Abstract
The transition state for the Trypanosoma cruzi uridine phosphorylase (TcUP) reaction has an expanded SN2 character. We used binding isotope effects (BIE’s) to probe uridine distortion in the complex with TcUP and sulfate to mimic the Michaelis complex. Inverse 1′-3H and 5′-3H BIE’s indicate a constrained bonding environment of these groups in the complex. Quantum chemical modeling identified a uridine conformer whose calculated BIE’s match the experimental values. This conformer differs in sugar pucker and uracil orientation from the unbound conformer and the transition-state structure. These results support ground-state stabilization in the Michaelis complex.
Ground-state destabilization has been proposed as a strategy employed by enzymes including triosephosphate isomerase, enoyl-CoA hydratase, and alkaline phosphatase, to accomplish catalysis.1–4 The process consists of enzyme-induced electronic strain of substrates in the Michaelis complex towards their transition-state configurations. However, some theoretical and experimental evidences have questioned this hypothesis in certain enzymes, for instance, orotidine 5′-decarboxylase and lysozyme.5–7
Kinetic isotope effects (KIE’s) measured by the internal competition method in enzymatic reactions report on V/K, probing events from free substrate in solution up to, and including, the first irreversible step, often assumed to be the release of the first product under initial rate conditions. KIE’s on V/K can be corrected for kinetic complexities, such as commitment factors, to yield intrinsic KIE’s, which reflect differences in bond vibration frequencies between the free substrate and the transition state for the chemical step, providing no information on the bonding environment in the Michaelis complex.2,8 Conversely, equilibrium binding isotope effects (BIE’s) arise from distinct bonding environments in the free substrate and in the Michaelis complex, prior to the chemical step.9 When both types of isotope effects are available for an enzyme reaction, the degree of distortion in the enzyme-bound substrate can be compared with that at the transition state, allowing an evaluation of ground-state destabilization.10,11
Here, the hypothesis of ground-state destabilization in the reaction catalyzed by Trypanosoma cruzi uridine phosphorylase (TcUP) (EC 2.4.2.3) is investigated by BIE analysis. TcUP catalyzes the reversible phosphorolysis of uridine to yield uracil and ribose 1-phosphate,12 and KIE studies using phosphate or arsenate as nucleophile indicated that the reaction proceeds via an expanded SN2-like transition state.13,14 If the C1′-N1 bond of uridine elongates in the Michaelis complex towards the transition state, the anomeric carbon develops partial sp2 character with increased freedom in the C1′-H1′ bond out-of-plane bending mode. Experimentally, this gives rise to a normal 1′-3H BIE. Scheme S1 depicts the atom numbering of uridine.
Tritium BIE’s were measured for the 1′-H and 5′-H2 positions of uridine, using [5′-14C]uridine as the light substrate in competitive radiolabel experiments. Briefly, 20 µM TcUP was incubated with 50 µM uridine ([5′-14C]uridine and either [1′-3H] or [5′-3H2]uridine), and 50 mM (NH4)2SO4 pH 7.5, in 100 mM HEPES pH 7.5. The incubation mixture (100 µL) was loaded onto an ultrafiltration apparatus, and an argon pressure of 30 psi was applied until approximately half the volume had passed through a semi-permeable membrane (10 kDa molecular weight exclusion). At this time, 30 µL were removed from the top and bottom of the ultrafiltration wells. Scintillation counting and spectral deconvolution were performed as previously reported.13 BIE’s were calculated with eq 1, where 14CT and 14CB are the total 14C counts in the top and bottom wells, respectively, and 3HT and 3HB are the total 3H counts in the top and bottom wells, respectively.15 BIE’s are expressed as averages and their standard errors for at least 10 replicates in 2 independent experiments. The effect of sulfate ion on the 1′-3H BIE was assessed by excluding (NH4)2SO4 from the incubation mixture. Enzyme was omitted in control experiments.
| (1) |
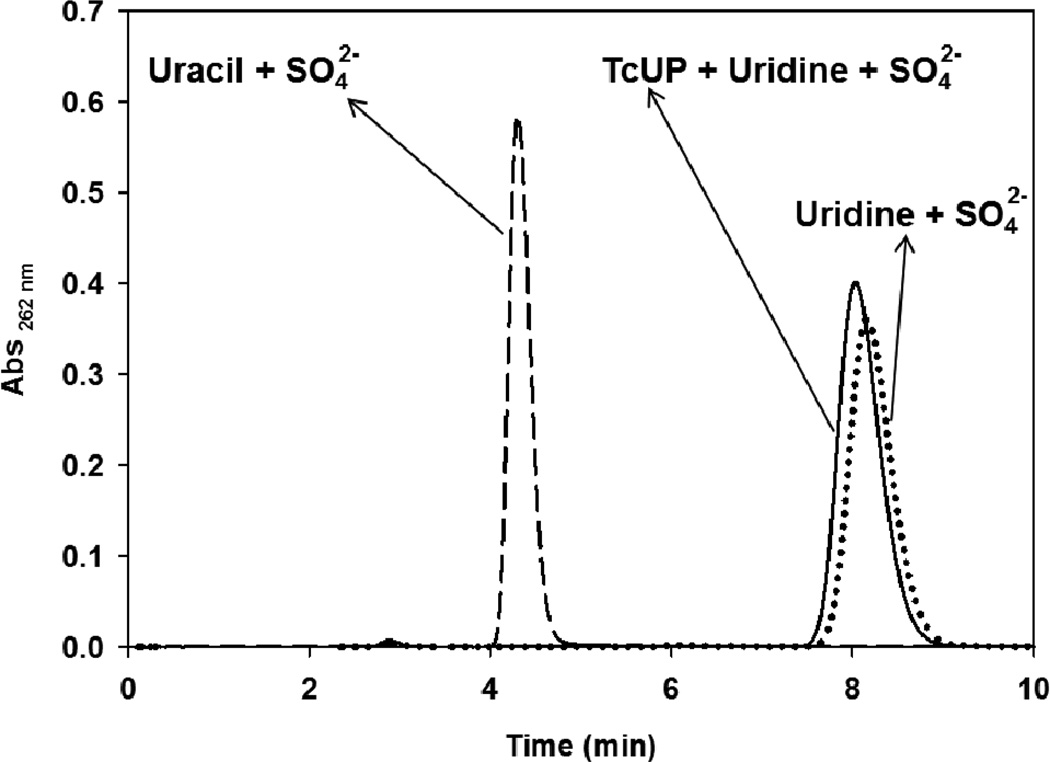
BIE values of 1′-3H = 0.947 ± 0.007 (−5.3%) and 5′-3H2 = 0.979 ± 0.006 (−2.1%) were obtained for the formation of the TcUP-uridine-sulfate ternary complex. A 1′-3H BIE = 0.953 ± 0.009 (−4.7%) was measured in the absence of (NH4)2SO4, indicating that the nucleophile mimic exerts negligible or no influence on the inverse isotope effect. The sulfate ion is known to occupy the phosphate binding sites in ribosyltransferases, and is employed as a non-reactive mimic of phosphate in ternary-complex crystal structures and BIE experiments.10,11,16 The lack of chemical activity of this complex was confirmed by incubating 50 mM (NH4)2SO4 pH 7.5, 10 µM TcUP, and 100 µM uridine in 100 mM HEPES pH 7.5 for 90 min, followed by reverse-phase HPLC separation as previously described.13 No uracil could be detected under this condition (Figure 1), which resembles that of the ultrafiltration experiments.
Figure 1.
Reverse-phase chromatograms of incubation mixtures containing TcUP, uridine, and sulfate (solid line), uracil and sulfate (dashed line), and uridine and sulfate (dotted line), in HEPES pH 7.5.
Inverse BIE’s occur when the bond to the tritium isotope is more constrained in the enzyme-bound state. Therefore, atoms H1′ and H(2)5′ experience a more constrained bonding environment when bound to TcUP. These values contrast with the intrinsic KIE’s of 1′-3H = 1.132 (13.2%) and 5′-3H2 = 1.041 (4.1%) for uridine arsenolysis by TcUP, which established increased bond vibrational freedom for these atoms at the transition state.13 Hence, ternary-complex interactions contribute isotope effects in the opposite direction to those arising from transition state formation. This finding is contrary to those found with P. falciparum and human orotate phosphoribosyltransferases (OPRT), where half of the normal intrinsic KIE values for 1′-3H are contributed by BIE’s,10 and with human thymidine phosphorylase (TP), where the 5′-3H2 BIE for thymidine is equivalent to and thus accounts for the entire intrinsic KIE.17 In summary, the experimental BIE data for TcUP support ground-state stabilization in both binary and ternary complexes.
Density-functional theory (DFT) calculations were performed to search for a conformational geometry of uridine in the Michaelis complex that could account for the BIE’s. Several conformations of uridine were optimized in vacuo at the B3LYP level of theory with the 6–31G** basis set, as implemented in Gaussian 09,18 by varying the O4′-C1′-N1-C6 dihedral angle with either 2′-endo-3′-exo or 2′-exo-3′-endo ribosyl pucker. All optimizations resulted in ground-state structures containing no imaginary frequencies. Isotope effects for each structure were calculated from scaled vibrational frequencies using ISOEFF98,19 and compared with the experimental BIE values.
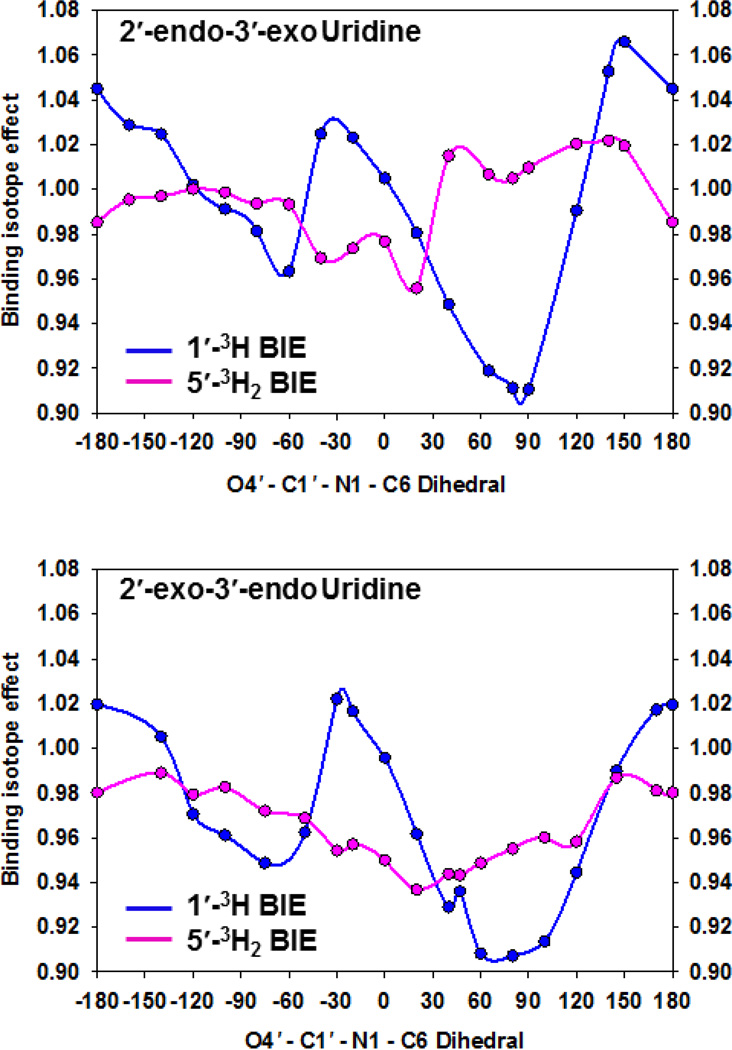
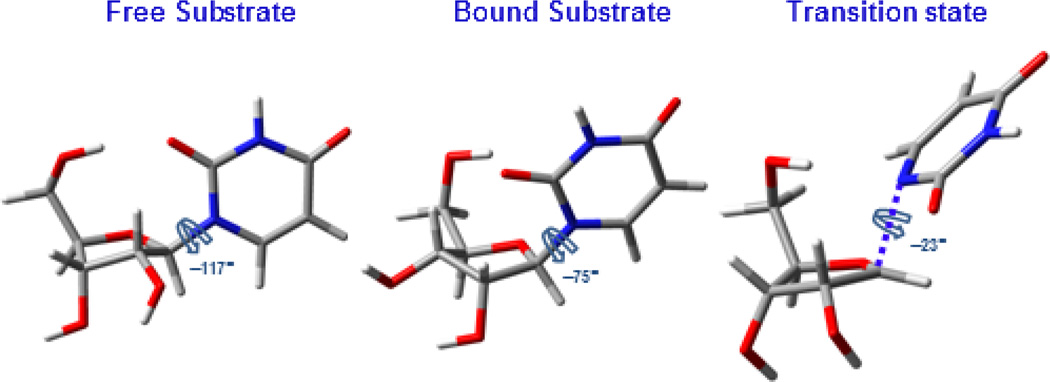
The 2′-endo-3′-exo and 2′-exo-3′-endo uridine conformations yielded distinct BIE’s throughout the range of O4′-C1′-N1-C6 dihedrals (Figure 2). The lowest energy conformation of free uridine, used as the reference upon which all BIE’s were calculated, adopts a 2′-endo-3′-exo pucker and an O4′-C1′-N1-C6 torsion angle of −117°13 (Figure 3, left). A 2′-exo-3′-endo conformer with O4′-C1′-N1-C6 dihedral angle of −75° (Figure 3, middle) generated calculated BIE’s on 1′-3H = 0.949 (−5.1%) and on 5′-3H2 = 0.972 (−2.8%), in excellent agreement with the experimental values of −5.3% and −2.1%, respectively. Inverse BIE’s were also calculated from other conformers, but none of them matched the experimental ones as closely (Table S1).
Figure 2.
Dependence of calculated 1′-3H (blue) and 5′-3H2 (pink) BIE’s on O4′-C1′-N1-C6 dihedral angles of 2′-endo-3′-exo (top) and 2′-exo-3′-endo (bottom) conformers of uridine.
Figure 3.
Stick models of DFT-optimized conformations of uridine as free substrate (left), in the Michaelis complex (middle), and at the transition state for arsenolysis (right), and their respective O4′-C1′-N1-C6 dihedral angles.
The 2′-exo-3′-endo ribosyl pucker differs from that of free uridine and also from the KIE-derived transition-state structure for the TcUP-catalyzed arsenolysis of uridine. At the transition state, a flatter 2′-endo-3′-exo sugar is predicted13 (Figure 3, right). The potential energy of the conformer proposed to be present in the Michaelis complex is approximately 3 kcal mol−1 higher than that of free uridine (Figure S1), an amount of energy stabilized, for example, by a modest hydrogen bond (H-bond).20 This could be explained by the loss of an intra-molecular H-bond between O(H)5′ and O2. This H-bond was predicted by DFT optimizations of ground-state uridine, as previously reported,13 and is likely disrupted as the O4′-C1′-N1-C6 torsion angle increases from −117° to −75° upon enzyme binding. At the transition state, this angle is further increased to −23°, favoring H-bond interactions with the side chain of an active site glutamine residue conserved in all known uridine phosphorylases.13,21 At present, structural data to predict interactions with the −75° dihedral conformation of uridine is lacking.
Distinct atomic charges are a prominent feature of transition-state structures of N-ribosyltransferase reactions.22 Ground state destabilization in the Michaelis complex is therefore expected to alter the charge distribution of bound uridine towards that of the transition state. Accordingly, natural bond orbital (NBO) charges were calculated for the proposed TcUP-bound uridine conformer, and compared with those reported for free substrate and transition state structures (Table 1).13 The only continuous trends in charge distribution among the three structures concern the subtle accumulation of negative charge at the N1 position and the small decrease in electron density at the H1′ position of bound uridine, both of which become more pronounced at the transition state. Position C1′ conserves its charge as uridine binds TcUP, and loses electron density only at the transition state as the N-ribosydic bond electrons migrate to the uracil moiety. Positions O4′, O5′, O2, Pro-R-H5′, and Pro-S-H5′ show altered NBO charges in the Michaelis complex in comparison with the free substrate. However, those charges change in the opposite direction as the system reaches the transition state.
Table 1.
Natural Bond Orbital Charges of Free Uridine (FU), Bound Uridine (BU), and Transition State (TS).
| Position | FU | BU | TS |
|---|---|---|---|
| C1′ | 0.25a | 0.25 | 0.36a |
| H1′ | 0.22 | 0.25 | 0.29 |
| N1 | −0.47a | −0.48 | −0.62a |
| O4′ | −0.59a | −0.61 | −0.51a |
| O5′ | −0.78 | −0.76 | −0.78 |
| Pro-R-H5′ | 0.22 | 0.23 | 0.22 |
| Pro-S-H5′ | 0.22 | 0.21 | 0.19 |
| O2 | −0.66 | −0.64 | −0.71 |
Previously published values.13
The inverse experimental 1′-3H and 5′-3H2 BIE’s presented in this work contrast with the normal KIE’s measured for TcUP-catalyzed arsenolysis of uridine. The uridine conformer whose calculated 1′-3H and 5′-3H2 BIE’s match the experimental values representing the conformation of uridine in the Michaelis complex does not incorporate characteristics of the transition-state structure. Neither the ribosyl pucker nor the NBO charge distribution pattern are altered to more closely resemble the transition state. These observations are consistent with ground-state stabilization in the Michaelis complex of TcUP. Ground-state stabilization here refers to local effects on the bound substrate, such as bonding environment, ribosyl pucker, and NBO charges, contrasting to those in the enzymatic transition state. Compared with previous reports of BIE’s for human and P. falciparum OPRT10 and human TP,17 the present work exemplifies the diverging strategies employed by different enzymes to catalyze similar chemical transformations.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH Research Grants GM41916 and AI049512 to V. L. S.
ABBREVIATIONS
- KIE
Kinetic Isotope Effect
- BIE
Equilibrium Binding Isotope Effect
- TcUP
Trypanosoma cruzi Uridine Phosphorylase
- DFT
Density-Functional Theory
- H-Bond
Hydrogen Bond
- NBO
Natural Bond Orbital
- OPRT
orotate phosphoribosyltransferase
- TP
thymidine phosphorylase
Footnotes
Supporting Information. Detailed description of materials and methods, Scheme S1, Table S1, Figure S1, and atomic coordinates for all optimized ground-state geometries of uridine. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Jencks WP. Catalysis in Chemistry and Enzymology. New York: Dover Publications; 1987. Chapter 5. [Google Scholar]
- 2.Anderson VE. Arch. Biochem. Biophys. 2005;433:27–33. doi: 10.1016/j.abb.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Belasco JG, Knowles JR. Biochemistry. 1980;19:472–477. doi: 10.1021/bi00544a012. [DOI] [PubMed] [Google Scholar]
- 4.Andrews LD, Deng H, Herschlag D. J. Am. Chem. Soc. 2011;133:11621–11631. doi: 10.1021/ja203370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warshel A, Strajbl M, Villa J, Florian J. Biochemistry. 2000;39:14728–14738. doi: 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- 6.Miller BG, Butterfoss GL, Short SA, Wolfenden R. Biochemistry. 2001;40:6227–6232. doi: 10.1021/bi0028993. [DOI] [PubMed] [Google Scholar]
- 7.Warshel A, Levitt M. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 8.Ruszczycky MW, Anderson VE. J. Theor. Biol. 2006;243:328–342. doi: 10.1016/j.jtbi.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Schramm VL. Curr. Opin. Chem. Biol. 2007;11:529–536. doi: 10.1016/j.cbpa.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Schramm VL. Biochemistry. 2011;50:4813–4818. doi: 10.1021/bi200638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murkin AS, Birck MR, Rinaldo-Matthis A, Wuxian S, Taylor EA, Almo SC, Schramm VL. Biochemistry. 2007;46:5038–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paege LM, Schlenk F. Arch. Biochem. Biophys. 1952;40:42–49. doi: 10.1016/0003-9861(52)90071-4. [DOI] [PubMed] [Google Scholar]
- 13.Silva RG, Vetticatt MJ, Merino EF, Cassera MB, Schramm VL. J. Am. Chem. Soc. 2011;133:9923–9931. doi: 10.1021/ja2031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva RG, Schramm VL. Biochemistry. 2011;50:9158–9166. doi: 10.1021/bi2013382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murkin AS, Tyler PC, Schramm VL. J. Am. Chem. Soc. 2008;130:2166–2167. doi: 10.1021/ja7104398. [DOI] [PubMed] [Google Scholar]
- 16.Canduri F, dos Santos DM, Silva RG, Mendes MA, Basso LA, Palma MS, de Azevedo WF, Jr, Santos DS. Biochem. Biophys. Res. Comm. 2004;313:907–914. doi: 10.1016/j.bbrc.2003.11.179. [DOI] [PubMed] [Google Scholar]
- 17.Birck MR, Schramm VL. J. Am. Chem. Soc. 2004;126:6882–6883. doi: 10.1021/ja0492642. [DOI] [PubMed] [Google Scholar]
- 18.Frisch MJ, et al. Gaussian 09, Revision A.02. Wallington, CT: Gaussian, Inc.; 2009. [Google Scholar]
- 19.Anisimov V, Paneth P. J. Math. Chem. 1999;26:75–86. [Google Scholar]
- 20.Lin J, Frey PA. J. Am. Chem. Soc. 2000;122:11258–11259. [Google Scholar]
- 21.Larson ET, Mudeppa DG, Gillespie JR, Mueller N, Napuli AJ, Arif JA, Ross J, Arakaki TL, Lauricella A, Detitta G, Luft J, Zucker F, Verlinde CL, Fan E, Van Voorhis WC, Buckner FS, Rathod PK, Hol WG, Merritt EA. J. Mol. Biol. 2010;396:1244–1259. doi: 10.1016/j.jmb.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schramm VL. Annu. Rev. Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.