(ACJ71670)).
Figure 3.
Active site of MCM and corresponding amino acids substitutions in acyl-CoA mutases. (A) MCM from P. shermanii is a αβ heterodimer (PDB file: 4REQ) and (B) close-up of active site of MCM, where two key active site residues (green) important for catalysis, are shown in stick representation.
A variant arrangement has been reported for MCM from Metallosphaera sedula that exhibits a split organization (Fig. 2) with one subunit being homologous to the substrate-binding domain and the second, to the AdoCbl-binding domain in the α-subunit of the canonical MCM (16–18). It is predicted to have an α2β2 heterotetrameric organization in analogy to other members of the acyl-CoA family in which the cofactor and substrate binding domains are encoded by separate subunits. Such an organization might also exist in other MCMs involved in the dicarboxylate-4-hydroxybutyrate pathway for CO2 assimilation found in members of the Archaeal orders Thermoproteales and Desulfurococcales (18), where the enzyme operates in the reverse direction (with respect to its use in mammals) converting succinyl-CoA to methylmalonyl-CoA. However, not all annotated mutases with this split organization in Archaea necessarily encode MCMs and a careful analysis of amino acid identities at key active site locations and enzyme activity determination are needed to make the correct identification (9). Another variant of the α2β2 organization in which the β subunit is fused to methylmalonyl-CoA epimerase is predicted in Bacillus tusciae (Fig. 2).
A split subunit organization is also seen in ICM and in HCM (Fig. 2) where subunit A (IcmA and HcmA, respectively) binds substrate and subunit B (IcmB and HcmB, respectively) binds AdoCbl (9, 19). In IcmF, the IcmA and B genes are fused to a third gene encoding the G-protein chaperone, MeaI (9). Hence, a single polypeptide houses the mutase active site and the GTPase active site, which gates loading of the AdoCbl cofactor to the apoenzyme (20). The native IcmF protein has an α2 oligomeric organization (11). Interestingly, in two bacteria (Saccharomonospora azurea and Niabella soli) only the B12-binding and G-protein chaperone domains are fused, whereas the substrate binding domains of ICM are found separately. In “stand-alone” mutases like MCM, a separate gene encodes the G-protein chaperone, MeaB, which associates tightly with the mutase (21, 22). Genes encoding putative G-protein chaperones are sometimes found adjacent to genes encoding the small AdoCbl-binding subunits of acyl-CoA mutases and await characterization. MeaH, an ortholog of MeaB is found in operons encoding HCM, and is postulated to have a function similar to MeaB (9). ECM, like IcmF and some MCMs, exists as an α2 homodimer (10). Based on our analysis of bacterial genomes, ECMs are not found adjacent to G-protein chaperones in contrast to bacterial α2 MCMs, in which MCM and MeaB are often encoded by the same operon.
Active Site Architecture of Acyl-CoA Mutases
The substrate-binding TIM barrel in MCM is composed of a core of 8 parallel (αβ)-repeats (15). The AdoCbl-binding Rossman domain in MCM (Fig. 3B) has an architecture that is very similar to the B12-binding domains in methionine synthase (23) and glutamate mutase (24). Cobalamin is bound to all these enzymes in the “base-off/His-on” conformation, in which the side chain of the histidine in a conserved DXHXXG motif, serves as the axial ligand to cobalt while the endogenous dimethylbenzimidazole base appended from the tetrapyrrolic ring, is displaced in a side pocket.
In MCM, the cofactor is bound at the interface between the substrate-and AdoCbl-binding domains (Fig. 3B) (15). The CoA tail of the substrate pierces through the TIM barrel positioning the “business” end of the substrate proximal to the 5′-deoxyadenosine moiety of the cofactor in preparation for catalysis (25). Two active site residues, Tyr89 and Arg207 (P. shermanii MCM numbering) are within hydrogen bonding distance to the carboxylate of the substrate and may play a role in orienting the substrate for initiation of radical-based chemistry. Tyr89 appears to play an additional role in labilizing the cobalt-carbon bond, wedging into the space occupied by the 5′-deoxyadenosine moiety of AdoCbl upon substrate binding (26).
Evolutionarily, the structural differences between the substrates for acyl-CoA mutases appear to have been accommodated by subtle modifications in a limited number of active site residues (Fig. 3B) (9, 11, 27). In both ICM and its variant, IcmF, Tyr→Phe and Arg→Gln substitutions accommodate the switch from a carboxylate in the substrate for MCM to a methyl group in the substrate for ICM and IcmF. In HCM, the analogous Tyr→Ile substitution accommodates the bulkier hydroxylated acyl-CoA substrate while in ECM, the Tyr and Arg residues found in MCM are retained. The conservation of the Tyr and Arg residues in MCM and ECM can be rationalized by the presence of a carboxylate moiety in the substrates for both these enzymes (10) (Fig. 1). The additional carbon in the substrate for ECM appears to be accommodated by sequence changes elsewhere. Based on sequence alignments, the His328→Gly and Asn366→Pro substitutions in the active sites of ECM (P. shermanii MCM numbering) are suggested to be important determinants of substrate specificity and for accommodating the bulkier ECM substrate (10). A structural perspective of the reaction mechanism of MCM, the prototype for the acyl-CoA mutases, has been reviewed recently (4) and is not discussed in this article.
An attempt to switch the substrate specificity of MCM to that of ICM was made by substituting Y89 and R207 with phenylalanine and glutamine respectively (27). Although this double mutant was able to bind n-butyryl-CoA, unlike wild-type MCM, it inactivated in the presence of n-butyryl-CoA in a suicidal internal electron transfer side reaction. These results point to additional roles played by key active site residues in guiding reactions along the reaction coordinate and suppressing unwanted side reactions.
Metabolic Roles and the Biotechnological Potential of Acyl-CoA Mutases
(i) MCM
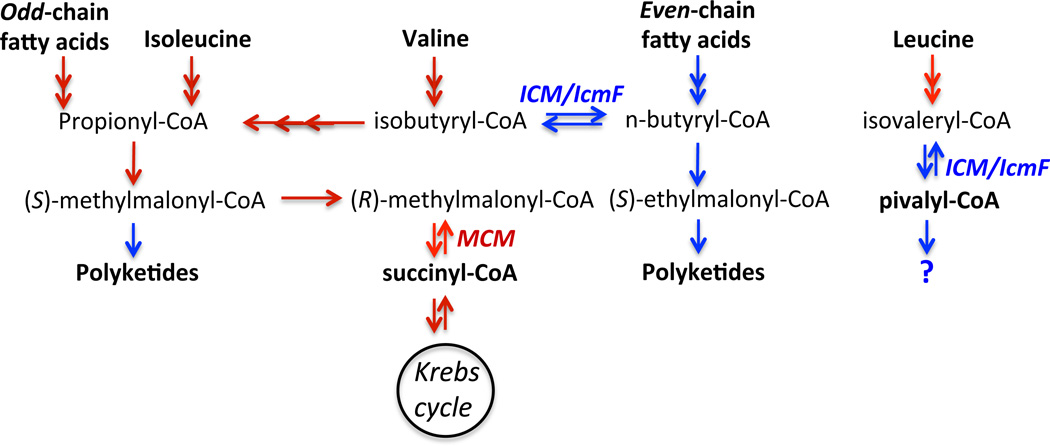
Of the known AdoCbl-dependent enzymes, MCM has the broadest distribution being found in organisms ranging from bacteria to humans (28). The physiological context of MCM is equally varied (Fig 4). In mammals, it is involved in catabolism of odd-chain fatty acids, cholesterol, thymine and the amino acids, valine, methionine, isoleucine and threonine. In humans, defects in MCM result in methylmalonic aciduria, an inborn error of metabolism inherited in an autosomal recessive fashion (29). In some microbes e.g. Streptomyces ssp, methylmalonyl-CoA extender units are used in the biosynthesis of polyketides such as the immunosuppressant, rapamycin, the insecticide, nanchangmycin (30), and in Saccharopolyspora erythraea, for the synthesis of the antibiotic, erythromycin. Methylmalonyl-CoA can be derived metabolically either via carboxylation of propionyl-CoA or by isomerization of succinyl-CoA, with the relative contributions of each route being dependent on the bacterial strain and on the fermentation conditions (Fig. 4). The heterologous production of complex polyketides in Escherichia coli can be limited by the availability of (S)-methylmalonyl-CoA. Hence, engineering E. coli to co-express methylmalonyl-CoA epimerase and MCM resulted in overproduction of methylmalonyl-CoA, which is derived from the Krebs cycle intermediate, succinyl-CoA. The epimerase interconverts the (S)- and (R)-isomers of methylmalonyl-CoA. Hence, under these conditions, MCM operates in the “reverse” direction (31). In a parallel strategy, duplication of the operon harboring the MCM and MeaB genes in addition to a regulatory gene, resulted in a 50% increase in erythromycin production in S. erythraea grown in a soybean oil-based medium (32). Under these growth conditions, MCM also operates in the reverse direction and methylmalonyl-CoA is primarily derived from succinyl-CoA in S. erythraea. However, when grown on soluble starch carbohydrate-based medium, the S. erythraea MCM operates in the direction from methylmalonyl-CoA to succinyl-CoA (33). In contrast, in Streptomyces hygroscopicus, the propionyl-CoA carboxylase pathway is the primary source of methylmalonyl-CoA (34). Hence, supplementing cultures overproducing the propionyl-CoA carboxylase and propionyl-CoA synthetase genes with the precursor, propionate enhances rapamycin production (34).
Figure 4.
Pathways in which MCM and ICM/IcmF participate. Reactions shown in red are found in both bacteria and higher organisms while those in blue have been found only in bacteria.
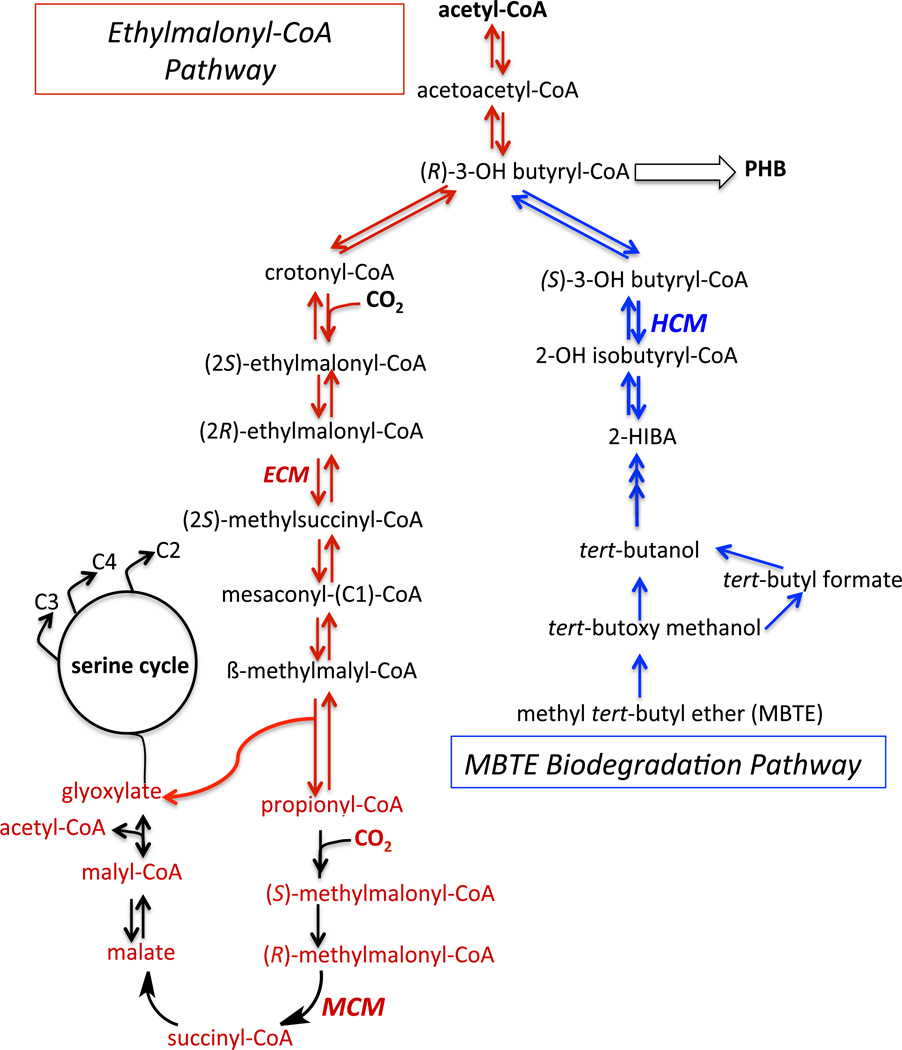
In the facultative methylotroph, Methylobacterium extorquens, MCM is a component of the glyoxylate regeneration pathway, which supports the serine cycle (Fig. 5, black arrows). The latter uses glyoxylate to assimilate formaldehyde derived from methanol oxidation and furnishes C2 (acetyl-CoA), C3 and C4 units for cell biosynthesis (35). Assimilation of acetyl-CoA occurs via the ethylmalonyl-CoA pathway (described in greater detail below), which utilizes the B12-dependent enzyme, ECM. Since intermediates are drained from the serine cycle to support cellular synthetic reactions, glyoxyate must be regenerated for the continued operation of this cycle. Glyoxylate regeneration is a component of acetyl-CoA assimilation by the ethylmalonyl-CoA pathway and results in the successive transformation of propionyl-CoA to methylmalonyl-CoA, succinyl-CoA, malate and malyl-CoA, respectively (35). The ability of M. extorquens to grow on methanol, which could be a major alternative carbon source in the future, has made it a target of considerable biotechnological interest. The exploitation of this organism as a biotechnological platform can be realized by understanding how its central carbon metabolism is wired and regulated and by identifying the bottlenecks that restrict flux. B12-dependent enzymes play an unusual role in M. extorquens in that they support primary carbon metabolism.
Figure 5.
Pathways involving ECM, MCM and HCM. Reactions shown in red represent the ethylmalonyl-CoA pathway for acetyl-CoA assimilation while those in black represent reactions within this pathway that lead to glyoxylate regeneration that supports the serine cycle. The pathway for degradation of MBTE is shown in blue.
(ii) ICM and IcmF
ICM was originally characterized in members of the Streptomyces genus where it is involved in the production of methylmalonyl-CoA for monensin synthesis. In this pathway, ICM generates isobutyryl-CoA, which is subsequently oxidized via methacrylyl-CoA, β-hydroxyisobutyryl-CoA and methylmalonyl-CoA semialdehyde to methylmalonyl-CoA (36–38). Metabolic labeling studies also implicate a role for the rearrangement of n- to iso-butyrate in other polyketide antibiotics including tylosin and narasin (39).
We have recently identified an ICM variant, IcmF (11), which was previously mis-annotated as MCM (21). Based on genomic sequences >220 IcmFs can be identified in different lineages of bacteria. A surprising source of IcmF is the sponge, Amphimedon queenslandica. During genome sequencing of this sponge, a dominant α proteobacterial symbiont was recovered begging the question as to whether IcmF is derived from the proteobacterial contaminant in the sample (40). The closest ortholog of the “sponge IcmF” with 65% amino acid identity is a protein from the proteobacterium, Cupriavidus taiwanensis.
ICM-like activity has been predicted in several sulfate-reducing bacteria based on NMR studies demonstrating isomerization of n-butyrate to isobutyrate (41, 42). The genome of one of these bacteria, Desulfarculis baarsii has been sequenced recently (43) and harbors two acyl-CoA mutases: (i) IcmF and (ii) and MCM. In the latter, the AdoCbl-binding and substrate-binding domains are encoded separately. The genomic data strongly suggests that in D. baarsii, IcmF is an active isobutyryl-CoA mutase.
We have recently demonstrated that purified IcmF can catalyze an additional reaction, i.e. the interconversion of isovaleryl-CoA and pivalyl-CoA (44) (Fig. 1). This is surprising since most acyl-CoA mutases exhibit fairly stringent substrate specificity. Several bacterial strains have been isolated that can mineralize pivalic acid (2,2-dimethylpropionic acid) (45). Although these strains, designated PIV strains, were not fully sequenced, their phylogenetic relationship to other bacteria was inferred based on 16S rRNA sequence analysis (45). Interestingly, the PIV-1 strain is closely related to Thauera selenatis, which encodes a stand-alone ICM, and Thauera sp. MZ1T, which contains IcmF. We speculate that IcmF might be used by some bacteria to generate the pivalyl-CoA building block for neo-fatty acids, neo-alkanes or their derivatives found in other natural products (46). The potential biotechnological exploitation of IcmF for bioremediation of pivalic acid found in pharmaceutical wastewaters (pivalic acid esters are used as pro-drugs) and/or for biosynthesis of a simple β-nonfunctional α-quarternary carboxylic acid, awaits development.
A potential metabolic pathway for the production of substrates for IcmF/ICM is degradation of the branched chain amino acids, leucine and valine (Fig. 4). Following transamination, catalyzed by the branched-chain amino acid transaminase, the resulting α-keto acids feed into the branched-chain alpha keto-acid dehydrogenase complex. The latter catalyzes an oxidative decarboxylation reaction generating isobutyryl-CoA from valine and isovaleryl-CoA from leucine.
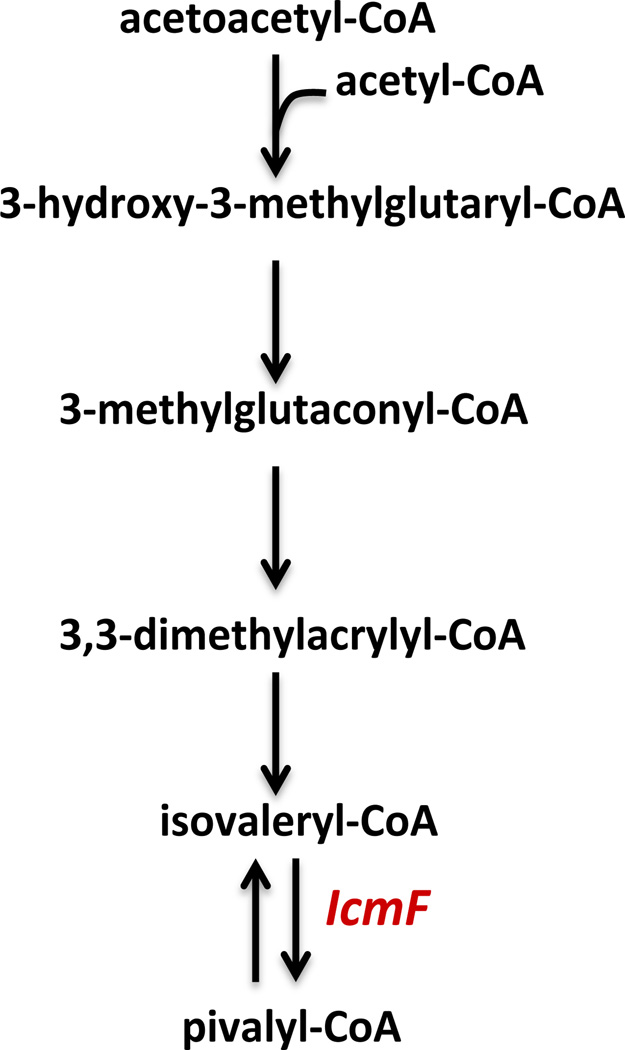
Recently, a pathway described in Myxococcus Xanthus, was shown to generate isovaleryl-CoA without the involvement of the branched-chain alpha keto-acid dehydrogenase complex (Fig. 6). In this alternative pathway, acetoacetyl-CoA is converted to isovaleryl-CoA via 3-hydroxy-3-methylglutaryl-CoA, 3-methylglutaconyl-CoA and 3,3-dimethylacrylyl-CoA (47). The genome of M. xanthus harbors the icmF gene, so the pathway proposed in Fig. 6, which result in pivalyl-CoA production might exist in this bacterium provided that IcmF is expressed together with isovaleryl-CoA biosynthesis pathway genes. Studies investigating the biological role of IcmF in M. xanthus are underway in our laboratory.
Figure 6.
An alternative pathway for isolvaleryl-CoA synthesis and a proposed pivalyl-CoA pathway. The pathway for biosynthesis of isovaleryl-CoA as shown was characterized in M. xanthus (47).
In several bacteria (e.g. Ralstonia solanacearum, R. metallidurans, Polaromonas sp. and Rhodoferax ferrireducens), the genes involved in degradation of branched-chain amino acids appear to be co-regulated with icmF (48). A DNA binding motif for the LiuR-/TetR-like transcriptional regulator is found just upstream of the icmF gene. Furthermore, the genes involved in degradation of branched-chain amino acids in these bacteria are also involved in the phenylacetate degradation pathway, which contains an alcohol dehydrogenase similar to 3-hydroxybutyryl-CoA dehydrogenase (44, 48–50). A second route for the supply of branched chain thioesters is β-oxidation of fatty acids. In fact, in some bacteria, icmF is found in the same operon as the genes involved in fatty acid catabolism (44). Although there appears to be a clear connection between IcmF and pathways for production of small branched-chain acyl-CoAs, the exact role of IcmF in bacterial metabolism remains to be elucidated. A potential application of ICM/IcmF is in metabolic engineering of pathways to produce branched C4 and C5 building blocks that can subsequently be converted to useful derivatives e.g. the corresponding alcohols, isobutanol and pivalyl alcohol.
(ii) ECM
It has been known for many years that MCM can use ethylmalonyl-CoA as an alternative, albeit poor, substrate, generating the rearranged product, methylsuccinyl-CoA (51). Furthermore, MCM can catalyze, although very inefficiently, two successive 1,2 rearrangements of glutaryl-CoA to methylsuccinyl-CoA and ethylmalonyl-CoA, respectively (52). Elucidation of the bacterial ethylmalonyl-CoA pathway for acetate assimilation has led to the recent discovery of a separate subclade of acyl-CoA mutases with ECM activity (10, 53). The ethylmalonyl-CoA pathway allows net synthesis of precursor metabolites for cell carbon biosynthesis (Fig. 5, red arrows). Initially described in Rhodobacter sphaeroides, this pathway appears to operate in organisms lacking a functional glyoxylate cycle for assimilation of acetyl-CoA into malate (53). The ethylmalonyl-CoA pathway converts three moles of acetyl-CoA and two moles of carbon dioxide to two moles of malate. The ECM gene is found in genomes of numerous bacteria belonging to α-proteobacteria and actinomycetes and in two Leptospira species. Based on genome analyses of 413 bacteria, it is estimated that ~7% utilize the ethylmalonyl-CoA pathway while another 1% use both the glyoxyate cycle and ethylmalonyl-CoA pathways for assimilating acetyl-CoA (54).
The presence of metabolically rare C5 intermediates in the ethylmalonyl-CoA pathway including (2R)- and (2S)-ethylmalonyl-CoA, (2S)-methylsuccinyl-CoA, mesaconyl-(C1)-CoA, and (2R, 3S)-methylmalyl-CoA and the catalysts that produce them, make it a treasure-trove of untapped biotechnological potential for generating novel value-added bioproducts (54). Carbon sources such as alcohols, carbohydrates, polyols or fatty acids can be used as starting materials for these C5 acids. Two organisms, which are potentially amenable to metabolic bioengineering, R. sphaeroides and M. extorquens, harbor the ethylmalonyl-CoA pathway.
(iii) HCM
In one direction, the substrate for HCM, 3-hydroxybutyryl-CoA, is a precursor of PHB (polyhydroxybutyrate), a well-known bacterial bioplastic (Fig. 5, blue arrows). In this direction, the product of the HCM-catalyzed isomerization is 2-hydroxyisobutyryl-CoA or the corresponding acid, 2-HIBA. The latter is an uncommon metabolite, rarely used in major metabolic pathways. 2-HIBA can also be generated via the oxidative breakdown of isobutane and isobutene or from the plant cyanoglycoside linamarin (9). However, bacteria capable of growing on hydroxyisobutyronitrile, the expected precursor of 2-HIBA formed during degradation of linamarin, have not been identified. Importantly, 2-HIBA is also formed during degradation of the fuel oxygenate, MBTE (methyl tert-butyl ether), a gasoline additive widely used to reduce CO emissions representing a >20 million ton market. The presence of MBTE in contaminated groundwater represents a significant environmental threat due to the recalcitrance of the tertiary butyl group to biodegradation. Hence, bioremediation of MBTE by bacterial strains such as Aquincola tertiaricarbonisis, Methylibium petroleiphilum PM1, Mycobacterium austroafricanum and Variovorax paradoxus CL-8 is of considerable interest (55).
The aerobic pathway for biomineralization of MBTE involves initial oxidation by a monooxygenase forming tert-butoxy methanol, which can either undergo spontaneous dismutation to tert-butanol or be oxidized to tert-butyl formate (Fig. 5). Subsequent hydroxylation and oxidation steps yield 2-HIBA, which following conversion to its CoA ester, is isomerized by HCM to the common intermediate, 3-hydroxybutyryl-CoA (56).
Although 2-HIBA is currently a specialty chemical, the methyl ester of methyacrylate, formed by dehydration of 2-HIBA, is used in the chemical industry for synthesis of acrylic glass and durable inks and coatings (57). Hence, the discovery of HCM opens doors to production of 2-HIBA from renewable biomass as a building block for the synthesis of polymers with an isobutene carbon skeleton. Isobutylene glycol and the chloro- and amino-derivatives of 2-HIBA are some other examples of building blocks used for polymer synthesis. Presently, the isobutane building block is chemically synthesized from C2 and C4 hydrocarbons derived from petroleum. For bio-based production of 2-HIBA, bioengineering of an organism that utilizes HCM and contains the pathway for PHB synthesis is an attractive starting point since the machinery for producing the precursor, 3-hydroxybutyrate would already exist in such a microbe. Inactivation of the PHB synthesis pathway would divert 3-hydroxybutyrate to HIBA production.
Future directions
Acyl-CoA mutases are the most rapidly growing subclass of AdoCbl-dependent enzymes with the discovery in recent years of new family members, which catalyze chemically tantalizing transformations with exciting biotechnological potential. Structurally, they are predicted to use the same architectural scaffold: a combination of a Rossman fold and a TIM barrel joined by a linker, which house the cofactor and substrate, respectively. Subtle differences in key active site residues positioned to interact with the substituents by which their respective substrates differ, appear to govern in large measure differences in their specificities. The propensity of acyl-CoA mutases to inactivation is potentially problematic for developing them commercially as valuable reagents and their G-protein chaperones (e.g. MeaB for MCM), which can protect against inactivation (58), might represent a solution to this problem. Elucidating the kinetics and regulation of the acyl-CoA mutases and their interactions with cognate G-protein chaperone partners represents an exciting area that is ripe for exploration as is the harnessing of these biocatalysts for generating novel reagents in metabolically engineered organisms.
Abbreviations Used
- AdoCbl
5′-deoxyadenosylcobalamin
- 5′-dAdo•
5′-deoxyadenosyl radical
- ECM
ethylmalonyl-CoA mutase
- HCM
hydroxyisobutyryl-CoA mutase
- ICM
isobutyryl-CoA mutase
- IcmF
isobutyryl-CoA mutase fused to its G-protein chaperone
- MCM
methylmalonyl-CoA mutase
- MBTE
methyl tert-butyl ether
- 2-HIBA
2-hydroxyisobutyrate
- TIM
triosephosphate isomerase
Footnotes
This work was supported by a grant from the National Institutes of Health (DK45776)
References
- 1.Matthews RG. Cobalamin- and corrinoid-dependent enzymes. Met Ions Life Sci. 2009;6:53–114. doi: 10.1039/BK9781847559159-00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 3.Frey PA. Radical mechanisms of enzymatic catalysis. Ann Rev Biochem. 2001;70:121–148. doi: 10.1146/annurev.biochem.70.1.121. [DOI] [PubMed] [Google Scholar]
- 4.Dowling DP, Croft AK, Drennan CL. Radical Use of Rossmann and TIM Barrel Architectures for Controlling Coenzyme B12Chemistry. Annu Rev Biophys. 2012;41:403–427. doi: 10.1146/annurev-biophys-050511-102225. [DOI] [PubMed] [Google Scholar]
- 5.Hay BP, Finke RG. Thermolysis of the Co-C bond in adenosylcorrins. 3. Quantifiation of the axial base effect in adenosylcobalamin by the synthesis and thermolysis of axial base-free adenosylcobinamide. Insights into the energetics of enzyme-assisted cobalt-carbon bond homolysis. J. Am. Chem. Soc. 1987;109:8012–8018. [Google Scholar]
- 6.Halpern J. Mechanisms of coenzyme B12-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee R. Radical peregrinations catalyzed by coenzyme B12-dependent enzymes. Biochemistry. 2001;40:6191–6198. doi: 10.1021/bi0104423. [DOI] [PubMed] [Google Scholar]
- 8.Toraya T. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem Rev. 2003;103:2095–2127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 9.Yaneva N, Schuster J, Schafer F, Lede V, Przybylski D, Paproth T, Harms H, Muller RH, Rohwerder T. Bacterial acyl-CoA mutase specifically catalyzes coenzyme B12-dependent isomerization of 2-hydroxyisobutyryl-CoA and (S)-3-hydroxybutyryl-CoA. J Biol Chem. 2012;287:15502–15511. doi: 10.1074/jbc.M111.314690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb TJ, Retey J, Fuchs G, Alber BE. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J Biol Chem. 2008;283:32283–32293. doi: 10.1074/jbc.M805527200. [DOI] [PubMed] [Google Scholar]
- 11.Cracan V, Padovani D, Banerjee R. IcmF is a fusion between the radical B12enzyme isobutyryl-CoA mutase and its G-protein chaperone. J Biol Chem. 2010;285:655–666. doi: 10.1074/jbc.M109.062182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebenau-Jehle C, Thomas M, Scharf G, Kockelkorn D, Knapp B, Schuhle K, Heider J, Fuchs G. Anaerobic metabolism of indoleacetate. J Bacteriol. 2012;194:2894–2903. doi: 10.1128/JB.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarling R, Sadeghi M, Drozdowska M, Lahme S, Buckel W, Rabus R, Widdel F, Golding BT, Wilkes H. Stereochemical investigations reveal the mechanism of the bacterial activation of n-alkanes without oxygen. Angew Chem Int Ed Engl. 2012;51:1334–1338. doi: 10.1002/anie.201106055. [DOI] [PubMed] [Google Scholar]
- 14.Froese DS, Kochan G, Muniz JR, Wu X, Gileadi C, Ugochukwu E, Krysztofinska E, Gravel RA, Oppermann U, Yue WW. Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation. J Biol Chem. 2010;285:38204–38213. doi: 10.1074/jbc.M110.177717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Bosecke P, Diat O, Evans PR. How coenzyme B12radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Structure. 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 16.Han Y, Hawkins AS, Adams MW, Kelly RM. Epimerase (Msed_ 0639) and Mutase (Msed_0638, Msed_2055) Convert (S)-Methylmalonyl-CoA to Succinyl-CoA in the Metallosphaera sedula 3-Hydroxypropionate/4-Hydroxybutyrate Cycle. App Environ Microbiol. 2012 doi: 10.1128/AEM.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menendez C, Bauer Z, Huber H, Gad'on N, Stetter KO, Fuchs G. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J Bacteriol. 1999;181:1088–1098. doi: 10.1128/jb.181.4.1088-1098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 19.Ratnatilleke A, Vrijbloed JW, Robinson JA. Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J Biol Chem. 1999;274:31679–31685. doi: 10.1074/jbc.274.44.31679. [DOI] [PubMed] [Google Scholar]
- 20.Padovani D, Banerjee R. A G-protein editor gates coenzyme B12loading and is corrupted in methylmalonic aciduria. Proc Natl Acad Sci U S A. 2009;106:21567–21572. doi: 10.1073/pnas.0908106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korotkova N, Lidstrom ME. MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation. J Biol Chem. 2004;279:13652–13658. doi: 10.1074/jbc.M312852200. [DOI] [PubMed] [Google Scholar]
- 22.Padovani D, Labunska T, Banerjee R. Energetics of interaction between the G-protein chaperone, MeaB and B12-dependent methylmalonyl-CoA mutase. J. Biol. Chem. 2006;281:17838–17844. doi: 10.1074/jbc.M600047200. [DOI] [PubMed] [Google Scholar]
- 23.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 24.Reitzer R, Gruber K, Jogl G, Wagner UG, Bothe H, Buckel W, Kratky C. Glutamate mutase from Clostridium cochlearium: the structure of a coenzyme B12-dependent enzyme provides new mechanistic insights. Structure Fold Des. 1999;7:891–902. doi: 10.1016/s0969-2126(99)80116-6. [DOI] [PubMed] [Google Scholar]
- 25.Mancia F, Evans P. Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. Structure. 1998;6:711–720. doi: 10.1016/s0969-2126(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 26.Vlasie MD, Banerjee R. Tyrosine 89 accelerates Co-carbon bond homolysis in methylmalonyl-CoA mutase. J Amer Chem Soc. 2003;125:5431–5435. doi: 10.1021/ja029420+. [DOI] [PubMed] [Google Scholar]
- 27.Vlasie MD, Banerjee R. When a spectator turns killer: suicidal electron transfer from cobalamin in methylmalonyl-CoA mutase. Biochemistry. 2004;43:8410–8417. doi: 10.1021/bi036299q. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee R, Chowdhury S. Methylmalonyl-CoA mutase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley and Sons; 1999. pp. 707–730. [Google Scholar]
- 29.Ledley FD, Rosenblatt DS. Mutations in Mut, methylmalonic acidemia: Clinical and enzymatic correlations. Hum Mutat. 1997;9:1–6. doi: 10.1002/(SICI)1098-1004(1997)9:1<1::AID-HUMU1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 30.Guo X, Liu T, Deng Z, Cane DE. Essential role of the donor acyl carrier protein in stereoselective chain translocation to a fully reducing module of the nanchangmycin polyketide synthase. Biochemistry. 2012;51:879–887. doi: 10.1021/bi201768v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry. 2002;41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- 32.Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM. Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab Eng. 2007;9:293–303. doi: 10.1016/j.ymben.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM. Effects of methylmalonyl-CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. J Ind Microbiol Biotechnol. 2006;33:600–609. doi: 10.1007/s10295-006-0094-3. [DOI] [PubMed] [Google Scholar]
- 34.Jung WS, Yoo YJ, Park JW, Park SR, Han AR, Ban YH, Kim EJ, Kim E, Yoon YJ. A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. App Microbiol Biotechnol. 2011;91:1389–1397. doi: 10.1007/s00253-011-3348-6. [DOI] [PubMed] [Google Scholar]
- 35.Smejkalova H, Erb TJ, Fuchs G. Methanol assimilation in Methylobacterium extorquens AM1: demonstration of all enzymes and their regulation. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds KA, O'Hagan D, Gani D, Robinson JA. Butyrate metabolism in streptomycetes. Characterization of an intramolecular vicinal interchange rearrangement linking isobutyrate and butyrate in Streptomyces cinnamonensis. J. Chem. Soc. Perkin Trans. 1988;1:3195–3207. [Google Scholar]
- 37.Vrijbloed JW, Zerbe-Burkhardt K, Ratnatilleke A, Grubelnik-Leiser A, Robinson JA. Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J Bacteriol. 1999;181:5600–5605. doi: 10.1128/jb.181.18.5600-5605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuzawa S, Kim W, Katz L, Keasling JD. Heterologous production of polyketides by modular type I polyketide synthases in Escherichia coli. Curr Opin Biotechnol. 2012 doi: 10.1016/j.copbio.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 39.Zerbe-Burkhardt K, Ratnatilleke A, Vrijbloed JW, Robinson JA. Isobutyryl-CoA mutase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley and Sons; New York: 1999. pp. 859–870. [Google Scholar]
- 40.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohwerder T, Müller H. New bacterial cobalamin-dependent CoA-carbonyl mutases involved in degradation pathways. In: CM E, editor. Vitamin B: new research. Nova Science Publishers; 2007. pp. 81–98. [Google Scholar]
- 42.Oude Elferink SJ, Lens PN, Dijkema C, Stams AJ. Isomerization of butyrate to isobutyrate by Desulforhabdus amnigenus. FEMS Microbiol Lett. 1996;142:237–241. [Google Scholar]
- 43.Sun H, Spring S, Lapidus A, Davenport K, Del Rio TG, Tice H, Nolan M, Copeland A, Cheng JF, Lucas S, Tapia R, Goodwin L, Pitluck S, Ivanova N, Pagani I, Mavromatis K, Ovchinnikova G, Pati A, Chen A, Palaniappan K, Hauser L, Chang YJ, Jeffries CD, Detter JC, Han C, Rohde M, Brambilla E, Goker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Land M. Complete genome sequence of Desulfarculus baarsii type strain (2st14) Stand Genomic Sci. 2010;3:276–284. doi: 10.4056/sigs.1243258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cracan V, Banerjee R. Novel coenzyme B12-dependent interconversion of isovaleryl-CoA and pivalyl-CoA. J Biol Chem. 2012;287:3723–3732. doi: 10.1074/jbc.M111.320051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Probian C, Wulfing A, Harder J. Anaerobic mineralization of quaternary carbon atoms: isolation of denitrifying bacteria on pivalic acid (2,2-dimethylpropionic acid) Appl Environ Microbiol. 2003;69:1866–1870. doi: 10.1128/AEM.69.3.1866-1870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dembitsky VM. Natural neo acids and neo alkanes: their analogs and derivatives. Lipids. 2006;41:309–340. doi: 10.1007/s11745-006-5103-9. [DOI] [PubMed] [Google Scholar]
- 47.Bode HB, Ring MW, Schwar G, Altmeyer MO, Kegler C, Jose IR, Singer M, Muller R. Identification of additional players in the alternative biosynthesis pathway to isovaleryl-CoA in the myxobacterium Myxococcus xanthus. Chembiochem. 2009;10:128–140. doi: 10.1002/cbic.200800219. [DOI] [PubMed] [Google Scholar]
- 48.Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol. 2009;191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds - from one strategy to four. Nat Rev Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 50.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 51.Retey J, Smith EH, Zagalak B. Investigation of the Mechanism of the Methylmalonyl-CoA Mutase Reaction with the Substrate Analogue: Ethylmalonyl-CoA. EurJBiochem. 1978;83:437–451. doi: 10.1111/j.1432-1033.1978.tb12110.x. [DOI] [PubMed] [Google Scholar]
- 52.Padmakumar R, Banerjee R. A Carbon Skeleton Walk: A novel double rearrangement of glutaryl-CoA catalyzed by the human methylmalonyl-CoA mutase. Biofactors. 1995–1996;5:83–86. [PubMed] [Google Scholar]
- 53.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: The ethylmalonyl-CoA pathway. Proc. Natl, Acad. Sci. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alber BE. Biotechnological potential of the ethylmalonyl-CoA pathway. App Microbiol Biotechnol. 2011;89:17–25. doi: 10.1007/s00253-010-2873-z. [DOI] [PubMed] [Google Scholar]
- 55.Schafer F, Muzica L, Schuster J, Treuter N, Rosell M, Harms H, Muller RH, Rohwerder T. Formation of alkenes via degradation of tert-alkyl ethers and alcohols by Aquincola tertiaricarbonis L108 and Methylibium spp. Appl Environ Microbiol. 2011;77:5981–5987. doi: 10.1128/AEM.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohwerder T, Breuer U, Benndorf D, Lechner U, Muller RH. The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl Environ Microbiol. 2006;72:4128–4135. doi: 10.1128/AEM.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohwerder T, Müller RH. Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb Cell Fact. 2010;9:13. doi: 10.1186/1475-2859-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Padovani D, Banerjee R. Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone. Biochemistry. 2006;45:9300–9306. doi: 10.1021/bi0604532. [DOI] [PubMed] [Google Scholar]